Abstract
Rad54 and Rad51 are important proteins for the repair of double-stranded DNA breaks by homologous recombination in eukaryotes. As previously shown, Rad51 protein forms nucleoprotein filaments on single-stranded DNA, and Rad54 protein directly interacts with such filaments to enhance synapsis, the homologous pairing with a double-stranded DNA partner. Here we demonstrate that Saccharomyces cerevisiae Rad54 protein has an additional role in the postsynaptic phase of DNA strand exchange by stimulating heteroduplex DNA extension of established joint molecules in Rad51/Rpa-mediated DNA strand exchange. This function depended on the ATPase activity of Rad54 protein and on specific protein:protein interactions between the yeast Rad54 and Rad51 proteins.
Keywords: heteroduplex DNA extension, double-strand break repair, recombination, yeast
Accurate repair of DNA double-strand breaks (DSB) is important for the survival and genomic stability of all organisms. Homologous recombination is an evolutionarily conserved process that is involved in DSB repair in all life forms (1). Central to this process is the homologous DNA pairing and DNA strand exchange that is performed by the Escherichia coli RecA protein or its eukaryal and archaeal homologs, Rad51 and RadA proteins, respectively (2). These proteins form nucleoprotein filaments with single-stranded DNA (ssDNA) of highly similar structure and function. The formation of the Rad51 (RecA, RadA) nucleoprotein filament is referred to as the presynaptic phase of homologous recombination and is stimulated by ssDNA binding proteins, like the eukaryotic Rpa (Replication protein A) (3, 4). The nucleoprotein filament performs the critical functions in recombination in the synaptic phase of the reaction: homology search and DNA strand exchange between the bound ssDNA and the homologous double-stranded DNA (dsDNA) partner (5). Heteroduplex DNA (hDNA) extension and branch migration occurs in the postsynaptic phase of the reaction (2). In E. coli, hDNA extension and branch migration is catalyzed by the RuvAB proteins (6). Finally, resolution of Holliday junctions is achieved in E. coli by the junction-specific RuvC endonuclease (6). A mechanism of hDNA extension that differs from the RuvAB paradigm has been described in bacteriophage T4 (7). Biochemical experiments have revealed activities that resemble bacterial RuvABC in fractionated mammalian cell extracts (8–10), but the responsible gene products have not been identified yet. Sequence analysis has failed to identify proteins with significant sequence homology to RuvABC proteins in eukaryotes (refs. 1 and 11; see Fig. 6). The mechanisms of hDNA extension, branch migration, and Holliday junction resolution in eukaryotes are poorly understood presently.
Figure 6.
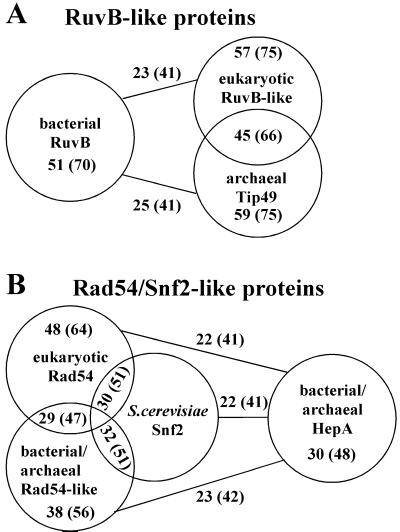
Comparison of proteins with homologies to RuvB and Rad54/Snf2. The protein sequences of E. coli RuvB and S. cerevisiae Rad54 were used in BLAST searches to define RuvB- and Rad54-like protein families. The GenBank accession numbers are given for each protein. The groups comprise the following proteins: (A) Bacterial RuvB proteins from Borrelia burgdorferi AAC66410, Bacillus halodurans BAB04944, Bacillus subtilis CAB75331, Campylobacter jejuni CAB73789, Chlamydia muridarum AAF39175, Chlamydophila pneumoniae BAA98598, Chlamydia trachomatis AAC67630, Deinococcus radiodurans AAF10176, E. coli BAA15671, Hemophilus influenzae AAC21975, Heliobacter pylori AAD08100, Mycoplasma pneumoniae AAB95954, Mycobacterium leprae AAA17098, Mycoplasma genitalium AAC71584, Mycobacterium tuberculosis CAB01285, Neisseria meningitidis AAF41624, Pseudomonas aeruginosa AAG04356, Rhizobium etli AAF36814, Rickettsia prowazekii CAA14843, Streptomyces coelicolor CAB70920, Synechocystis sp. BAA10350, Thermus thermophilus BAA76480, Thermotoga maritima AAB03727, Treponema pallidum AAC65150, Vibrio cholerae AAF94993, Xylella fastidiosa AAF84708; eukaryotic RuvB-like proteins from Arabidopsis thaliana CAB66921 and BAB08471, Drosophila melanogaster AAF43412 (reptin) and AAF43411 (pontin), Homo sapiens BAA28169 RuvBL1) and BAA76708 (RuvBL2), Mus musculus BAA76297, Rattus norvegicus BAA76313, Saccharomyces cerevisiae CAA88704 (Rvb1) and CAA97952 (Rvb2); archaeal Tip49 proteins from Archaeoglobus fulgidus AAB89434, Aeropyrum pernix BAA79281, Pyrococcus abyssi CAB49285, Pyrococcus horikoshii BAA30923, Sulfolobus solfataricus (see http://niji.imb.nrc.ca/sulfolobus). (B) Eukaryotic Rad54 proteins from Arabidopsis thaliana BAB02963, Caenorhabditis elegans CAA22254 (W06D4.6), Drosophila melanogaster AAC24577 (OKR), Gallus gallus AAB54115 and AAG09308 (Rad54B), Homo sapiens CAA66379 and AAD34331 (Rad54B), Mus musculus CAA66380, Neurospora crassa BAA93079, S. cerevisiae AAA34949 and CAA85017 (Rdh54/Tid1), Schizosaccharomyces pombe CAA82750 (Rhp54); S. cerevisiae Swi2/Snf2 AAA35059; bacterial Rad54-like proteins from Bacillus cereus CAA67095, Bacillus halodurans BAB06632, Bacillus subtilis CAB15645, Chlamydia muridarum AAF73609 and AAF73530, Chlamydophila pneumoniae AAF38809 and AAF73724, Chlamydia trachomatis AAC68157 and AAC68303, Deinococcus radiodurans AAF10831, Mycoplasma genitalium AAC71234, Mycoplasma pneumoniae AAB95782, Mycobacterium tuberculosis CAA17284, Pseudomonas aeruginosa AAG04188, Streptomyces coelicolor CAB60181 and CAB82825, Synechocystis sp. BAA18659; archaeal Rad54-like protein from Sulfolobus solfataricus (see http://niji.imb.nrc.ca/sulfolobus); bacterial HepA-like proteins form Bacillus halodurans BAB06536, Bacillus subtilis BAA12545, Dich elobacter nodosus AAC33384, Deinococcus radiodurans AAF12565, E. coli AAC73170, Hemophilus influenzae AAC22275, Thermotoga maritima AAD36069, Vibrio cholerae AAF95648; archaeal HepA-like proteins from Archaeoglobus fulgidus AAB91314, Aeropyrum pernix BAA79369, Halobacterium sp. AAG20812, Pyrococcus abyssi CAB49794, Pyrococcus horikoshii BAA29994. All possible pairwise alignments with RuvB and Rad54 homologs were carried out by using BLAST. The numbers indicate the average identities and similarities (in brackets) inside the different groups as well as between the groups. Comparison of an unrelated protein Xrn1 from S. cerevisiae (AAA35219) to all bacterial RuvB proteins resulted in 23% identity and 41% similarity. Comparison of a random shuffled E. coli RuvB sequence to all bacterial RuvB proteins (including E. coli RuvB protein) resulted in 23% identity and 39% similarity.
