Abstract
We attached the pathogen associated molecular pattern Kdo2-Lipid A (the lipopolysaccharide (LPS) from Escherichia coli (E. coli)) to QDs by hydrophobic interactions to synthetically mimic the surface of E. coli. QD-LPS conjugates bind, are taken up and activate effectively macrophages in vitro and they have potent immunostimulatory activity in vivo.
Biological chemistry uses many types of small synthetic molecules to probe and understand facets of biology. Important examples include the range of organic dyes which as fluorescent probes enable the behaviour of individual biomolecules to be tracked in vitro and in vivo.1 Also important are synthetic compounds which, resembling the structures and functions of active sites, provide a chemical approach to understand enzyme catalysis.2 However, it is important that we progressively move to understanding more complex biological systems and for this, new nanomaterials offer unique opportunities.
Colloidal quantum dots (QDs) have become important materials in biology as alternatives to traditional organic and genetically-encoded fluorophores due to their unique optical properties.3 So far they have been used to track individual biomolecules, but for this application a widespread concern is that biomolecules can loose activity when they are attached to QDs because these are multivalent and large.4 Thus, recent attention has turned toward labeling strategies which enable site-specific recognition5 and controlling the number of molecules that can be attached to a single QD down to a single molecule.6 However, multivalency is important for regulating a wide range of biological processes.7 It has been shown that the ability of multivalent ligands to cluster cell surface receptors for the initiation of downstream signal transduction responses can lead to increased activity over monovalent ligands.7 Thus, the nanometer-size and multivalency of QDs can become useful features for some applications, as some studies are beginning to show.3e,8
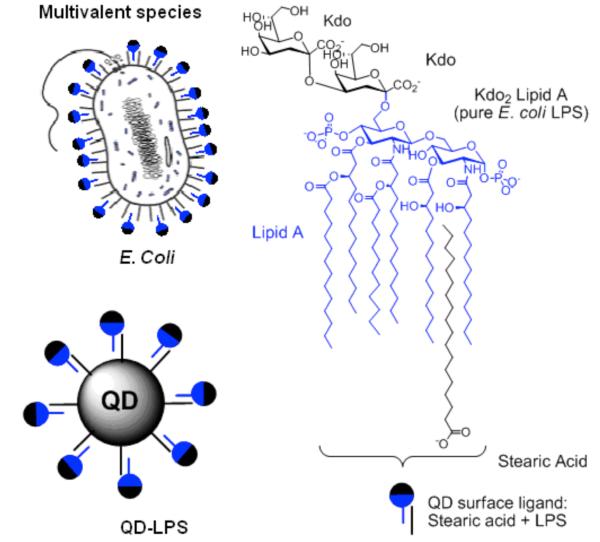
Attachment of pathogen associated molecular patterns (PAMPs; evolutionary conserved, pathogen-derived motifs which the host uses to discriminate self from non-self9) to QDs opens up the door to synthetic mimics of bacteria and viruses. The creation of these new tools will enable us to elucidate how microbial pathogens are processed by the immune system, and therefore to gain new perspectives on how to combat infectious diseases more effectively. Here we focus on lipopolysaccharides (LPS) –a PAMP which decorates the surface of Gram-negative bacteria. Immune responses to LPS play a key role in septic shock – the most common cause of mortality in intensive care units worldwide10 – and can be tailored to give clinically useful immune responses to enhance the efficiency of vaccines.11 The primary immunostimulatory component of LPS is the lipid A core (also known as endotoxin, Fig. 1), which activates Toll like receptor 4 (TLR4) on the extracellular side of the membrane of cells of the innate immune system. TLR4 is required for signaling through a group of Toll/interleukin-1 receptor (TIR)-adaptors (TIRAP, MyD88, TRIF and TRAM).12 However, it is known that other receptors are involved in LPS-induced cell activation and that these form receptor clusters. LPS binding protein (LBP), CD14, MD2, the macrophage scavenger receptor (SR-A) and β2 leukocyte integrins CD11b/CD18 have been shown to participate in LPS-induced cell activation.12,13 LPS has amphipathic properties, and although it varies among different bacterium in most cases, two 2-keto-3-deoxyoctonate (kdo) units are linked to lipid A (Fig. 1). Recently, we discovered that metal surfaces decorated with long-chain hydrophobic molecules can act as effective pattern recognition receptors for the hydrophobic core of LPS for electrochemical detection of LPS.14 Here we attach LPS to QDs by hydrophobic interactions to synthetically mimic the surface of Escherichia coli (E. coli) with a brightly fluorescent and chemically controllable nanoparticle (Fig. 1). We show that the QD-LPS conjugates bind, are taken up and activate effectively antigen presenting cells and have potent immunostimulatory activity in vivo.
