Abstract
Although the beneficial role of silicon (Si) in stimulating the growth and development of many plants is generally accepted, our knowledge concerning the physiological and molecular mechanisms underlying this response remains far from comprehensive. Considerable effort has been invested in understanding the role of Si on plant disease, which has led to several new and compelling hypotheses; in unstressed plants, however, Si is believed to have no molecular or metabolic effects. Recently, we have demonstrated that Si nutrition can modulate the carbon/nitrogen balance in unstressed rice plants. Our findings point to an important role of Si as a signaling metabolite able to promote amino acid remobilization. In this article we additionally discuss the agronomic significance of these novel observations and suggest Si nutrition as an important target in future attempts to improve yields of agronomic crops.
Keywords: silicon (Si), signaling, metabolite profile, amino acid remobilization
Although silicon (Si) is not considered an essential element for higher plants, it has been proven to be beneficial for the healthy growth and development of many plant species, particularly graminaceous plants such as rice and sugarcane as well as some cyperaceous plants.1-4 Indeed, earlier studies on Si nutrition have reported the ability of Si to alleviate abiotic and biotic stresses by acting as a physical barrier against pathogens (and possibly insects) and also by inducing active defense mechanisms.5,6 In addition, the effect of Si on promoting disease resistance is well documented and biochemical explanations for the observed resistance have been revealed.7-9 The beneficial effects of Si are, therefore, most obvious in plants encountering stress situations,10 whereas in unstressed plants Si is believed to have a negligible effect on metabolism, suggesting a nonessential role for this element. However, accumulating evidence suggests a role for Si nutrition also in unstressed plants, as noted by the enhanced agronomic yields of crops such as rice upon Si fertilization.11
We have recently demonstrated that Si nutrition impacts the source-sink relationship and stimulates amino acid remobilization in rice.12 We demonstrated that Si-treated plants have reduced levels of several amino acids, a fact linked with an augmentation of the sink strength which was clearly mediated by Si nutrition with little, if any, impact on growth per se.12 Although Si did not affect the actual rate of 14C incorporation into amino acids (Table 1), our data support a role for Si nutrition in orchestrating amino acid remobilization. We observed a strong correlation between Si concentration and the levels of compounds intimately associated with respiration (isocitrate and 2-oxoglutarate), and a handful of amino acids (alanine, arginine, glutamine, ornithine, isoleucine, methionine and valine).12 When taken together, we clearly demonstrated that, at least under the conditions explored in our study, Si nutrition plays an important role in modulating the rate of flux from 2-oxoglutarate into amino acid metabolism, supporting the emergent view that amino acid metabolism is a tightly and intricately controlled network.13,14 Accordingly, an increased incorporation of amino acids in storage proteins could be anticipated. This may also lead to transcriptional upregulation of plastidial branched-chain amino acid transaminase together with other aminotransferases, as previously observed.15 However, it should also be noted that all these amino acids have recently been demonstrated to be efficient alternative respiratory substrates during carbon starvation and other situations of biotic stress.16,17.
Table 1. The effects of silicon (Si) supply (0 or 2 mM: –Si or +Si, respectively) and grain load (0 or full grain burden: –G and +G, respectively) on the actual rate of 14C incorporation into amino acids and enzyme activities (expressed as µmol g−1 FW min−1) in flag leaves of two rice genotypes [cv ‘Oochikara’ (WT) and the lsi1 mutant defective for Si uptake] grown in nutrient solutions. With the exception of PFK1 and PGA kinase, Si nutrition did not affect significantly all the parameters analyzed.
