Abstract
During the past few years, the development of effective, empirical technologies for treatment of cardiac arrhythmias has exceeded the pace at which detailed knowledge of the underlying biology has accumulated. As a result, although some clinical arrhythmias can be cured with techniques such as catheter ablation, drug treatment and prediction of the risk of sudden death remain fairly primitive. The identification of key candidate genes for monogenic arrhythmia syndromes shows that to bring basic biology to the clinic is a powerful approach. Increasingly sophisticated experimental models and methods of measurement, including stem cell-based models of human cardiac arrhythmias, are being deployed to study how perturbations in several biologic pathways can result in an arrhythmia-prone heart. The biology of arrhythmia is largely quantifiable, which allows for systematic analysis that could transform treatment strategies that are often still empirical into management based on molecular evidence.
Introduction
Effective treatment is now available for many arrhythmias.1,2 Devices and sophisticated catheters, together with computerised-mapping systems that allow for ablation therapy, have brought some remarkable advances. Such technologies have transformed clinical electrophysiology into one of the most rapidly expanding cardiology subspecialties. Pacemakers are the accepted standard of care for those with bradycardia, and if facilities are available, patients with Wolff-Parkinson-White syndrome or similar arrhythmias should be referred for ablation.3 Many complex atrial and ventricular arrhythmias are also amenable to potential cure with these approaches.1,2 These successes are all causes for celebration.
However, knowledge of the underlying biology has not kept up with technical developments, and major questions about clinical management remain. First, although we know some of the general factors that predispose to arrhythmias, the precision of our analysis is not always sufficient to justify prophylaxis or intervention.4 Second, if we want to suppress arrhythmia not amenable to ablation, we have few options. The range of available drugs has scarcely expanded in the past 30 years, and available drug treatments have proarrhythmic risk, other toxic effects, low tolerability, and variable (although never better than modest) efficacy.5 The generally poor outcomes with contemporary drug therapy are understandable, since most agents were developed in the absence of molecular targets and without precise understanding of the mechanisms of proarrhythmic and antiarrhythmic drug actions. Finally, we are incapable of accurately identifying those at risk of sudden cardiac death (which is almost always caused by arrhythmia), which leaves the effective—but crude and expensive—option of implantable cardioverter defibrillators as the only viable choice for many.2,4 Thus, although some parts of contemporary arrhythmia care can be successful and provide seemingly definitive solutions, others will be seen in retrospect as fairly primitive.4
However, recent developments suggest that much arrhythmia biology is tractable.6 Advances in the relevant genetics and genomics, and the availability of new model systems are encouraging.7,8 The emerging picture is one of many molecular perturbations that come together and interact in individuals to generate arrhythmia-prone hearts, expressed through the phenotypic variability familiar to clinicians. In this Series paper, we review, in a necessarily selective way, the present state of arrhythmia biology, with a focus on general principles. We also provide a description of how the necessary translation from experimental findings to effective, individualised clinical advances can be delivered.
Defining arrhythmia phenotypes
Elucidation of the underlying mechanisms of the heart rhythm is intuitively appealing, since many features are amenable to quantification.6,9,10 Fundamental descriptors, such as rate and rhythm, or the patterns of myocardial conduction and repolarisation, can all be measured fairly straightforwardly, and with reasonable precision, at essentially every biological level.6 As such, in preclinical settings the relations between genetic variants, cellular and biophysical outputs, and integrated heart function have been reported.11–13 In population studies, the epidemiology of arrhythmias has advanced greatly and is likely to become even more sophisticated once phenotypic categories are more strictly defined and the underlying genetics and genomics are better understood.4,14
However, substantial limitations remain. Symptoms of individuals with arrhythmias are widely divergent and their effects are highly variable.15 Most of us could be wholly unaware of an occasional upset of normal heart rhythm, whereas others with fairly minor, benign arrhythmias have very severe symptoms. The causes of such differences in perception are unclear, but these differences preclude the use of symptoms alone for assessment.15 Additionally, by their very nature many arrhythmias are paroxysmal and seemingly sporadic. The inability to observe symptoms at the time of each occurrence also adds to a blurring of clinical phenotypes. If an arrhythmia occurs when a patient is not being observed then the precise nature of their problem, and the putative underlying mechanism, will remain obscure, and treatment will necessarily be empirical. Advances in monitoring will hopefully allow the heart rhythm to be observed continuously.16 Wide application of such technical advances would allow for phenotypic boundaries to be more strictly defined, and would enhance our knowledge of the natural history of arrhythmia and the design of clinical trials.