The yeast Saccharomyces cerevisiae is an excellent system to study DSB repair by homologous recombination. The genes of the RAD52 epistasis group (RAD50, XRS2, MRE11, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, RFA1) define this recombinational repair pathway (1). Numerous interactions occur between the encoded proteins, and they have provided a basis for understanding the specific function(s) of each protein during homologous recombination. The central role of Rad51 protein is supported by its numerous interactions with itself (12, 13), Rad52 (12, 14), Rad55 (13, 15), and Rad54 proteins (16, 17). In addition to its interaction with Rad51 protein, Rad52 protein also interacts with Rpa (18, 19). Rad55 and Rad57 proteins form a stable heterodimer (20). In addition, Rad50, Mre11, and Xrs2 proteins form a heterotrimer with nuclease activity, believed to be involved in DSB processing (1).
Functional studies with the eukaryotic RAD52 group proteins have provided insights into the mechanism of recombinational repair. Rpa, Rad55/57 heterodimer, Rad52, Rad54, and its homolog Tid1/Rdh54 proteins have been found to stimulate Rad51 protein-mediated in vitro recombination. By eliminating secondary structures in ssDNA, Rpa stimulates the presynaptic phase and enhances the formation of the presynaptic filament (3, 4). Rad52 protein and the Rad55/57 heterodimer stimulate the presynaptic phase by mediating the exchange of an Rpa-ssDNA filament for a Rad51 protein-ssDNA filament (21–23). Human Rad52 protein was shown to stimulate the human Rad51 protein in an Rpa-independent mode (24). Finally, Rad54 protein was found to stimulate Rad51 protein-mediated in vitro recombination reactions (25, 26) in the synaptic phase of the reaction by specifically interacting with the established Rad51 nucleoprotein filament (27–29). Topological remodeling of the dsDNA by Rad54 was proposed as a mechanism for the observed stimulation (26–28, 30). The Rad54-related Tid1 protein was also found to stimulate Rad51-mediated in vitro recombination in a fashion similar to that of Rad54 protein, probably involving topological remodeling of the duplex DNA as well (31).
Mutations in the RAD54 gene in S. cerevisiae confer a strong DSB-repair defect and also affect other aspects of DNA metabolism, consistent with an important function during homologous recombination (1). The gene is evolutionarily conserved and plays a similar role in vertebrates (32–34). Rad54 protein is a member of the Snf2/Swi2 protein family of DNA-dependent/stimulated ATPases that modulate protein:DNA interactions in transcription, DNA repair, and recombination (35). Rad54 protein possesses a dsDNA-specific ATPase activity that is important for its in vivo and in vitro functions (25–29, 36, 37). The energy of ATP-hydrolysis is required for Rad54 protein to topologically remodel duplex DNA (26, 30) by introducing unconstrained negative and positive supercoils (28). This activity is probably responsible for the stimulation of Rad51 protein-mediated in vitro recombination (27, 28).
Previous work with yeast recombination proteins has established that Rad54 protein functions in the synaptic phase of Rad51 protein-mediated in vitro recombination, but did not address directly a possible role during postsynapsis (25–29). We found that Rad54 protein, in addition to its synaptic role, also functions postsynaptically by stimulating the extension of hDNA in established joint molecules in the Rad51 protein-mediated DNA strand exchange between circular ssDNA and linear dsDNA. This stimulation depended on the ATPase activity of Rad54 protein, and on its specific interaction with the cognate S. cerevisiae strand exchange protein Rad51. Our data reveal an aspect of the critical role of yeast Rad54 protein in homologous recombination and supply evidence for a protein involved in the postsynaptic phase of recombinational repair in eukaryotes.
Materials and Methods
Proteins and DNA.
S. cerevisiae Rad51 protein and Rpa as well as Rad54 and Rad54-K341R proteins were purified as described (4, 22, 29). The Rad54 proteins were used as GST-fusion proteins and the wild-type fusion was fully active in vivo and in vitro (29). The ATPase activity measured at 37°C (1,100–1,300 min−1) is in accord with previously published data (1,270 min−1; ref. 25). ΦX174 and M13 DNAs were purchased from New England Biolabs.
DNA Strand Exchange and hDNA Extension Assay.