| Parameters | WT |
|
lsi1 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| –Si |
|
+Si |
|
–Si |
|
+Si |
|||||
| –G | +G | –G | +G | –G | +G | –G | +G | ||||
|
14C uptake (kBq g−1 h−1) |
241 ± 8 |
224 ± 10 |
|
206 ± 12 |
210 ± 8 |
|
234 ± 6 |
196 ± 10 |
|
186 ± 8 |
222 ± 14 |
| Rubisco (initial activity) |
5.0 ± 0.4 |
5.3 ± 0.4 |
|
4.8 ± 0.5 |
8.5 ± 0.1 |
|
6.1 ± 0.1 |
5.5 ± 0.1 |
|
5.2 ± 0.8 |
4.7 ± 0.5 |
| Rubisco (max. activity) |
8.2 ± 0.4 |
9.6 ± 0.3 |
|
8.8 ± 0.4 |
10.9 ± 0.7 |
|
9.4 ± 0.7 |
8.8 ± 0.2 |
|
8.2 ± 0.6 |
7.5 ± 0.4 |
| NADP-GAPDH1 |
1.4 ± 0.1 |
1.2 ± 0.1 |
|
1.4 ± 0.2 |
1.7 ± 0.1 |
|
0.76 ± 0.1 |
1.1 ± 0.10 |
|
0.82 ± 0.0 |
0.73 ± 0.1 |
| NAD-GAPDH |
3.4 ± 0.4 |
3.8 ± 0.5 |
|
4.0 ± 0.3 |
3.9 ± 0.4 |
|
2.4 ± 0.3 |
2.5 ± 0.3 |
|
2.2 ± 0.1 |
2.9 ± 0.9 |
| PGM (x10−3) |
177 ± 59 |
473 ± 131 |
|
284 ± 49 |
410 ± 61 |
|
358 ± 68 |
329 ± 64 |
|
198 ± 78 |
327 ± 17 |
| PGI |
5.3 ± 0.8 |
14.4 ± 4.0 |
|
8.2 ± 0.7 |
9.4 ± 1.9 |
|
8.6 ± 1.9 |
8.1 ± 1.8 |
|
8.0 ± 1.8 |
8.2 ± 0.4 |
| PFK (x10−3) |
116 ± 9 |
92 ± 6 |
|
92 ± 12 |
73 ± 8 |
|
84 ± 10 |
89 ± 10 |
|
83 ± 8 |
61 ± 12 |
| PGA Kinase (x10−3) |
1.6 ± 0.1 |
1.6 ± 0.2 |
|
1.8 ± 0.1 |
2.1 ± 0.1 |
|
1.6 ± 0.1 |
1.5 ± 0.1 |
|
1.6 ± 0.1 |
1.8 ± 0.1 |
| NAD-MDH |
11.2 ± 1.1 |
23.1 ± 4.5 |
|
13.5 ± 1.6 |
19.4 ± 2.7 |
|
6.7 ± 1.6 |
15.0 ± 3.3 |
|
7.3 ± 2.1 |
18.4 ± 0.9 |
| Aldolase (x10−3) |
4.0 ± 0.4 |
4.6 ± 0.5 |
|
4.6 ± 0.3 |
6.3 ± 0.5 |
|
3.9 ± 0.2 |
4.5 ± 0.4 |
|
3.9 ± 0.2 |
5.4 ± 0.48 |
| Transaldolase (x10−3) |
26.1 ± 8 |
13.6 ± 3 |
|
30.1 ± 10 |
15.4 ± 1 |
|
35.2 ± 6 |
33.7 ± 7 |
|
45.0 ± 10 |
30.6 ± 7 |
| TPI (x10−3) | 0.42 ± 0.04 | 0.39 ± 0.02 | 0.47 ± 0.04 | 0.49 ± 0.03 | 0.45 ± 0.04 | 0.47 ± 0.03 | 0.39 ± 0.02 | 0.48 ± 0.07 | |||
1GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGM, phosphoglucomutase; PGI, phosphoglucose isomerase; PFK, phosphofructokinase; PGA, phosphoglycerate; MDH, malate dehydrogenase; TPI, triose-phosphate isomerase.