Systematic approaches to cardiac arrhythmias
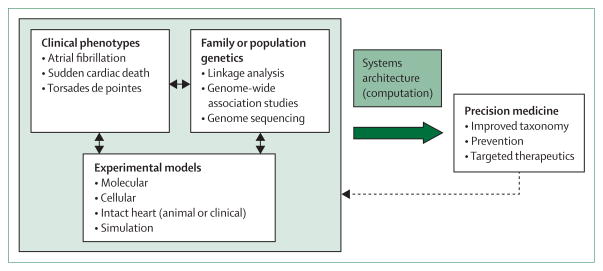
The definition of systems biology remains contentious and is a subject of continued discussion.17 For practical purposes, we define systems biology as an analytical framework that is characterised by integrated descriptions of several biological processes that are based on systematic measurements.6,9,10 Applied to medicine, the approach necessitates as a starting point large, high-quality datasets that describe the phenotypes and natural history of a disease, in this case cardiac arrhythmia. If necessary, revised diagnostic categories might be used to describe specific entities more precisely.15,18 These data are combined with results of genetic, genomic, or other molecular analyses and complemented with work from experimental systems, ideally of human origin, which can model functional consequences.7,8,14,19 The properties of such models can then be examined in response to either potentially deleterious changes or interventions (such as drugs) intended to rescue the disease phenotype.20–22 Logistical issues arise because of the large amounts of data that are generated by such a systems approach, which require computational models to elucidate the underlying network architecture.6,9,10,18,23–25 Nevertheless, such strategies allow the heartbeat to be characterised in both health and disease. Such work should mean that, in due course, knowledge gaps will be closed, which will allow for clinical advances and new capabilities for risk stratification and identification of drug targets (figure 1).5,6,26 The needs are urgent, since arrhythmias have such a substantial effect on morbidity and mortality (figure 2).4,14,27–31
Figure 1. A systems approach to analysis of arrhythmia susceptibility.
A major objective of a systems approach to arrhythmia susceptibility is to develop a new taxonomy that is clinically useful and that integrates multiparametric data. Clinical and genetic inputs are used in conjunction with appropriately designed mechanistic studies in clinical and model systems. Iterative computer-based modelling provides an architectural framework that leads to improved taxonomy and allows for tailoring of medical treatment (precision medicine).
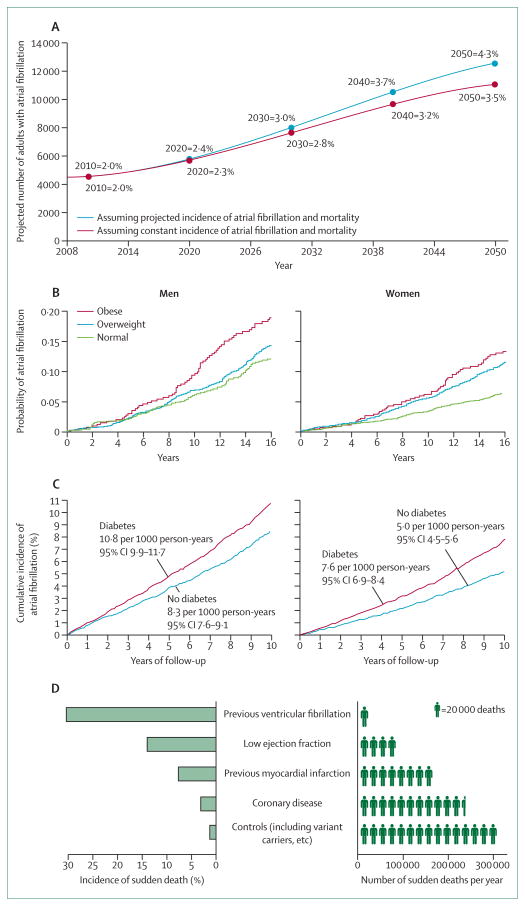
Figure 2. Population-based risk of cardiac arrhythmias.
Projected increase in population burden of atrial fibrillation in Iceland, 2008–2050 (A), based on community-based findings from Reykjavik (1991–2008),27 shows a trend that is essentially identical to that reported from Olmsted County, MN, USA.28 Obesity (B) and diabetes (C) are associated with increased probability of atrial fibrillation in men and women.29,30 Incidence of sudden cardiac death as a percentage of some specific clinical populations and estimated numbers of sudden cardiac deaths each year in each population in the USA (D) show an inverse relation, with most deaths in those not known to have a clinical problem.31 This situation raises questions about appropriate screening. (A) is reproduced from reference 27, by permission of Oxford University Press; (B) is reproduced from reference 29, by permission of the American Medical Association; (C) is reproduced from reference 30, by permission of the American Diabetes Association; (D) is reproduced from reference 31, by permission of Wolters Kluwer Health.