DNA strand exchange was performed as described (29). The conditions were derived from previous work (4, 38). Briefly, reactions (21 μl) contained 30 mM Tris acetate, pH 7.5, 1 mM DTT, 50 μg/ml BSA, 20 mM ATP, 20 mM MgOAc, 4 mM spermidine, an ATP-regenerating system consisting of 20 mM creatine phosphate and 1.2 μg creatine kinase, 33 μM (nt) ΦX174 ssDNA, and 10.3 μM Rad51 or RecA protein (as indicated). After a 15-min incubation at 30°C, 1.8 μM Rpa was added and the reaction was kept at 30°C for 30 min. The time course was started by the addition of 33 μM (nt) ΦX174 dsDNA linearized with PstI and labeled at the 5′ ends by using T4 polynucleotide kinase (New England Biolabs), 4 mM spermidine, and storage buffer, Rad54 or Rad54-K341R protein (0.2 μM), respectively. The final salt concentration was 30 mM NaCl. For hDNA extension assays (modified from ref. 39), 6-μl portions of the reactions were digested with the restriction endonucleases StuI (100 units), SacII (30 units), or SspI (15 units) in a total volume of 40 μl. After incubation at 37°C for 1 h, the samples were deproteinized with 4 μl of stop buffer (0.7% SDS, 4 mg/ml proteinase K, 350 mM EDTA) for 20 min at 37°C, mixed with 5 μl of alkaline loading buffer (50 mM NaOH, 1 mM EDTA, 2.5% Ficoll, 0.025% bromcresol green), and subjected to denaturing gel electrophoresis on 1% agarose gels at 25 V over night. The gels were dried and quantified on a phosphorimager by using imagequant software. Rates of hDNA extension were calculated from normalized graphs as shown in Fig. 3, dividing the time needed for 50% of the joint molecules moving from one restriction site to the other by the length of the interval (see Fig. 4 for intervals).
Figure 3.
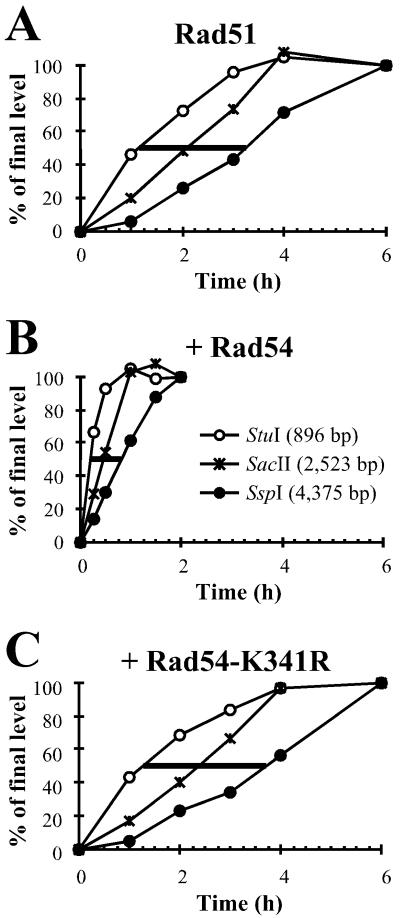
Rad54 protein stimulates joint molecule formation and subsequent hDNA extension. DNA from DNA strand exchange reactions as described in Fig. 1A was digested with StuI (○), SacII (*), or SspI (●) and subjected to denaturing gel electrophoresis. All bands were quantified on a phosphorimager and the relative amount corresponding to the uncut fragment arising from hDNA extension past the restriction site was calculated (see Fig. 1 B and C). All data were corrected for the contribution to the accumulation of uncut fragment by the relocalization of Rad51 protein as described in Fig. 2. Reactions contained S. cerevisiae Rad51 protein and Rpa, to which storage buffer (Rad51 control; A), Rad54 (B), or Rad54-K341R protein (C) was added. The data were normalized by setting the endpoints of each reaction to 100%. This was possible because all reactions were at or very near to their endpoints and achieved very similar final yields (25–30% products). The lines at 50% represent the time needed for half of the population of joint molecules to migrate from the StuI site to the SspI site.
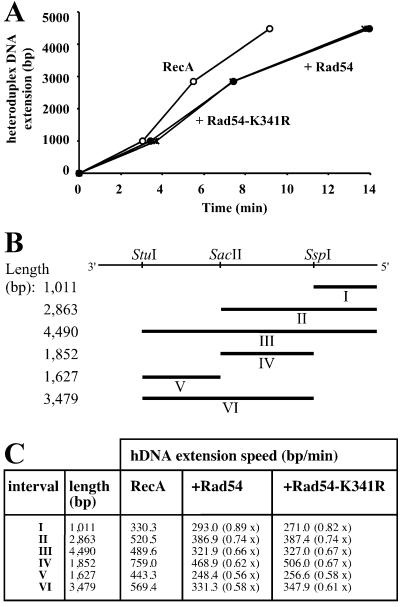
Figure 4.
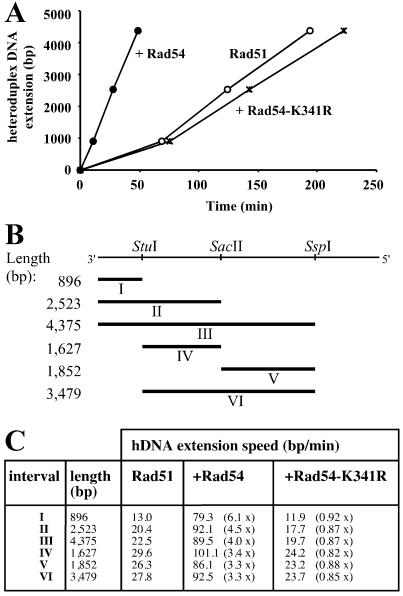
Rad54 protein stimulates the rate of hDNA extension in Rad51 protein-mediated DNA strand exchange. (A) Graphic representation of the effect of Rad54 protein on hDNA extension derived from the data shown in Fig. 3 A–C. Plotted are the time points when 50% of the joint molecules have passed the physical distance defined by the three restriction sites. Reactions contained S. cerevisiae Rad51 protein and Rpa as well as storage buffer (Rad51 control; ○), Rad54 (●), or Rad54-K341R (*). (B) Intervals in DNA strand exchange. ΦX174 dsDNA linearized with PstI is represented on top and has a total length of 5,386 bp. The direction refers to the virion DNA (plus strand). The restriction sites used for this analysis are indicated. Rates for hDNA extension could be calculated for intervals I–VI. (C) Rad54 protein stimulates the rate of hDNA extension. The intervals are illustrated in B. The numbers in parentheses represent the fold stimulation in comparison to control reactions containing Rad51 protein to which protein storage buffer was added.