Fig. 1.
Analogy between E. coli and LPS (Kdo2-lipid A)-coated QDs.
Kdo2-lipid A (pure E. coli LPS) was attached to core-shell CdSe-ZnS QDs coated with stearic acid – a non-toxic naturally produced fatty acid commonly used in pharmaceuticals and cosmetics. These QDs showed an absorption peak due to the first excitonic transition at 628 nm and a photoluminescence maximum at 642 nm (fwhm = 42 nm) and had a diameter of ~ 5.8 nm. By high-resolution transmission electron microscopy (HRTEM) the QDs appeared nanocrystalline and mostly spherical in shape (Fig. S1, ESI). QD-LPS micelles were prepared by mixing the hydrophobic QDs with excess of Kdo2-lipid A by self-assembly of the biopolymer chains around the QD hydrophobic core (see ESI). Dynamic light scattering (DLS) studies revealed that the QD-LPS micelles are between 11 and 15 nm in diameter in solution (Fig. S2, ESI). This result correlates nicely with the size of the QD (the 5.8 nm of the semiconductor nanocrystal and 2 × 2.0 nm due to stearic acid gives a spherical particle of 9.8 nm) and Kdo2-lipid A (3 nm).15
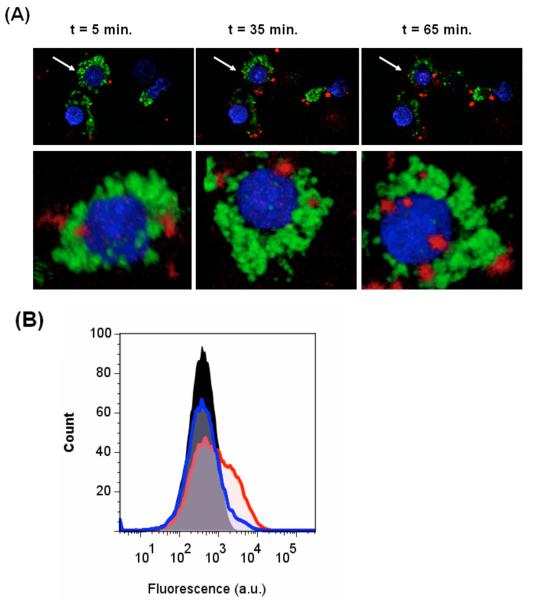
Confocal microscopy studies revealed that after 30 min incubation QD-LPS micelles (10 nM) at 37 °C had been taken up by macrophages (mouse macrophage cell line J774, Fig. 2A), presumably by receptor induced endocytosis (for LPS, a clathrin-mediated process).16 In contrast, control QD micelles containing n-poly(ethyleneglycol) phosphatidylethanolamine (PEG-PE) did not bind, and those containing PEG-PE and the naturally ocurring phospholipid phosphatidylcholine (PC) did bind but were taken up more slowly even at higher concentrations (Fig. S3, ESI). Flow cytometric analysis of the macrophages co-cultured with the QD-LPS micelles for 3 h confirmed QD-LPS binding, even at the low concentration of 0.65 nM (Fig. 2B).
Fig. 2.
(A) Confocal images of J774 macrophages after staining with Hoechst 33343 (blue, nuclei) and DiOC18 (green, membrane) at 5 min, 35 min, and 65 min after beginning incubation with 10 nM QD-LPS (red) containing medium at 37 °C (top). The 3D reconstituted images the cell marked with an arrow (bottom). (B) Flow cytometry analysis of macrophages before (black) and after incubation with QD-LPS (0.65 nM, red line; 65 pM, blue line).