Population-based risk of atrial fibrillation and sudden cardiac death
Atrial fibrillation is the most common sustained arrhythmia. It is age-related, with a life-time risk estimated at 25% for a 40-year-old person.14 The most serious chronic disease sequelae of atrial fibrillation include stroke, heart failure, and dementia—all of which have devastating effects on an individual’s health and high costs for families and society.14 Two broad categories of atrial fibrillation are seen in clinical settings. One is a small group of usually comparatively young patients, who often have a family history of the disease and might also have a history of high-intensity exercise.14 However, most patients belong to the second group of usually older individuals, the numbers of whom are growing.14,27 Although the prevalence of atrial fibrillation in the USA is currently estimated to be in the range of 2·7–6·1 million, it is expected to rise substantially to an estimated 16 million by 2050.28,32 This upward trend has also been reported elsewhere (figure 2A).27
The increased incidence is driven partly by ageing populations, but other factors are also implicated.14 Although hypertension remains the most well described risk, metabolic factors also play a part.14,30 For example, investigators of the Framingham Heart Study estimated that obesity was associated with a 50% increase in risk of atrial fibrillation (figure 2B).29 In Olmsted County, MN, USA, obesity might account for about 60% of the enhanced, age-adjusted and sex-adjusted increase in incidence.28 Furthermore, investigators of the Women’s Health Study documented a linear association between body-mass index and atrial fibrillation and also reported that short-term increases in body mass contributed substantially to risk of the arrhythmia.33 Although some of the effects of obesity might be haemodynamic (eg, through impaired ventricular relaxation or atrial stretch), more direct metabolic effects seem likely.29,33 Diabetes is also independently associated with atrial fibrillation (figure 2C).30 Additionally, comparative epidemiological data for prevalence of atrial fibrillation in racial groups and various geographical locations provide evidence of intrinsic (presumably genetic) interactions.14 Black people, for example, have a higher prevalence of hypertension and metabolic disease but a lower incidence of atrial fibrillation than a comparable white population.14
The other major public health effect of arrhythmia is sudden cardiac death,4,34 for which the difficulties in accurately assessing numbers are also well known—an issue not helped by the absence of uniform definitions.34 Therefore, as for atrial fibrillation, incidence is usually expressed in broad ranges.28,32,34 For example, incidence in the USA is between 180 000 and 450 000 cases per year (figure 2D),34 and the global incidence has been estimated at 4·5 million cases per year.4 Incidence undoubtedly increases as the population ages,4 and associations with metabolic factors have been reported, such as a link with diabetes.35
Both atrial fibrillation and sudden cardiac death are phenotypically very heterogeneous, with increasingly recognised subcategories of disease and risk states,14,31 which complicates their epidemiological study. Their appearance in later life lends support to a generally held view that they are expressions of an acquired disorder. With the characterisation of the genetic contribution to arrhythmia from the study of rare monogenic disorders,36,37 increasing evidence now strongly supports the idea that substantial genetic predisposition exists for other arrhythmias, including atrial and ventricular fibrillation in all patient groups.14,31
Monogenic ion-channel diseases
The molecular elucidation of the long QT syndromes established that discrete genetic variants could of themselves cause arrhythmias.38,39 These findings were an important milestone both for cardiology and for the use of systems approaches for the analysis of arrhythmia susceptibility.36
The long QT syndrome phenotype, which often presents in childhood, of presyncope, syncope, seizures, or, as the first manifestation, sudden cardiac death, has been well described.2,40 The symptoms, the occurrence of which might be mutation-specific, most often arise as a result of a ventricular arrhythmia (torsade de pointes), on the background of electrocardiographic QT prolongation.2,40 Inheritance is usually autosomal dominant, but proximal mechanisms were unclear until reports of mutations in candidate ion-channel genes, SCN5A39 and KCNH2.38,41 These pivotal studies relied on linkage analysis based on the assembly of large family groups.36 Several disease genes have since been implicated, which encode not only ion channels (the prime candidates for modulation of cellular excitability and impulse propagation), but also channel-associated proteins such as ankyrins, syntrophins, and channel-β subunits.37,40