Results
Assay for hDNA Extension and Controls.
The assay used to measure the extension of hDNA in the postsynaptic phase of S. cerevisiae Rad51/Rpa-mediated DNA strand exchange is described in Fig. 1. It is based on previous work on hDNA extension stimulated by the helicases in bacteriophage T4 uvsX protein-mediated DNA strand exchange (7, 39). We term the postsynaptic phase in the three-strand DNA strand exchange reaction “hDNA extension,” and not branch migration as in some previous studies (2, 7, 39, 40). Branch migration is referred to as the postsynaptic phase in four-stranded reactions. Restriction enzyme sites on the linear dsDNA were used to monitor hDNA as it passes the respective recognition site. The accumulation of uncleaved, [32P] end-labeled ssDNA (the upper strand of the duplex substrate, Fig. 1A) can easily be quantified as a measure of the extent of hDNA following gel electrophoresis on denaturing gels. An example of the data are shown in Fig. 1 B and C. Using three, instead of one (7, 39), restriction sites, we were able to measure the rates of hDNA extension in three internal intervals that did not contain the starting point of the reaction (see Fig. 4). Because the reaction showed a clear directionality (as shown in Fig. 1A), the internal intervals allowed the measurement of effects of the S. cerevisiae Rad54 protein on postsynapsis (hDNA extension) independently of its synaptic role in forming joint molecules (as discussed in detail later). This is important because Rad54 protein had been previously shown to specifically stimulate the formation of joint molecules in DNA strand exchange (29).
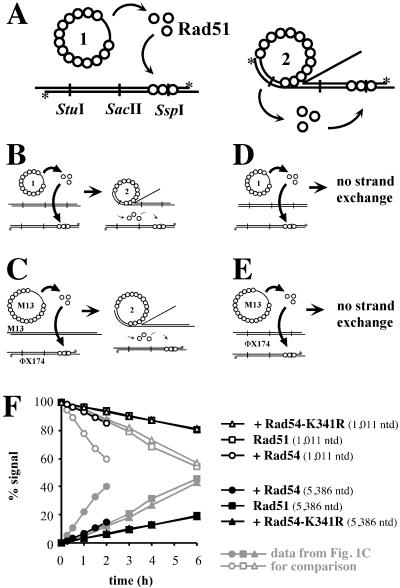
Figure 1.
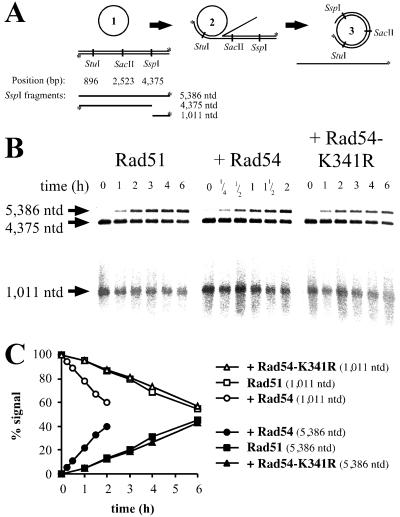
DNA strand exchange and assay for hDNA extension. (A) Schematic representation of the hDNA extension assay (modified from ref. 39). DNA strand exchange with circular ssDNA and linear dsDNA substrates (1). The joint molecules (2) are resolved by hDNA extension to nicked circular and displaced ssDNA end products (3). Digestion with different restriction endonucleases followed by denaturing gel electrophoresis is used to observe hDNA extension. Note that the displaced strand cannot be digested once hDNA extends over the restriction site. The restriction endonucleases used and the positions of the corresponding restriction sites along the dsDNA are indicated. Radioactively labeled 5′ ends are marked with an asterisk. The expected fragments after digestion with SspI are indicated and data for this restriction enzyme are shown in B and C. (B) Denaturing agarose gels analyzing DNAs from DNA strand exchange reactions (A) containing Rad51 protein and Rpa. Storage buffer (Rad51 control), Rad54, or Rad54-K341R protein was added as indicated. All reactions shown were digested with SspI restriction endonuclease. The band corresponding to the full-length displaced strand (5,386 nt) appears in a time-dependent manner. At the same time, the amount of the corresponding shorter fragment (1,011 nt) declines. The sizes of the fragments are indicated on the left. The incubation times of the DNA strand exchange assays are shown for each lane. Note the shorter incubation times for the reactions containing Rad54 protein. A minor loss of label that was not dependent on Rad54 protein was observed at late time points. This loss of label did not affect the quantitation, as the relative amount of the 5,386-nt product was determined in comparison to the digested 1,011-nt fragment (see C). (C) Graphical representation of data shown in B. The 5,386-nt (●, ■, ▴) and 1,011-nt (○, □, ▵) bands were quantified by phosphorimaging, and the relative amounts of both fragments are displayed. At 0 h the signal for the 1,001-nt fragment was defined as 100% and the signal for the 5,386-nt fragment as 0%. Reactions contained S. cerevisiae Rad51 protein and Rpa, as well as storage buffer (Rad51 control; □, ■), Rad54 (○, ●), or Rad54-K341R (▵, ▴) protein. The relative decrease and increase of both fragments after quantitation by a phosphorimager is more evident than by visual inspection of the scanned image.
