Abstract
The bZIP proteins, GBF1, HY5 and HYH, play important regulatory roles in Arabidopsis seedling development. Whereas GBF1 plays a dual regulatory role, HY5 and HYH act as positive regulators of photomorphogenesis. The molecular and functional relations of GBF1 with HY5 and HYH in photomorphogenesis have recently been demonstrated. However, the possible interaction of bZIP domain of each of these proteins remains to be investigated. In this study, our results suggest that bZIP domains of HY5 and HYH are able to interact with the bZIP domain of GBF1. Taken together with the earlier study,9 these results suggest that the N-terminal domain of GBF1 has an inhibitory effect on its interaction with HY5 and HYH.
Keywords: photomorphogenesis, GBF1, HY5, HYH, heterodimerization, bZIP
Plant growth is controlled by integration of several cues including environmental, developmental and metabolic signals. Light is one of the most important environmental factors for plant growth and development. Arabidopsis seedlings grow with two distinct developmental patterns, skotomorphogenesis in dark and photomorphogenesis in light. During dark to light transition, light signal induces a developmental arrest upon which autotrophic growth and adult development can initiate. Many positive and negative regulators involved in this transition have been characterized, and functional relationships among them have been established.1 COP1, an E3 ubiquitin ligase, suppresses photomorphogenesis in the darkness by targeted degradation of photomorphogenesis promoting factors.2,3 HY5, a bZIP transcription factor and master positive regulator of photomorphogenesis, binds to regulartory regions of large number of genes to promote photomorphogenesis.4,5 HY5 homolog, HYH, is also a positive regulator, however is specifically involved in blue light-mediated photomorphogenesis.6 Another bZIP protein, GBF1 plays a dual regulatory role in photomorphogenesis: it promotes cotyledon expansion, however acts as a negative regulator of light mediated inhibition of hypocotyl elongation.7 Whereas HY5 and HYH are degraded by COP1 in the dark, GBF1 is likely to be degraded by a proteasomal pathway independent of COP1 in the darkness.8 Further, COP1 is required for the optimum level of accumulation of GBF1 in light.8
Very recently it has been shown that GBF1 heterodimerizes with both HY5 and HYH and regulates photomorphogenesis in an interdependent manner.9 Domain-wise protein-protein interaction studies have revealed that although bZIP domain of GBF1 is sufficient to mediate its interaction with full length HY5 and HYH proteins, the bZIP domain of HY5 or HYH is unable to interact with full-length GBF1.9 Here, in this study, we have investigated the possible reason of inability of bZIP domain of HY5 or HYH to interact with full-length GBF1 protein using BiFC experiments.
N-terminal-Truncated GBF1 Can Interact with bZIP Domain of HY5 or HYH
In our previous study we found that the N-terminal, proline-rich region of GBF1 is not necessary in its heterodimerization with HY5 and HYH proteins, and more importantly, our DNA-protein interaction studies have revealed that the bZIP domain of GBF1 forms much stronger heterodimer with HY5 than the full-length GBF1.9 Therefore, we ask whether the proline-rich N-terminal domain of GBF1 plays any negative regulatory role in its heterodimerization with the bZIP domains of HY5 and HYH. This hypothesis is further strengthened by the fact that the full-length GBF1 protein is unable to interact with bZIP domain of HY5 or HYH.9 In order to examine this hypothesis, we removed the N-terminal proline-rich region of GBF1, and then analyzed interaction of this truncated version of GBF1 with bZIP domains of HY5 and HYH through BiFC experiments. As shown in Figure 1B and C, the truncated GBF1 was able to form heterodimers with bZIP domains of HY5 and HYH. Thus, our results indicate that the N-terminal proline rich region of GBF1 is likely to play an inhibitory role in heterodimerization with the bZIP domains of HY5 and HYH proteins.
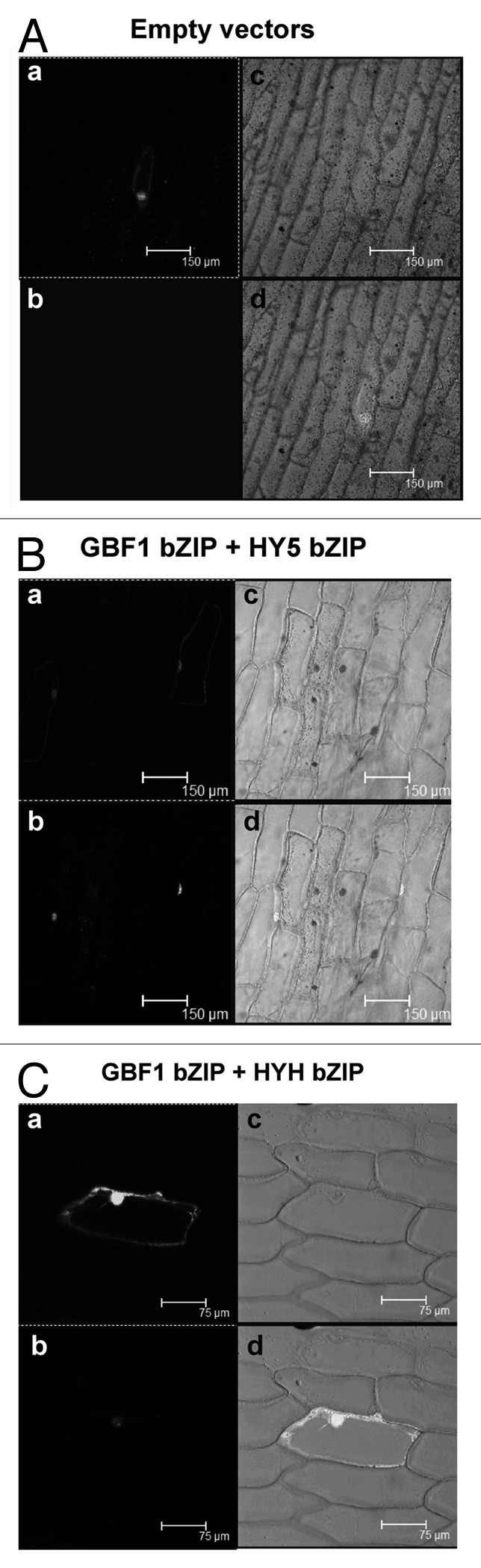
Figure 1. The bZIP domain of GBF1 forms heterodimers with bZIP domain of HY5 and HYH. In (A–C), (a) shows GFP channel fluorescence produced by GFP of pCAMBIA-1302 vector, serving as a positive control of transformation; (b) shows the YFP channel image produced by reconstruction of YFP; (c) shows the respective bright field image. (d) Shows merged image of (a, b and c). (A) Empty BiFC vectors and pCAMBIA-1302 vector (GFP) were co-transformed into onion epidermal cells. (B) bZIP domain (aa 220–315) of GBF1 (fused with YFP Nter) and bZIP domain (aa 77–168) of HY5 (fused with YFP Cter) were co-expressed into onion epidermal cells along with GFP. (C) bZIP domain (aa 220–315) of GBF1 (fused with YFP Cter) and bZIP domain (aa 74–149) of HYH (fused with YFP Nter) were co-expressed into onion epidermal cells along with GFP.