The genetic characterisation of long QT syndrome and its subtypes has provided a framework for much arrhythmia research.6,37,42 Disease gene products have been subject to wide-ranging laboratory investigation, including molecular modelling42 and generation of genetically modified animals7,11,13 and cellular models. Such models include those that use human cardiomyocytes derived from induced pluripotent stem (iPS) cells.20,43,44 Clinical studies have emphasised variable penetrance; gene carriage can occur without a concomitant electrocardiogram (ECG) phenotype,40 so the disorders are more prevalent than previously realised.37 Variable penetrance, which substantially reflects contributions by modifier genes, can result in strikingly diverse phenotypes—eg, QT intervals that range from 406 to 676 ms in a South African long QT syndrome founder population that harbours variant Ala341Val in KCNQ1.45 The term monogenic should therefore be used with this complexity in mind, since other variants beyond those that cause the disease are likely to contribute to clinical phenotypes.46
Identification of long QT syndrome disease genes provided an impetus for the detailed descriptions of other disorders characterised by familial sudden cardiac death, with each associated with particular phenotypic features.36,37 These disorders include Brugada syndrome,47 catecholaminergic polymorphic ventricular tachycardia (CPVT),48 short QT syndrome, some forms of early repolarisation,49 and various cardiomyopathies50,51—all of which share some common features with long QT syndromes.2 For example, a baseline, possibly unstable, ECG phenotype might exist, families are often small so linkage analysis is not usually possible, and genes are usually autosomal dominant but with widely variable penetrance.37,45 These features and others have caused difficulties for the implication of specific genetic variants in the generation of clinical phenotypes.19 The application of next-generation resequencing of candidate genes and exome sequencing could provide some resolution.52 Intermediate phenotypes have started to be described with such approaches. For example, the complementary application of single-nucleotide polymorphism genotyping and whole-gene sequencing implicated the previously unidentified arrhythmia susceptibility gene MYH6 in sinus node disease.53
Molecular understanding of long QT syndrome has undoubtedly been important for the management of individual patients and has facilitated family screening.37 For example, gene-specific disease behaviour, natural history, and treatment preferences for long QT syndrome were recognised soon after the initial gene variant identification.40 More recently, mutant-specific ion-channel characteristics have been reported and proposed as a means to guide therapeutic choices.54,55 That environmental factors can trigger the first expression of disease in later life has been widely recognised. One important example is drug-induced (acquired) long QT syndrome, a disorder that has had a major effect on drug development and regulation.41,56 5–15% of affected individuals with long QT syndrome that appeared for the first time after exposure to an implicated drug have plausible disease-causing variants in relevant candidate genes,56 and high-density single-nucleotide polymorphism approaches have started to provide evidence of common susceptibility variants, such as KCNE1 Asp85Asn.57 The monogenic ion-channel diseases therefore provide a framework that shows how knowledge of the molecular basis of disease can be used to rationalise management.19 However, the most important effect for integrated heart-rhythm management might have been on how we regard the biology that underlies more complex arrhythmias.
Genetics of population-based risk
Electrocardiography remains the essential diagnostic and phenotypic method for classification of disorders of the heartbeat. Changes of rate and rhythm and variations in the morphology of ECG complexes are all relevant to a full description of an individual’s phenotype. Morphological variants might suggest acquired disease or the presence of an ion-channel disease.40,49,58 ECGs are rich in readily quantifiable data and are widely available from well characterised populations, so, perhaps unsurprisingly, they provided an attractive target for genome-wide association studies intended to identify the genetic determinants of cardiac conduction and repolarisation (figure 3).31,59–61
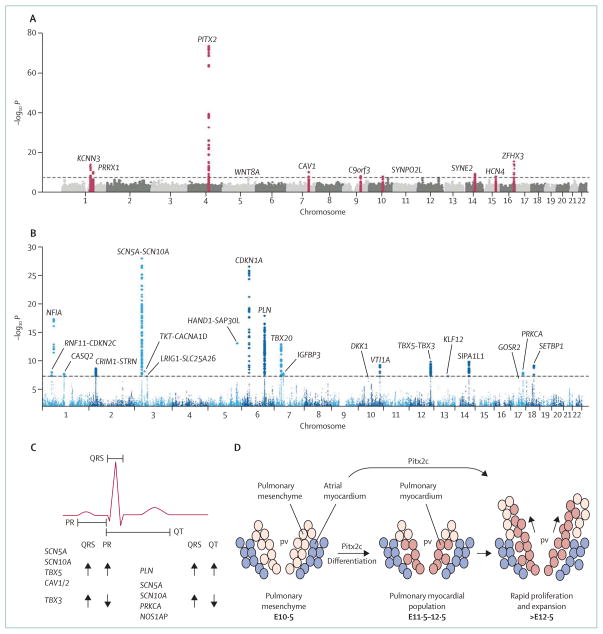
Figure 3. Genome-wide association studies for arrhythmias and cardiac conduction.