An interesting caveat in this assay is that proteins, in particular Rad51 protein, may relocate onto the dsDNA and inhibit restriction enzyme cleavage (Fig. 2A). This relocalization of Rad51 protein was monitored by four control reactions measuring restriction enzyme inhibition by protein transfer from the ssDNA filament to the dsDNA in the presence and absence of DNA strand exchange (Fig. 2 B–E). The relocalization of Rad51 protein was monitored by measuring the inhibition of all restriction enzymes used (StuI, SacII, and SspI). No significant difference could be observed between reactions with or without DNA strand exchange, indicating that the observed relocalization of Rad51 protein was not stimulated by an ongoing reaction. The amount of inhibition observed in all four types of control experiments for all restriction endonucleases (StuI, SacII, and SspI) was very similar and reached 12–20% of total products in a linear, time-dependent fashion. Addition of wild-type, but not mutant, Rad54 protein to such control reactions enhanced the observed relocalization of Rad51 protein in comparison to controls containing only Rad51 protein, but never exceeded 20% of the total signal.
Figure 2.
Control experiments for hDNA extension assay. Control reactions to measure the amount of inhibition of the restriction endonucleases due to the relocalization of Rad51 protein to the dsDNA. (A) Relocalization of Rad51 protein from ssDNA to dsDNA might occur either spontaneously (Right) or may be enhanced during DNA strand exchange (Left). (B–E) To determine the amount of signal in the hDNA extension assay, which corresponds to uncleaved dsDNA analyzed on a denaturing gel because of interference by Rad51 protein relocating on dsDNA, four types of control reactions were performed: in the presence of ongoing DNA strand exchange (B + C) and in the absence of DNA strand exchange (D + E). (B + D) Reactions contained blunt-ended dsDNA (linearized with FspI and labeled at the 5′ ends by polynucleotide kinase as indicated with asterisks), which could not participate in DNA strand exchange (ref. 58 and data not shown). The unlabeled dsDNA was either digested with PstI (B) to enable DNA strand exchange or with FspI (D) to preclude DNA strand exchange. (C + E) Control experiments with heterologous M13mp19 ssDNA. (C) Reactions with ongoing DNA strand exchange contained circular M13mp19 ssDNA and PstI linearized M13mp19 dsDNA. (E) Reactions without DNA strand exchange contained circular M13mp19 ssDNA and PstI linearized ΦX174 dsDNA. The relocalization of Rad51 protein was monitored with a radioactively end-labeled (*) ΦX174 dsDNA linearized with PstI. (F) Graphical representation of data from control reactions as in E by using SspI restriction endonuclease. To reactions with Rad51 protein and Rpa, storage buffer (Rad51 control) (□, ■), Rad54 (○, ●), or Rad54-K341R (▵, ▴) protein was added. All results were obtained in the same manner as in Fig. 1, and the data from Fig. 1C are indicated in gray for comparison.
All experimental data (Figs. 3 and 4) were corrected for the contribution of Rad51 protein relocalization to the overall signal in the hDNA extension assay. The results from the control reactions with M13mp19 ssDNA and linear ΦX174 dsDNA (Fig. 2E) containing S. cerevisiae Rad51 protein and Rpa supplemented with storage buffer (control), Rad54, or Rad54-K341R protein (Fig. 2F) were subtracted from the experimental data for each corresponding time point for each of the three restriction enzymes (StuI, SacII, and SspI) used. No inhibition of restriction enzyme cleavage of dsDNA could be observed to be due to Rpa or Rad54 protein (data not shown). The progress of the reaction from left to right also argues against another potential artifact in this assay, that the restriction enzymes were inhibited by internal (paranemic) joint formation.
Rad54 Protein Stimulates hDNA Extension in an ATP-Dependent Fashion.
Previous experiments have shown that the ATPase activity of Rad54 protein is quickly inactivated at 37°C, but not at 30°C (25, 26, 29). Therefore, we performed all experiments at 30°C, to be able to measure a possible postsynaptic (late) effect of Rad54 protein. DNA strand exchange reactions were performed with S. cerevisiae Rad51 protein and Rpa. Product formation was measured as the accumulation of the uncleaved 5,386-nt fragment deriving from the linear ΦX174 dsDNA after electrophoresis on denaturing agarose gels. All reactions reached their endpoint, which was about 25–30% of product formation, defined as the accumulation of uncleaved ssDNA for each of the three restriction enzymes (data not shown). The reaction proceeded from the left to the right (Figs. 1–4), which corresponds to a 3′ to 5′ direction with respect to the displaced strand of the dsDNA. This is consistent with the previously determined polarity of the full-length S. cerevisiae and human Rad51 proteins, which is opposite to that of RecA protein (38, 41, 42). The addition of wild-type S. cerevisiae Rad54 protein greatly shortened the time needed for the accumulation of uncleaved product. To calculate the rates of hDNA extension, the absolute data were normalized (Fig. 3 A–C). This allows for the determination of the time point at which 50% of the possible joints contained hDNA that spanned the restriction site examined. The stimulation of joint molecule formation could account for the faster accumulation of hDNA that reached the first restriction site (StuI; Fig. 3). In addition to this effect, the time for 50% of the formed branches to move from the first to the last site (StuI to SspI) was reduced in the presence of Rad54 protein (compare the horizontal lines at 50% in Fig. 3). Plotting the time points at which 50% of the joint molecules had hDNA that spanned the restriction sites shows a strong increase in the rate of hDNA extension in the presence of Rad54 protein (Fig. 4A). A summary of the calculated rates of hDNA extension is shown in Fig. 4C. Wild-type S. cerevisiae Rad54 protein stimulated hDNA extension about 3-fold in the three internal intervals (intervals IV, V, and VI in Fig. 4C). The effect on intervals including the left end of the dsDNA, where joint formation initiates (Fig. 1A), was 4- to 6-fold. These results show that Rad54 protein stimulates hDNA extension independently of its effect on the formation of joint molecules.
The Rad54-K341R mutant protein severely reduces ATPase activity and is defective in vitro and in vivo (26–29, 37). Here, the mutant protein served as a control to demonstrate that the effect of wild-type Rad54 protein depends on its ATPase activity. Rad54-K341R protein did not stimulate hDNA extension, and all reactions containing the mutant Rad54 protein resulted in slightly lower rates of hDNA extension than the control reactions (Figs. 3 and 4). This is consistent with the inhibitory effect of Rad54-K341R protein on nicked circle formation observed previously (29).