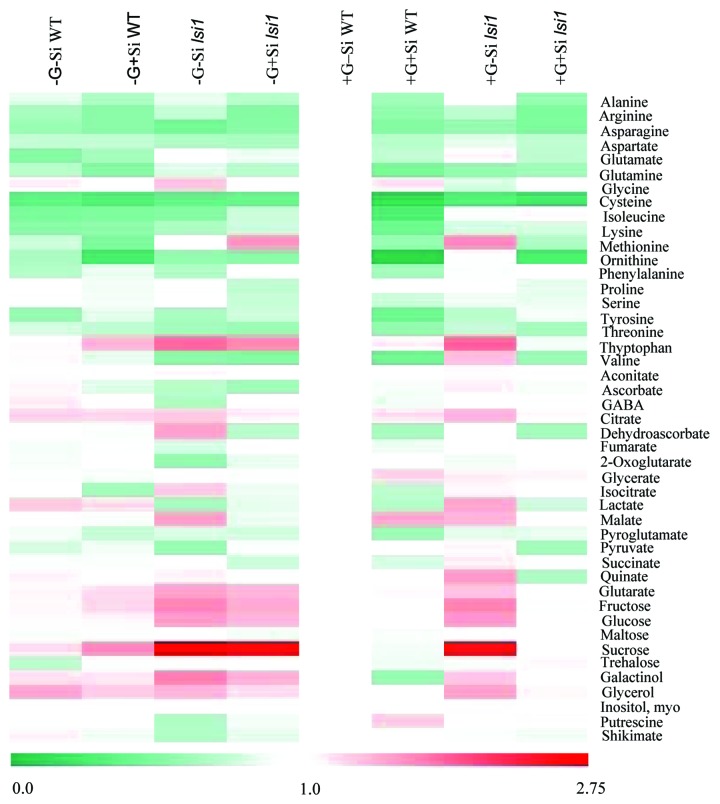
Our recent results demonstrated that several amino acids (e.g., alanine, aspartate, ornithine and threonine), as well as the sugars glucose and fructose, accounted for by the main changes observed in primary metabolism of rice plants grown in presence of Si. It has also been suggested that amino acid export (as observed in our study) can be regulated by sucrose transport or metabolism18 since both sucrose and amino acid export to the sieve tube depend on the photosynthetic metabolism of the source cell.19 Thus, given the changes in the metabolite profile (Fig. 1) as well as that Si was able to increase photosynthetic rates it seems reasonable to assume that Si may directly impact the metabolite profile of rice, as noted for the low-Si mutant defective in Si uptake (lsi1), where a clear separation of the effects of Si from the effects of grain load on the metabolite profile could be demonstrated using multivariate canonical analysis. It is important to mention, however, that in many cases, reductions of enzyme/protein levels do not lead to significant metabolic alterations, probably due to the induction of compensatory mechanisms.20 However, this seems not to be the case in our study since in the lsi1 knockdown lines the leaf metabolite content was much more affected than in the corresponding WT controls. Indeed, the changes observed in leaf metabolism cannot directly be associated with changes in the transcript levels of enzymes21 or enzyme activities associated with Si nutrition (Table 1), suggesting either post-transcriptional regulation or mass-action/ allosteric regulation of enzyme activities are major factors underlying the metabolic changes observed in our recent study.12 By sharp contrast, K deficiency has been associated with enzyme regulation at the levels of both mRNA and protein by maintaining carbon flux into amino acids and proteins and decreasing the carbon/nitrogen ratio of the total amino acid pools.22 Returning to Si, our data suggest that this mineral may act as a signal to promote amino acid remobilization similarly to the situation observed with diseased rice plants, in which Si nutrition may trigger mechanisms of host resistance via alterations in plant metabolism.23 Support for this assumption comes from a recent study demonstrating that Si nutrition can modulate the expression of a leucine-rich repeat (LRR) family protein24 and can play a central role in perceiving the, as yet uncharacterized, Si signal. LRR proteins belong to the receptor-like kinase (RLK) family, a major protein family with more than 1100 members in rice.25 Notably, plant RLKs characterized to date have been implicated in diverse biological processes, including development, the self-incompatibility response, pathogen responses, as well as responses to several environmental stresses.26 In addition, a considerable impact of Si supply was previously observed on the transcript levels of LRR-RLK genes,24 highlighting the possibility that this regulatory protein plays a central role either in Si signal perception, or in the promotion of metabolic alterations following this event, or both. Collectively, this information highlights the importance of Si nutrition as a potential target to enhance the agronomic yields of crops such as rice.