(A) Results of a meta-analysis of genome-wide association studies of patients with lone atrial fibrillation.59 In this Manhattan plot, the x-axis represents the genomic coordinates of each single-nucleotide polymorphism and the y-axis is the negative logarithm of the association p value (the higher the number, the greater the significance). The dashed line shows the threshold for genome-wide significance (p<5 × 10−8). So far, three major genes (KCNN3, PITX3, and ZFHX3) and seven additional loci close to genes for ion channels, or associated with cardiopulmonary development or signal transduction, have been identified.59 (B) Manhattan plot of association of single-nucleotide polymorphisms with QRS duration in genome-wide association meta-analysis of 40 407 individuals from 14 studies.60 22 loci reached genome-wide significance. (C) Effects of genome-wide study-identified variants on the electrocardiogram.60 Many variants that prolong the PR interval are also associated with increased QRS duration, whereas variants associated with prolonged QRS are associated with a shortened QT interval. (D) Pitx3, an atrial fibrillation candidate gene, is necessary for the development of the pulmonary myocardium. Pulmonary mesenchyme differentiates into myocardium and expands to form a sheet around the pulmonary vein branches (E10·5–12·5, mouse gestation in days).61 (A) is reproduced from reference 59, by permission of Patrick Ellinor and Nature Publishing Group; (B) and (C) are reproduced from reference 60, by permission of Nona Sotoodehnia and Nature Publishing Group; (D) is reproduced from reference 61, by permission of Wolters Kluwer Health. pv=pulmonary vein. Pitx2c=Pitx2 isoform c.
Genome-wide association has been used to assess the causes of variability in normal range QT intervals and in atrioventricular conduction (as revealed in the PR interval of the ECG) and intraventricular conduction (QRS duration).31 In each case, variants in several loci have been implicated;62–65 some are in known candidate genes, whereas others are in genes or pathways not previously implicated in cardiac electrophysiology.66 Such studies identify loci at which DNA polymorphisms contribute to variability in a trait, but the specific causative polymorphisms are not usually identified.19 For the PR interval of the ECG, polymorphisms in SCN5A (which encodes the major sodium channel isoform in heart) and in unanticipated candidates such as SCN10A (which encodes another sodium channel), TBX5 (which encodes a T-box transcription factor), and CAV1 (which encodes caveolin 1, a scaffolding protein) have been implicated.62,63 Such polymorphisms provide new starting points for the elucidation of gene networks that control both normal electrophysiology and arrhythmia susceptibility.18 Epidemiological research had already identified the PR interval as a potential intermediate phenotype for atrial fibrillation,67 and candidate genetic determinants of the PR interval might also predispose individuals to the arrhythmia.14 These results from genome-wide studies are broadly reproducible, and have also been seen in unselected patients whose characteristics have been recorded in electronic medical records.68
Results of twin and family studies have previously suggested a substantial genetic contribution to QRS duration, with heritability estimates of up to 40%.31 Genome-wide association studies have now identified 22 loci associated with QRS duration, with SCN10A unexpectedly represented (figure 3B, 3C).60,62,64 A genome-wide association study of the QT interval65 has similarly implicated several genetic loci, with some overlap with PR and QRS determinants (eg, SCN5A) and some others independent of these (such as NOS1AP, a nitric oxide synthase subunit); variants in NOS1AP have been implicated as modulators of the congenital long QT syndrome clinical phenotype and of risk of sudden cardiac death in populations.66 More generally, variability in PR, QRS, and QT has been associated with arrhythmia risk in population studies.31,65 Genes that modulate these traits are therefore expected to have variants that also modulate risk of arrhythmia and sudden cardiac death.