To determine whether this postsynaptic effect of Rad54 protein depended on species-specific protein:protein interactions, as previously shown for the synaptic effect (25–29), we tested the effect on RecA protein-mediated DNA strand exchange. Rad54 protein was unable to stimulate hDNA extension in reactions carried out by E. coli RecA protein (Fig. 5). The polarity of strand exchange was opposite to the experiments carried out with Rad51 protein (Fig. 1A), consistent with the polarity determined previously for RecA protein (2). The rate of hDNA extension was 330–759 bp/min in reactions containing storage buffer (control). Previous measurements of RecA-promoted hDNA extension were between 120 and 600 bp/min (2, 40), with an upper limit of 1,200 bp/min (43). Thus, our estimate is in general agreement with these previous studies that were done by using different assays systems and assay conditions. The addition of Rad54 or Rad54-K341R protein to RecA protein-mediated DNA strand exchange slightly reduced the rates of hDNA extension (Fig. 5B). A slight inhibition of RecA protein-mediated DNA strand exchange by Rad54 protein has been noticed (29). This observation also underlines the importance of species-specific interactions between the Rad54 and Rad51 proteins during hDNA extension.
Figure 5.
Rad54 protein does not stimulate the rate of hDNA extension in RecA-mediated DNA strand exchange. (A) Graphic representation of the effect of Rad54 protein on hDNA extension derived from data similar to those shown in Fig. 3 for Rad51 protein. Plotted are the time points when 50% of the joint molecules have passed the physical distance defined by the three restriction sites. Reactions contained E. coli RecA protein and Rpa as well as storage buffer (RecA control; ○), Rad54 (●), or Rad54-K341R (*). (B) Intervals in RecA-mediated DNA strand exchange. The restriction sites used for this analysis are indicated. Rates for hDNA extension speeds could be calculated for intervals I–VI. Note that the directionality of the reaction is opposite to that described in Fig. 4B. (C) Rad54 protein is unable to stimulate hDNA extension of joint molecules formed by RecA. The intervals are illustrated in B. The numbers in parentheses represent the fold stimulation in comparison to control reactions containing RecA protein to which protein storage buffer was added.
Discussion
Rad54 protein is an important component of the recombinational repair pathway, and cells lacking Rad54 protein exhibit defects in DNA repair and homologous recombination (1, 32, 33). Purified Rad54 protein has been previously shown to stimulate in vitro recombination reactions catalyzed by Rad51 protein (25, 26). Additional studies found that Rad54 protein enhanced Rad51 protein-mediated DNA strand exchange and D-loop formation in the synaptic phase of the reaction (27–29). A possible role of Rad54 protein in postsynapsis (i.e., during the hDNA extension phase) was suggested indirectly by results in a previous study (29) of the role of Rad54 protein during Rad51 protein-mediated DNA strand exchange. Most importantly, the ATPase-defective Rad54-K341R protein specifically inhibited nicked circle, but not the formation of joint molecules or high molecular weight networks in DNA strand exchange, showing that the protein can have an effect on later stages of the reaction. In addition, heteroduplex DNA analysis by using S1 nuclease experiments suggested a contribution of Rad54 protein to heteroduplex DNA length (29).
The role of Rad54 protein during DNA strand exchange is unique in that it stimulates DNA strand exchange and D-loop formation by specifically interacting with the established Rad51 nucleoprotein filament, which targets Rad54 protein to the homologous pairing site and stimulates its DNA remodeling activity (25–29). Here, we find that in addition to this role in synapsis, Rad54 protein stimulates the extension of hDNA after the initial synapsis. This effect was demonstrated as a more than 3-fold enhancement of the rate of hDNA extension in established joint molecules in internal intervals of the duplex DNA substrate during Rad51 protein-mediated DNA strand exchange. By focusing on internal intervals the synaptic effect was excluded, as the reaction starts at the end of the duplex DNA. As expected, the effect on terminal intervals was even higher (up to 6-fold), because of the additional contributions of Rad54 to stimulating the synaptic phase of DNA strand exchange. In comparison, RuvAB complex stimulates hDNA extension in RecA protein-mediated reactions about 5-fold (5, 44, 45). Because the stimulation of hDNA extension by Rad54 protein is ATP-dependent and Rad54 protein is using the energy derived from ATP hydrolysis to topologically remodel dsDNA (26–28, 30), we suspect that continued unwinding of the dsDNA by Rad54 protein is critical. Because Rad54 protein does not stimulate RecA-mediated DNA strand exchange, we conclude that the stimulation of hDNA extension depends on species-specific protein:protein interaction between Rad54 and the strand exchange protein Rad51. This conclusion is consistent with the specific in vivo and in vitro interactions documented for both proteins (16, 17, 25).
There are different possible mechanisms for how Rad54 might stimulate hDNA extension, and the exact mechanism remains to be established. Rad54 protein stimulates synapsis by its specific interaction with the Rad51 nucleoprotein filament that targets the Rad54 protein DNA remodeling activity to the site of homologous pairing (27, 29). The continued unwinding of dsDNA by Rad54 protein after forming a stable joint could be the basis for its stimulation of hDNA extension in the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Alternatively, hDNA extension could be viewed as a succession of individual joints forming along the Rad51 nucleoprotein filament, which are stimulated each by Rad54 protein, as previously proposed for hDNA extension and heterology bypass in RecA-mediated DNA strand exchange (see ref. 2). More work is needed to define the exact mechanism by which Rad54 stimulates hDNA extension and whether Rad54 protein needs to be continually present to exert an effect during postsynapsis.