Figure 1. Changes in metabolite content in flag leaves of two rice genotypes [cv ‘Oochikara’ (WT) and the lsi1 mutant defective for Si uptake] under the effects of Si supply (0 and 2 mM: –Si and +Si, respectively) and grain load (0 or full grain burden: -G and +G, respectively). Overlay heat map of the metabolite profiles represents the changes in relative metabolite contents. The colors indicate the proportional content of each putatively identified metabolite among the samples, as determined by the average peak response area after normalization against the +G–Si WT plants. The lowest normalized value receives green, and the highest receives red (see color bar at the bottom).
In summary, we provided compelling evidence that Si likely plays an as yet unknown function in rice metabolism, even under unstressed conditions. Our recent results12 suggest that Si-mediated metabolic alterations are a complex phenomenon that is clearly worthy of more detailed analysis at both molecular and biochemical levels.
Acknowledgments
This research was supported by the Foundation for Research Assistance of the Minas Gerais State, Brazil (FAPEMIG, Grant APQ-02260–11) and by the National Council for Scientific and Technological Development, Brazil (CNPq, Grant 302605/2010–0) to FMD.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22523
References
- 1.Epstein E. The anomaly of silicon in plant biology. Proc Natl Acad Sci USA. 1994;91:11–7. doi: 10.1073/pnas.91.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang Y. Effects of silicon on enzyme activity and sodium, potassium and calcium concentration in barley under salt stress. Plant Soil. 1999;209:217–24. doi: 10.1023/A:1004526604913. [DOI] [Google Scholar]
- 3.Epstein E. SILICON. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:641–64. doi: 10.1146/annurev.arplant.50.1.641. [DOI] [PubMed] [Google Scholar]
- 4.Liang Y, Wong JWC, Wei L. Silicon-mediated enhancement of cadmium tolerance in maize (Zea mays L.) grown in cadmium contaminated soil. Chemosphere. 2005;58:475–83. doi: 10.1016/j.chemosphere.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Ma JF. Role of silicon in enhancing the resistance of plants to biotic and abiotic stresses. Soil Sci Plant Nutr. 2004;50:11–8. doi: 10.1080/00380768.2004.10408447. [DOI] [Google Scholar]
- 6.Fauteux F, Rémus-Borel W, Menzies JG, Bélanger RR. Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol Lett. 2005;249:1–6. doi: 10.1016/j.femsle.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues FA, Jurick WM, Datnoff LE, Jones JB, Rollins JA. Silicon influences cytological and molecular events in compatible and incompatible rice-Magnaporthe grisea interactions. Physiol Mol Plant Pathol. 2005;66:144–59. doi: 10.1016/j.pmpp.2005.06.002. [DOI] [Google Scholar]
- 8.Ghareeb H, Bozsó Z, Ott PG, Repenning C, Stahl F, Wydra K. Transcriptome of silicon-induced resistance against Ralstonia solanacearum in the silicon non-accumulator tomato implicates priming effect. Physiol Mol Plant Pathol. 2011;75:83–9. doi: 10.1016/j.pmpp.2010.11.004. [DOI] [Google Scholar]
- 9.Shetty R, Fretté X, Jensen B, Shetty NP, Jensen JD, Jørgensen HJL, et al. Silicon-Induced changes in antifungal phenolic acids, flavonoids, and key phenylpropanoid pathway genes during the interaction between miniature roses and the biotrophic pathogen Podosphaera pannosa. Plant Physiol. 2011;157:2194–205. doi: 10.1104/pp.111.185215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein E. Silicon: its manifold roles in plants. Ann Appl Biol. 2009;155:155–60. doi: 10.1111/j.1744-7348.2009.00343.x. [DOI] [Google Scholar]