Genetics of atrial fibrillation and sudden cardiac death
The familial occurrence of atrial fibrillation is well established; having an affected parent doubles an individual’s risk.14 Ion-channel genes are strongly implicated in some families,14,69 and patients with ion-channel diseases have an increased risk of atrial fibrillation.14,70 However, the clinical heterogeneity of atrial fibrillation has made the investigation of underlying genetics difficult.15 Whereas in some, atrial fibrillation is continuous and readily observed, in others it is transient, with acute triggers, and can regress or even not be detected at all.14,15 As such, strict definition of affected and unaffected people in families or populations can be problematic. However, despite such practical difficulties, susceptibility alleles have been identified, the first of which was reported to be at chromosomal location 4q25 (figure 3A, 3D).71 Genetic sequence variants at that site have been strongly associated with atrial fibrillation in several populations, with the main implicated candidate gene being PITX2 (figure 3A).
PITX2 isoform c (PITX2c) is a homeobox transcription factor involved in assignment of laterality and in the development of the pulmonary myocardium that is implicated in the differentiation of the muscular sleeves around pulmonary veins.61 Although primary causal involvement of PITX2c is not proven, since Pitx2c+/− mice have inducible atrial fibrillation and PITX2 seems to be downregulated in patients with atrial fibrillation, the gene clearly could be functionally important.72,73 Both candidate-gene approaches and genome-wide association studies have identified other candidate genes, the importance of which remain largely unknown in this context (figure 3A).14,59
Familial susceptibility to sudden cardiac death is well established, even in those people who have underlying coronary artery disease.31 For example, the Paris Prospective Study followed the course of 1746 asymptomatic middle-aged men for up to 23 years; individuals both of whose parents had died suddenly had a nine times greater risk of sudden cardiac death than the mean.74 Several potentially relevant genetic variants have been proposed. Variants of slight effect size at chromosomal location 21q21 modulate risk of ventricular fibrillation immediately after myocardial infarction,75 and variants in genes previously implicated in ion-channel diseases are associated with a broad-based risk of sudden cardiac death.76,77 A common variant at chromosomal location 9p21 (robustly linked to myocardial infarction, but in a region devoid of protein-coding genes) might have an independent effect on risk.78 However, up to now, common genetic variants seem, perhaps unsurprisingly, unable to predict which of those patients with implantable cardioverter defibrillators are at risk of ventricular fibrillation episodes.79
Despite such substantial advances, variants identified in genome-wide association studies account for only a small proportion of the risk of atrial fibrillation and sudden cardiac death.14,19,31,59 The so-called missing heritability could be accounted for by rarer variants with larger effect size19,80 or by interactions between several genetic variants and environmental factors. High-throughput parallel sequencing of coding regions (exomes) or entire genomes in large populations19,52 are potential approaches for the investigation of these possibilities.
Conventional physiological principles and models
A systems approach to assessment of emerging genetic data necessitates their incorporation into physiological models (figures 1, 4).6,9,81–83 In the conventional conceptual view of cardiac arrhythmogenesis, a triggering beat (or beats) interacts with a predisposed cardiac muscle (the substrate).83–85 Heterogeneities in excitability and refractoriness are the essential features of most substrates.84
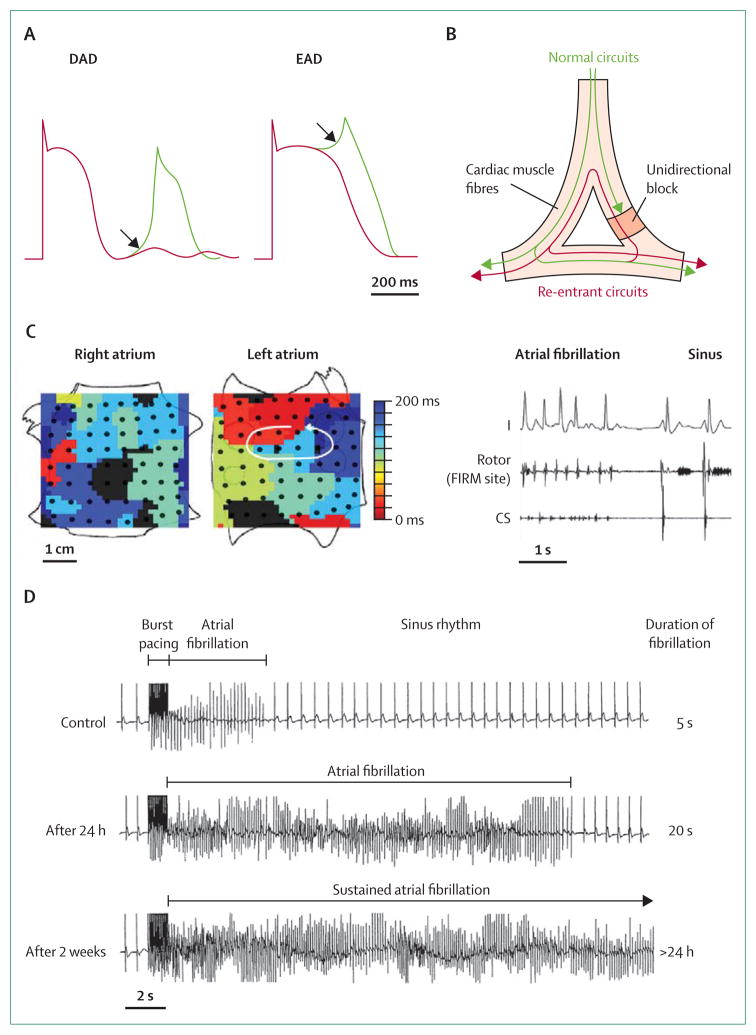
Figure 4. Conventional physiological mechanisms of arrhythmia.