Presently, very little is known about the later stages of homologous recombination, hDNA extension, branch migration, and junction resolution in eukaryotes. In E. coli, the mechanism of hDNA extension and branch migration by the RuvAB proteins is well understood (6). RuvB protein is a dsDNA-stimulated ATPase and DNA helicase (46). It forms a hexameric ring motor that can catalyze hDNA extension of Holliday junctions in conjunction with RuvA protein (47–49). Rad54 protein is a dsDNA-specific ATPase (25, 29, 30) and as a member of the Swi2/Snf2 family shares sequence motifs with DNA helicases (35). Despite these similarities between RuvB and Rad54 proteins, both proteins are very different. Whereas RuvB protein exhibits DNA helicase activity in standard assays, Rad54 protein does not (ref. 25 and 26; C. Bornarth and S. C. Kowalczykowski, personal communication; K. Kiianitsa and W.-D.H., unpublished observation). Unlike Rad54 protein, the RuvAB proteins do not stimulate the synaptic phase of in vitro recombination, and the RuvAB proteins show no evident physical interaction with RecA protein as Rad54 protein does with Rad51 protein (5, 6, 16, 17, 27–29, 44). RuvB protein performs hDNA extension by specific interaction with the Holliday junction mediated by its tethering to RuvA protein independent of the DNA strand exchange protein (6, 47–49). We do not expect Rad54 protein to specifically interact with junctions, like RuvAB proteins. The effect of Rad54 protein will most likely depend on its interactions with Rad51 protein. The latter notion is supported by the absence of Rad54 protein-mediated effects in RecA protein-mediated hDNA extension.
In bacteriophage T4, a mechanism for hDNA extension different form RuvAB has been documented. The gene 41 protein, a DNA helicase that also functions in phage replication, drives polar hDNA extension in this system. It is loaded onto the displaced strand by the gene 59 and gene 32 proteins and mediates hDNA extension by moving along the displaced ssDNA (7). We do not expect Rad54 to have a similar role, because its ATPase activity is strictly dependent on dsDNA and it does not exhibit helicase activity (25, 26, 29, 30).
The existing paradigms for hDNA extension and branch migration in bacteria and bacteriophage T4 can probably not be generalized to all organisms. Gene 41 protein, which carries out hDNA extension (7) and functions as a replicative helicase, seems to be unique to bacteriophage T4. RuvB, which carries out hDNA extension and branch migration in E. coli (6), is well conserved in most, but not all, bacteria (Fig. 6A). Sequence homologs to RuvB cannot be found in the completely sequenced genomes of the bacteria Aquifex aeolicus, Buchnera sp., and Ureaplasma urealytica. Sequence comparisons of RuvB with RuvB-like proteins in eukaryotes and archaea revealed rather low homologies, which are only slightly higher than the homology of RuvB protein to a randomly shuffled sequence with the same amino acid content or with an unrelated protein (Fig. 6 A and legend). These homologies are essentially restricted to the ATPase motifs that are common to all proteins in the AAA family (ATPases associated with various cellular activities; ref. 50). Eukaryotic and archaeal RuvB-like proteins have been found to interact with TATA-binding protein (TBP) and are also known as Tip49 (TBP-interacting proteins). They were shown to be associated with RNA polymerase II holoenzyme (51, 52). Both observations make these proteins unlikely candidates to carry out hDNA extension and branch migration during homologous recombination and rather suggest an involvement in transcription.
Rad54 protein is well conserved among eukaryotes (Fig. 6B) and shows significant sequence homology with Snf2/Swi2-like proteins, which is restricted to the seven ATPase/helicase motifs (Fig. 6B). Snf2/Swi2-like proteins have been found in all three kingdoms of life. Rad54/Snf2-like proteins in archaea and bacteria share essentially only the ATPase/helicase motifs and show little conservation with Rad54 proteins outside these domains (Fig. 6B). The HepA family of proteins (helicase-like proteins) contains conserved Rad54/Snf2-like motives in the N-terminal part of the protein and has low homology to the Rad54 and Snf2 proteins (Fig. 6B). HepA from E. coli was purified as a protein associated with RNA polymerase (53), suggesting a possible role during transcription. The other family of archaeal and bacterial Rad54-like proteins has ATPase/helicase domains located in the C terminus similarly to Rad54, but is equally homologous to eukaryotic Rad54 proteins and S. cerevisiae Snf2 (Fig. 6B). It is not clear whether these proteins are involved in recombination like Rad54 protein, in transcription like the Snf2 or Mot1 proteins, or in other processes in archaea and bacteria. Rad54-like sequences were not found in several complete bacterial (A. aeolicus, Buchnera sp., N. meningitidis, R. prowazekii, T. pallidum, U. urealytica, X. fastidiosa) and archaeal (Methanobacterium thermoautotrophicum, Thermoplasma acidophilum) genomes.
The mechanisms of hDNA extension and branch migration in E. coli, in bacteriophage T4, and S. cerevisiae may differ. The RuvAB paradigm of hDNA extension and branch migration can possibly not be applied to all organisms, as indicated by the apparent absence of obvious RuvB homologs in some bacterial species, in eukaryotes, and in archaea. The eukaryotic-specific Rad54 protein might provide a unique mechanism of hDNA extension in eukaryotes. This does not exclude that other mechanisms of hDNA extension and branch migration are present in eukaryotes as well. Such mechanisms may include proteins that evolutionarily derived from E. coli RuvB, but that have evolved beyond a point where homology can be recognized by sequence alignment or yet unknown proteins (10). One such protein may be the Bloom's syndrome helicase, which has been shown to promote hDNA extension of Holliday junctions in vitro (54).
Further experiments will be needed to show that a role of Rad54 protein in hDNA extension is biologically important and whether Rad54 protein is also able to promote branch migration in four-stranded reactions in vitro. Several genetic observations are consistent with a role of Rad54 protein in postsynapsis because they suggest that Rad54 protein acts after Rad51 protein during recombination. First, the synthetic lethality of rad54 srs2 is suppressed by eliminating Rad51 filament formation (55). This result suggests that in the absence of Rad54 protein the Rad51 filament produces a lethal intermediate, possibly a blocked joint. Second, cytological observations suggest that Rad54 protein is needed for turnover of Rad51 protein-containing foci (56). Third, pathway analyses support the view that Rad54 acts after Rad51 in recombinational repair (17, 57). Although the basis for these results is not fully understood, these observations are consistent with a possible role of Rad54 in postsynapsis after initial joint formation by the Rad51 filament.
Acknowledgments
We are very grateful to Vladimir Bashkirov, Carole Bornarth, Edwin Haghnazari, Konstantin Kiianitsa, Alexander Mazin, Jim New, Mike Rolfsmeier, Tomohiko Sugiyama, and especially Steve Kowalczykowski for helpful discussions and comments on the manuscript. Rad51, Rpa, and RecA proteins were kindly supplied by the Steve Kowalczykowski laboratory. The communication of unpublished results by Carole Bornarth and Steve Kowalczykowski was highly appreciated. This work was supported by grants from the Human Frontiers Science Program (RG63) and the National Institutes of Health (GM-58015) to W.-D.H.