It is tempted to speculate that during the interaction of full-length proteins, the inhibitory effect of N-terminal region of GBF1 might be neutralized by N-terminal domains of HY5 and HYH proteins, and thereby the full-length GBF1 protein is able to interact with full-length HY5 and HYH proteins, however not with N-terminal-removed truncated HY5 and HYH proteins. At lower intensities of light, GBF1 works downstream to HY5 in the regulation of photomorphogenesis; at moderate intensities of light, the proteins work antagonistic to each other and at higher intensities of light, HY5 works downstream to GBF1.9 In order to establish such dynamic epistatic relationships, it is essential that protein partners should readily form heterodimers (for antagonistic function) as well as break existing heterodimers to form homodimers (to work epistatic to each other). Thus, it is likely that the heterodimerization-favoring domain (bZIP domains) as well as non-favoring (N-terminal domain of GBF1)-domain, are crucial to establish the dynamic epistatic relationships in the regulation of photomorphogenesis.
Working Model of Interaction of GBF1 with HY5 and HYH
Based on the results obtained in this study as well as our previous study,9 the model (Fig. 2A) summarizes the role of various domains of GBF1 in its interaction with HY5 and HYH. The leucine zipper domain mediates the heterodimerization, whereas the N-terminal proline-rich domain inhibits this heterodimerization. The basic region is important for DNA binding and nuclear localization. Figure 2B summarizes the observed interactions in this study as well as the previous study.9 Full-length GBF1 interacts with full-length HY5 and HYH proteins. Similarly, the bZIP domain of GBF1 interacts with the bZIP domains of HY5 and HYH. The bZIP domain of GBF1 also interacts with full-length HY5 and HYH proteins, but the bZIP domain of HY5 or HYH doesn’t interact with full-length GBF1 protein (because of presence of inhibitory N-terminal in GBF1).
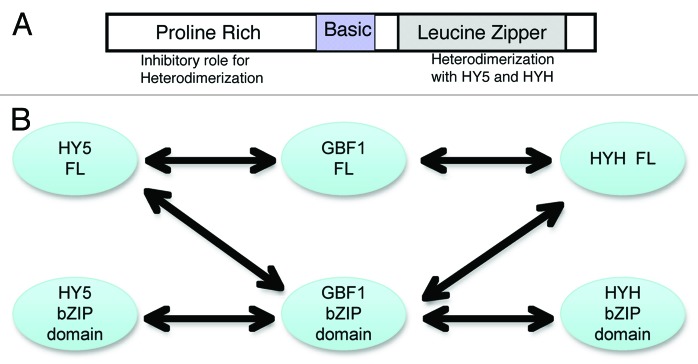
Figure 2. Domain-wise interaction of GBF1 with HY5 and HYH. (A) Various domains of GBF1 in its interaction with HY5 and HYH. (B) Full-length GBF1 as well as its bZIP domain, but not the N-terminal domain interact with full-length HY5 and HYH proteins. The bZIP domains of HY5 and HYH are able to interact with bZIP domain of GBF1, however not with the full-length GBF1.
Materials and methods used in this study are in accordance with the previous study.9
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22703
References
- 1.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–30. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 2.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–6. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 3.Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–7. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–95. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, He H, Wang X, Wang X, Yang X, Li L, et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65:346–58. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 6.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–59. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallappa C, Yadav V, Negi P, Chattopadhyay S. A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J Biol Chem. 2006;281:22190–9. doi: 10.1074/jbc.M601172200. [DOI] [PubMed] [Google Scholar]
- 8.Mallappa C, Singh A, Ram H, Chattopadhyay S. GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. J Biol Chem. 2008;283:35772–82. doi: 10.1074/jbc.M803437200. [DOI] [PubMed] [Google Scholar]
- 9.Singh A, Ram H, Abbas N, Chattopadhyay S. Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J Biol Chem. 2012;287:25995–6009. doi: 10.1074/jbc.M111.333906. [DOI] [PMC free article] [PubMed] [Google Scholar]