(A) Afterdepolarisations are membrane depolarisations that occur late in or after the completion of the action potential. Delayed afterdepolarisations (DADs) occur after full repolarisation and early afterdepolarisations (EADs) during late repolarisation. (B) Circus-movement re-entry is characterised by an activation pattern that travels along a preferred anatomical structure to reactivate previously excited tissue. Such re-entry is dependent on unidirectional block and is supported by slow conduction and short refractory periods. (C) Acute termination of atrial fibrillation (AF) to sinus rhythm by FIRM (focal impulse and rotor modulation) ablation. Left: left-atrial rotor with counter-clockwise activation (red to blue) and disorganised right atrium during AF in a 60-year-old man. Right: FIRM ablation at left-atrial rotor terminated AF to sinus rhythm in less than 1 min, with ablation artifact recorded at centre of rotor. The patient is AF-free on implanted cardiac monitor after more than 1 year.81 The demonstration of spiral waves and their functional importance in human AF potentially provides a widely applicable rational approach to AF ablation. (D) AF-induced changes (remodelling) that promote AF.82 Work done in paced goats provides strong evidence for an evolution of the substrate. In control goats, high-frequency burst pacing induced only 5 s of AF, whereas after 24 h of artificially maintained AF the duration increased to 20 s. After 2 weeks, episodes of fibrillation last more than 24 h. (A) is reproduced from reference 83, by permission of the American Society for Clinical Investigation; (B) is reproduced from reference 36, by permission of Elsevier; (C) is reproduced from reference 81, by permission of Elsevier; (D) is reproduced from reference 82, by permission of Wolters Kluwer Health. CS=coronary sinus electrocardiogram.
The interaction between trigger and substrate causes initiation and perpetuation of arrhythmias.84 We are generally protected because the substrate, even in diseased hearts, is resistant to a breakdown in the normal sequence of activation. As such, only a small proportion of the ectopic beats (potential triggers) recorded by continuous electrocardiography will lead to sustained arrhythmia.16,83 Experimental models also strongly support the view that in well coupled cardiac tissue, the heart is protected from the consequences of sporadic depolarisations that might otherwise lead to sustained arrhythmia.85
The arrhythmogenic substrate might have been acquired in those with coronary disease, inflammatory disorders, or idiopathic dilated cardiomyopathy,50,51 but genetic factors almost always contribute.31,75 Although various sophisticated methods for imaging,86 physiological investigation,11,20,43 and omics techniques87 are becoming available and are applicable to the analysis of substrates, a major limitation is the availability of human tissue.6 As such, our understanding about how human arrhythmia substrates develop against a background of genetic, epigenetic,80 and other factors is fairly basic, and most experimental physiology has relied on work done in animals.
Triggered activity results from membrane oscillations (afterdepolarisations) during or after otherwise normal action potentials and in many circumstances provides the triggers that initiate arrhythmias (figure 4A).83,85 Delayed afterdepolarisations occur after full repolarisation and are favoured by cellular calcium loading.20,22,48,88 A key molecule in membrane depolarisations (and in the generation of a delayed after de polarisation), is the electrogenic sodium–calcium exchanger, which allows diastolic calcium leak from the sarcoplasmic reticulum.84,88 Delayed afterdepolarisations are an important mechanism in CPVT and in focal arrhythmia caused by ischaemia or adrenergic stress.84 Early afterdepolarisations occur before full completion of repolarisation—in which the leading candidate mechanism might involve window currents through L-type calcium channels (figure 4A).84 Early afterdepolarisations seem to occur most often when there is prolongation of action potentials, as is seen in patients with long QT syndrome or with heart failure.