Abbreviations
- DSB
double-strand break
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- Rpa
Replication protein A
- hDNA
heteroduplex DNA
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Paques F, Haber J E. Microbiol Molec Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianco P R, Tracy R B, Kowalczykowski S C. Front Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 3.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 4.Sugiyama T, Zaitseva E M, Kowalczykowski S C. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 5.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 7.Salinas F, Kodadek T. Cell. 1995;82:111–119. doi: 10.1016/0092-8674(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 8.Elborough K M, West S C. EMBO J. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyde H, Davies A A, Benson F E, West S C. J Biol Chem. 1994;269:5202–5209. [PubMed] [Google Scholar]
- 10.Constantinou A, Davies A A, West S C. Cell. 2001;104:259–268. doi: 10.1016/s0092-8674(01)00210-0. [DOI] [PubMed] [Google Scholar]
- 11.Wood R D, Mitchell M, Sgouros J, Lindahl T. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 12.Donovan J W, Milne G T, Weaver D T. Genes Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- 13.Hays S L, Firmenich A A, Berg P. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R D, Symington L S. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Xie Y Q, Houston P, Stemke-Hale K, Mortensen U H, Rothstein R, Kodadek T. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 17.Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer W D. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J, Rothstein R. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hays S L, Firmenich A A, Massey P, Banerjee R, Berg P. Mol Cell Biol. 1998;18:4400–4406. doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 21.Sung P. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 22.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Nature (London) 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 23.Shinohara A, Ogawa T. Nature (London) 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 24.Benson F E, Baumann P, West S C. Nature (London) 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 25.Petukhova G, Stratton S, Sung P. Nature (London) 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 26.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 27.Mazin A V, Bornarth C J, Solinger J A, Heyer W-D, Kowalczykowski S C. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 28.van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 29.Solinger J A, Lutz G, Sugiyama T, Kowalczykowski S C, Heyer W-D. J Mol Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- 30.Tan T L R, Essers J, Citterio E, Swagemakers S M A, de Wit J, Benson F E, Hoeijmakers J H J, Kanaar R. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 31.Petukhova G, Sung P, Klein H. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bezzubova O, Silbergleit A, YamaguchiIwai Y, Takeda S, Buerstedde J M. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 33.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, deWit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 34.Kanaar R, Troelstra C, Swagemakers S M A, Essers J, Smit B, Franssen J H, Pastink A, Bezzubova O Y, Buerstedde J M, Clever B, et al. Curr Biol. 1996;6:828–838. doi: 10.1016/s0960-9822(02)00606-1. [DOI] [PubMed] [Google Scholar]
- 35.Pazin M J, Kadonaga J T. Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 36.Swagemakers S M A, Essers J, deWit J, Hoeijmakers J H J, Kanaar R. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 37.Clever B, Schmuckli-Maurer J, Sigrist M, Glassner B, Heyer W-D. Yeast. 1999;15:721–740. doi: 10.1002/(SICI)1097-0061(19990630)15:9<721::AID-YEA414>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 38.Sung P, Robberson D L. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 39.Kodadek T, Alberts B M. Nature (London) 1987;326:312–314. doi: 10.1038/326312a0. [DOI] [PubMed] [Google Scholar]
- 40.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baumann P, West S C. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumann P, West S C. J Mol Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- 43.Cox M M, Morrical S W, Neuendorf S K. Cold Spring Harbor Symp Quant Biol. 1984;49:525–533. doi: 10.1101/sqb.1984.049.01.059. [DOI] [PubMed] [Google Scholar]
- 44.Tsaneva I R, Müller B, West S C. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 45.West S C, Connolly B. Mol Microbiol. 1992;6:2755–2759. doi: 10.1111/j.1365-2958.1992.tb01454.x. [DOI] [PubMed] [Google Scholar]
- 46.Tsaneva I R, Muller B, West S C. Proc Natl Acad Sci USA. 1993;90:1315–1319. doi: 10.1073/pnas.90.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hiom K, West S C. Cell. 1995;80:787–793. doi: 10.1016/0092-8674(95)90357-7. [DOI] [PubMed] [Google Scholar]
- 48.Muller B, Tsaneva I R, West S C. J Biol Chem. 1993;268:17179–17184. [PubMed] [Google Scholar]
- 49.Parsons C A, Stasiak A, Bennett R J, West S C. Nature (London) 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 50.Iwasaki H, Han Y W, Okamoto T, Ohnishi T, Yoshikawa M, Yamada K, Toh T, Daiyasu H, Ogura T, Shinagawa H. Mol Microbiol. 2000;36:528–538. doi: 10.1046/j.1365-2958.2000.01842.x. [DOI] [PubMed] [Google Scholar]
- 51.Qiu X-B, Lin Y-L, Thome K C, Pian P, Schlegel B P, Weremowicz S, Parvin J D, Dutta A. J Biol Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- 52.Lim C R, Kimata Y, Ohdate H, Kobuko T, Kikuchi N, Horigome T, Kohno K. J Biol Chem. 2000;275:22409–22417. doi: 10.1074/jbc.M001031200. [DOI] [PubMed] [Google Scholar]
- 53.Muzzin O, Campbell E A, Xia L, Severinova E, Darst S A, Severinov K. J Biol Chem. 1998;273:15157–15161. doi: 10.1074/jbc.273.24.15157. [DOI] [PubMed] [Google Scholar]
- 54.Karow J K, Constantinou A, Li J L, West S C, Hickson I D. Proc Natl Acad Sci USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. . (First Published May 23, 2000; 10.1073/pnas.100448097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schild D. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shinohara M, Gasior S L, Bishop D K, Shinohara A. Proc Natl Acad Sci USA. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rattray A J, Symington L S. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Namsaraev E, Berg P. Mol Cell Biol. 1997;17:5359–5368. doi: 10.1128/mcb.17.9.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]