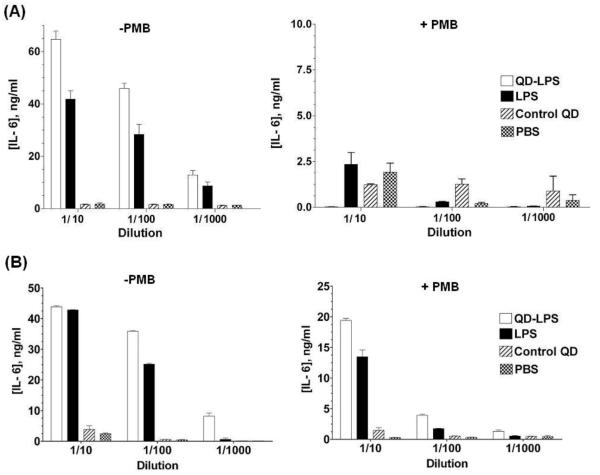
We tested the immunostimulatory activity of the QD-LPS micelles with the mouse macrophage cell line J774 and bone-marrow derived dendritic cells (BMDCs) from wild-type mice. Controls we used were PBS, uncoupled Kdo2-lipid A and PC and PEG-PE coated QDs.17 After incubation for 16-24 h, supernatants were harvested and cytokine IL-6, a pro-inflammatory cytokine secreted by macrophages and BMDCs upon PAMP stimulation,18 was quantified by ELISA. Remarkably, cytokine production for macrophages treated with 1.5 nM QD-LPS conjugates was higher than with 250 nM LPS alone (1/10 dilution, Fig. 3A), and similar for BMDCs (Fig. 3B). If we consider the dimensions of Kdo2-lipid A15 (surface area of ~ 2 nm2) and that the QD at the point of interaction with the LPS chains has a radius of 4 nm, then a QD cannot hold more than 100 LPS molecules. The actual number of LPS molecules per QD was determined using the zinc dipicolylamine (Zn-DPA) complex which Hamachi et al.19 originally developed for fluorescence detection of phosphorylated peptides (see ESI). Recent studies have shown that this zinc complex selectively targets and fluorecescently labels the surface of bacterial cell walls and LPS-modified surfaces via recognition of the anionic phosphate esters of LPS.14,20 The QD-LPS micelles used in this study captured ca. 130 molecules of this phosphate binding zinc(II) complex (see ESI). Because each Kdo2-lipid A has two dianionic phosphate ester groups, we estimate that these QDs therefore carry around 65 LPS molecules each. Thus, the immunostimulatory activity of QD-LPS micelles is considerably greater than that of LPS as 0.15 nM QD-LPS shows more potent IL-6 induction than 250 nM LPS alone. Release of cytokines was strongly inhibited by the LPS antagonist polymyxin B (PMB), and control QDs lacked any activity; thus the immunostimulatory properties of the QD-LPS micelles are LPS-induced. Similar results were seen with TNF-α (data not shown); another cytokine secreted by macrophages following their stimulation with LPS.
Fig. 3.