- 11.Tamai K, Ma J. Reexamination of silicon effects on rice growth and production under field conditions using a low silicon mutant. Plant Soil. 2008;307:21–7. doi: 10.1007/s11104-008-9571-y. [DOI] [Google Scholar]
- 12.Detmann KC, Araújo WL, Martins SCV, Sanglard LMVP, Reis JV, Detmann E, et al. Silicon nutrition increases grain yield, which, in turn, exerts a feed-forward stimulation of photosynthetic rates via enhanced mesophyll conductance and alters primary metabolism in rice. New Phytol. 2012;196:752–62. doi: 10.1111/j.1469-8137.2012.04299.x. [DOI] [PubMed] [Google Scholar]
- 13.Sweetlove LJ, Fernie AR. Regulation of metabolic networks: understanding metabolic complexity in the systems biology era. New Phytol. 2005;168:9–24. doi: 10.1111/j.1469-8137.2005.01513.x. [DOI] [PubMed] [Google Scholar]
- 14.Less H, Galili G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 2008;147:316–30. doi: 10.1104/pp.108.115733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meitzel T, Radchuk R, Nunes-Nesi A, Fernie AR, Link W, Weschke W, et al. Hybrid embryos of Vicia faba develop enhanced sink strength, which is established during early development. Plant J. 2011;65:517–31. doi: 10.1111/j.1365-313X.2010.04450.x. [DOI] [PubMed] [Google Scholar]
- 16.Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell. 2010;22:1549–63. doi: 10.1105/tpc.110.075630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011;16:489–98. doi: 10.1016/j.tplants.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Barneix AJ. Physiology and biochemistry of source-regulated protein accumulation in the wheat grain. J Plant Physiol. 2007;164:581–90. doi: 10.1016/j.jplph.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Winter H, Lohaus G, Heldt HW. Phloem transport of amino acids in relation to their cytosolic levels in barley leaves. Plant Physiol. 1992;99:996–1004. doi: 10.1104/pp.99.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodges M. Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J Exp Bot. 2002;53:905–16. doi: 10.1093/jexbot/53.370.905. [DOI] [PubMed] [Google Scholar]
- 21.Fauteux F, Chain F, Belzile F, Menzies JG, Bélanger RR. The protective role of silicon in the Arabidopsis-powdery mildew pathosystem. Proc Natl Acad Sci USA. 2006;103:17554–9. doi: 10.1073/pnas.0606330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y. Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 2009;150:772–85. doi: 10.1104/pp.108.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dallagnol LJ, Rodrigues FA, DaMatta FM, Mielli MVB, Pereira SC. Deficiency in silicon uptake affects cytological, physiological, and biochemical events in the rice-Bipolaris oryzae interaction. Phytopathology. 2011;101:92–104. doi: 10.1094/PHYTO-04-10-0105. [DOI] [PubMed] [Google Scholar]
- 24.Fleck AT, Nye T, Repenning C, Stahl F, Zahn M, Schenk MK. Silicon enhances suberization and lignification in roots of rice (Oryza sativa) J Exp Bot. 2011;62:2001–11. doi: 10.1093/jxb/erq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morillo SA, Tax FE. Functional analysis of receptor-like kinases in monocots and dicots. Curr Opin Plant Biol. 2006;9:460–9. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant. 2008;1:732–50. doi: 10.1093/mp/ssn047. [DOI] [PubMed] [Google Scholar]