(A) IL-6 release from macrophages treated at 1/10 dilution with: [QD-LPS] = 1.5 nM; [LPS] = 250 nM; [control QD] = 2.5 nM. PMB is the LPS inhibitor polymyxin B; [PMB] = 250 μM. PBS = phosphate-buffered saline, pH 7.4. The control QD is coated with PC and PEG-PE. Incubation time = 16 h. (B) IL-6 release from bone-marrow derived dendritic cells from wild-type mice treated at 1/10 dilution with: [QD-LPS] = 1.5 nM; [LPS] = 250 nM; [control QD] = 2.5 nM. PMB is the LPS inhibitor polymyxin B; [PMB] = 250 μM. PBS = phosphate-buffered saline, pH 7.4. The control QD is coated with PC and PEG-PE. Incubation time = 24 h
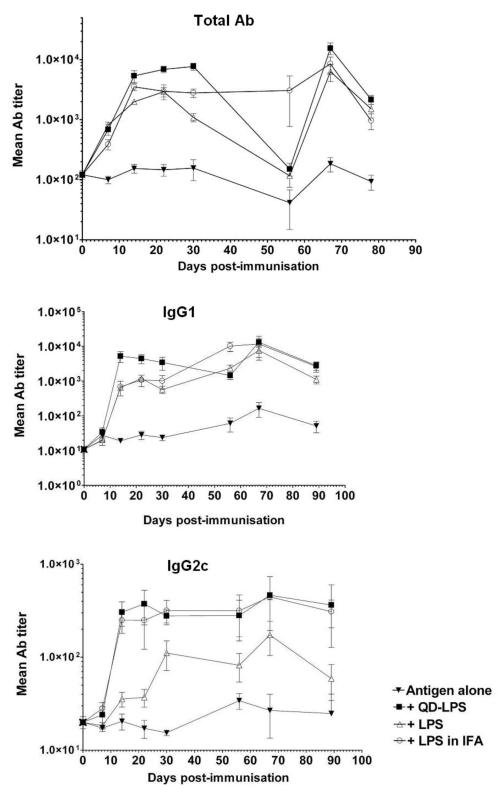
To determine if the potent immunomodulatory activity of QD-LPS micelles observed in vitro translated to in vivo immune responses we carried out a series of immunisations. Groups of five mice were immunised intraperitoneally (i.p.) with dinitrophenylated-ovalbumin (DNP-OVA) as model antigen, alone and co-administered with LPS, QD-LPS micelles and sub-cutaneously (s.c.) administered LPS in incomplete Freund’s adjuvant (IFA is a widely used and potent adjuvant).21 Mice were bled before immunisation and at 7 day intervals, and anti-DNP serum antibodies quantified by indirect ELISA. We quantified total antibodies and the isotypes IgG2c and IgG1. In the immune response to an injected antigen in mice and humans CD4+ T helper cells 1 and 2 (Th1 and Th2) play an important role and can have a significant impact on the overall protection a vaccine provides.22 In mice production of IgG2c is recognized as characteristic of a Th1 immune response,23 whereas a Th2 response is associated with the induction of IgG1. The QD-LPS conjugates (10 pmol of QD, ~ 1.5 μg of LPS based on 65 molecules of LPS per QD) showed the highest adjuvant potency (i.e. ability to improve the immune response to vaccine antigens); greater than LPS alone (4 μg) and even than LPS isolated from E. coli (10 μg) emulsified in IFA (Fig. 4). Attachment of LPS to QDs enhances its immunostimulatory activity in vivo, acting as potent adjuvant. No signs of toxicity were detected over the 3-4 months following immunization. The demonstration that QD-LPS has potent adjuvant activity, even at low LPS doses, is an exciting one as finding new adjuvants has become an important target and bottleneck in vaccine development.21
Fig. 4.
(A) Antibody (Ab) responses (total Ab, IgG1 and IgG2c) to antigen (DNP-OVA, 100 μg) alone and co-administered (i.p.) with QD-LPS (10 pmol), LPS (4 μg) and E. coli LPS (10 μg) emulsified in IFA (s.c. injected); boosting with DNP-OVA after 56 days. Values plotted are means based on titers from five individual mice and representative of two different experiments.
In summary, we have attached Kdo2-lipid A to QDs to synthetically mimic the surface of E. coli with a brightly fluorescent nanomaterial. The immunostimulatory activity of this important biomolecule was found to be higher attached to the QD than alone, both in vitro and in vivo. Although many different types of biomolecules have been attached to QDs, we are not aware of another study in which biological activity increased by attachment to QDs. Despite the focus being on LPS and micelles which are smaller than typical pathogens, it is clear that other microbial products can be attached to QDs and that several QDs can be self-assembled to form larger structures. In doing so, it should be possible to better mimic the multivalency and ‘cocktail’ of biomolecules encountered by the immune system when interacting with microbial pathogens in vivo. Thus, we anticipate that QD-PAMP conjugates could become important model materials to investigate how bacteria and viruses interact and are processed by the immune system, and to learn how to fight infectious diseases more effectively. We are continuing our studies to develop synthetic materials which can be easily tracked in vitro and in vivo and are capable of setting off the alarms of the immune system in a predictable way because of their biological cargos.
We are grateful to Cancer Research UK and EaStCHEM for PhD studentships to M. K and M. G. The work was partially funded by a grant to D.G. from the Wellcome Trust and a Moray Endowment Award to J.C.M-R. We thank Dr. Wuzong Zhou and Ross Blackley for the HRTEM studies, and Dr David Kelly for help with the confocal studies.
Supplementary Material
Footnotes
Electronic Supplementary Information (ESI) available: Full experimental details and additional characterization data. See DOI: 10.1039/b000000x/
Notes and references
- 1.(a) Zhang J, Campbell RE, Ting AY, Tsien RY. Nat. Rev. 2002;3:906. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]; (b) Haugland RP. The Handbook—A Guide to Fluorescent Probes and Labeling Technologies. 10th ed Molecular Probes; Eugene: 2005. [Google Scholar]
- 2.(a) Breslow R. Acc. Chem. Res. 1995;28:146. [Google Scholar]; (b) Kirby AJ, Hollfelder F. From Enzyme Models to Model Enzymes. RSC publishing; Cambridge: 2009. [Google Scholar]
- 3.(a) Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. Curr. Opin Biotech. 2005;16:63. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]; (c) Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat. Mater. 2005;4:435. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]; (d) Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Nat. Methods. 2008;5:763. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]; (e) Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Nat. Biotech. 2004;22:198. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 4.(a) Parak WJ, Pellegrino T, Plank C. Nanotechnology. 2005;16:R9. doi: 10.1088/0957-4484/16/2/R01. [DOI] [PubMed] [Google Scholar]; (b) Huo Q. Coll. Surf. B. 2007;59:1. doi: 10.1016/j.colsurfb.2007.04.019. [DOI] [PubMed] [Google Scholar]; (c) Howarth M, Liu W, Puthenveetil S, Zheng S, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY. Nat. Methods. 2008;5:397. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Zhang Y, So MK, Loening AM, Yao H, Gambhir SS, Rao J. Angew. Chem. Int. Ed. 2006;45:4936. doi: 10.1002/anie.200601197. [DOI] [PubMed] [Google Scholar]; (b) Chen I, Howarth M, Lin W, Ting AY. Nat. Methods. 2005;2:99. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]; (c) Gupta M, Caniard A, Touceda-Varela A, Campopiano DJ, Mareque-Rivas JC. Bioconjugate Chem. 2008;19:1964. doi: 10.1021/bc800273j. [DOI] [PubMed] [Google Scholar]; (d) Kim J, Park HY, Kim J, Ryu J, Kwon Y, Grailhe R, Song R. Chem. Comm. 2008;16:1910. doi: 10.1039/b719434j. [DOI] [PubMed] [Google Scholar]
- 6.Sperling RA, Pellegrino T, Li JK, Chang WA, Parak WJ. Adv. Func. Mater. 2006;16:943. [Google Scholar]
- 7.(a) Mammen M, Choi S–K, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; (b) Kiessling LL, Gestwicki JE, Strong LE. Curr. Opin. Chem. Biol. 2000;4:696. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]; (c) Bertozzi CR, Kiessling LL. Science. 2001;291:2357. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 8.(a) Menéndez G, Roberti MJ, Sigot V, Etchehon M, Jovin TM, Jares-Erijman EA. Proc. SPIE. 2009;7189:71890P. [Google Scholar]; (b) Roberti MJ, Morgan M, Menendez G, Pietrasanta LI, Jovin TM, Jares-Erijman EA. J. Am. Chem. Soc. 2009;131:8102. doi: 10.1021/ja900225w. [DOI] [PubMed] [Google Scholar]
- 9.(a) O’Neill LA. Curr. Opin. Immunol. 2006;18:3. [Google Scholar]; (b) Oda K, Kitano H. Mol. Syst. Biol. 2006;2:2006.0015. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Beutler B, Rietschel ET. Nat. Rev. Immunol. 2003;3:169. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]; (b) Cohen J. Nature. 2002;420:885. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 11.Mata-Haro V, Cekic C, Martin M, Chilton PM, Casella CR, Mitchell TC. Science. 2007;316:1628. doi: 10.1126/science.1138963. [DOI] [PubMed] [Google Scholar]
- 12.Watts C. Nat. Immunol. 2008;9:343. doi: 10.1038/ni0408-343. [DOI] [PubMed] [Google Scholar]
- 13.(a) Hampton RY, Golenbock DT, Penman M, Krieger M, Raetz CR. Nature. 1991;352:342. doi: 10.1038/352342a0. [DOI] [PubMed] [Google Scholar]; (b) Peiser L, Gough PJ, Kodama T, Gordon S. Infect. Immun. 2000;68:1953. doi: 10.1128/iai.68.4.1953-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ingalls RR, Monks BG, Savedra R, Christ WJ, Delude RL, Medvedev AE, Espevik T, Golenbock DT. J. Immunol. 1998;161:5413. [PubMed] [Google Scholar]
- 14.Ganesh V, Bodewits K, Bartholdson SF, Natale D, Campopiano DJ, Mareque-Rivas JC. Angew. Chem. Int. Ed. 2009;48:356. doi: 10.1002/anie.200804168. [DOI] [PubMed] [Google Scholar]
- 15.Obst S, Kastowsky M, Bradaczek H. Biophys. J. 1997;72:1031. doi: 10.1016/S0006-3495(97)78755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Husebye H, Halaas Ø, Stenmark H, Tunheim G, Sandanger Ø, Bogen B, Brech A, Latz E, Espevik T. EMBO J. 2006;25:683. doi: 10.1038/sj.emboj.7600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Dubertret B, Skourides P, Norris DJ, Noireaux AH, Brivanlou AH, Libchaber A. Science. 2002;298:1759. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]; (b) Carion O, Mahler B, Pons T, Dubertret B. Nat. Protocols. 2007;2(10):2383. doi: 10.1038/nprot.2007.351. [DOI] [PubMed] [Google Scholar]
- 18.(a) Kishimoto T, Akira S, Taga T. Science. 1992;258:593. doi: 10.1126/science.1411569. [DOI] [PubMed] [Google Scholar]; (b) Zanoni I, Ostuni R, Capuano G, Collini M, Caccia M, Ronchi AE, Rochetti M, Mingozzi F, Foti M, Chirico G, Costa B, Zara A, Ricciardi-Castagnoli P, Granucci F. Nature. 2009;460:264. doi: 10.1038/nature08118. [DOI] [PubMed] [Google Scholar]
- 19.(a) Ojida A, Mito-oka Y, Inoue M-A, Hamachi I. J. Am. Chem. Soc. 2002;124:6256. doi: 10.1021/ja025761b. [DOI] [PubMed] [Google Scholar]; (b) Yamaguch S, Yoshimura I, Kohira T, Tamaru S-I, Hamachi I. J. Am. Chem. Soc. 2005;127:11835. doi: 10.1021/ja052838y. [DOI] [PubMed] [Google Scholar]
- 20.(a) Leevy WM, Johnson JR, Lakshmi C, Morris J, Marquez M, Smith BD. Chem. Commun. 2006:1595. doi: 10.1039/b517519d. [DOI] [PubMed] [Google Scholar]; (b) Johnson JR, Fu N, Arunkumar E, Leevy WM, Gammon ST, Piwnica-Worms D, Smith BD. Angew. Chem., Int. Ed. 2007;46:5528. doi: 10.1002/anie.200701491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pashine A, Valiante NM, Ulmer JB. Nat. Medicine. 2005;11:S63. doi: 10.1038/nm1210. [DOI] [PubMed] [Google Scholar]
- 22.Singh M, O’Hagan D. Nat. Biotech. 1999;17:1075. doi: 10.1038/15058. [DOI] [PubMed] [Google Scholar]
- 23.Martin RM, Brady JL, Lew AM. J. Immunol. Methods. 1998;212:187. doi: 10.1016/s0022-1759(98)00015-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.