Re-entrant tachycardias caused by propagation of waves through large circuits of excitable tissue are the cause of several well defined, acquired clinical disorders, including atrial flutter and many ventricular tachycardias.1–3 Fixed re-entrant circuits that result in anatomical re-entry might also be mainly congenital and genetic (eg, Wolff-Parkinson-White syndrome).3,89 The functional characteristics of these macro-reentrant circuits have to allow for unidirectional block, and they will incorporate areas of slowed conduction, changed refractoriness, and an excitable gap, and the circuit can be entrained (figure 4B).2,84 In addition to such anatomically defined circuits, re-entrant excitation can also use functional circuits—ie, circuits that are not anatomically defined but arise (often transiently) because of other perturbations. These functional circuits can result in patterns such as spirals or scrolls, the location of which might meander through the heart with successive depolarisations.81,84,90 Much work now lends support to the suggestion that such spiral-wave re-entry has a role in both atrial and ventricular fibrillation (figure 4C).81,90
Essentially, once an arrhythmia has been triggered, its course within a substrate is likely to conform to a reentrant pattern of activation that can have a stable course,2,3 or be less stable and so take the form of a spiral or scroll wave.2,81,84 Restitution describes the rate-dependent interaction between the trigger and the substrate and helps to account for transitions between rapid, relatively benign rhythms and fibrillation.10,91
Mechanisms of atrial fibrillation
Issues of initiation and perpetuation have attracted particular attention in atrial fibrillation because of their clinical relevance.1,92 Two principal hypotheses have been advanced to account for the behaviour of clinical arrhythmia.83,84 The multiple wavelet hypothesis, which is based on mapping in dogs and patients, holds that several, non-localised interacting circuits meander around the atria.83,84 Alternatively, the localised source hypothesis proposes that rapidly conducted beats (eg, from focal, delayed afterdepolarisation-mediated sources88 or electrical spiral waves [rotors]) interact with an amenable substrate.83,84 Since the pulmonary veins were first implicated,1 localised sources have come to be regarded as of increased importance. The direct clinical relevance of localised sources has been lent support not only by the efficacy of pulmonary vein isolation in many patients,1 but also, more recently, by the apparently effective targeted ablation of spiral waves (figure 4C).1,81
Atrial fibrillation substrates change with time. Initially, the cardiac substrate is thought to be characterised by shortened action potentials and abbreviated refractoriness.24 Later, in many patients, structural changes will ensue, with scarring and fibrosis.84 This temporal evolution is described as remodelling, and during this process the relative contributions of functional and anatomical features of the substrate to arrhythmia will change.83,93 Remodelling shows wide variation between individuals, which is governed by many factors, including genetic factors, individual behaviour, and treatment.5,83 Rapid rates undoubtedly promote remodelling and have been used to generate atrial fibrillation-prone animals (figure 4D).82 However, the underlying biology of remodelling remains poorly understood, but is of great interest and seems likely to account for much of the clinical variability of atrial fibrillation.5,93 Some patients progress from infrequent runs of self-terminating fibrillation to more persistent patterns of arrhythmia in a fairly short time, whereas in others clinical evidence of arrhythmia can even regress.93
Substantial experimental evidence suggests that the interaction between the trigger and substrate in atrial fibrillation is governed by the principles we have described. Accordingly, the importance of the pulmonary veins in most patients is universally accepted, and these provide the trigger for initiation of the arrhythmia.1 The substrate is receptive to the emerging trigger, with steep restitution reported around the pulmonary veins.94 Additionally, exaggerated shortening of refractoriness in the left atrium95 and the patterns of left-atrial ion-channel expression further promote wave break96 and the initiation of re-entrant excitation (clinically manifest as atrial fibrillation).90
Tractable model systems
Computer models of heart function that incorporate equations to describe activity of individual ion channels or transporters are well established.6,9,25 However, collection of high-quality data from the full range of relevant animal species to populate the models has been difficult.6 Ideally, these data should include descriptions of the function, expression, interactions, and metabolic status of individual proteins.9 Because human tissue is difficult to obtain,6 heterologous expression of ion channels in non-human cells12 with complementary studies in wild type97 and genetically modified animals11,21,97,98 have had to suffice for detailed functional studies.
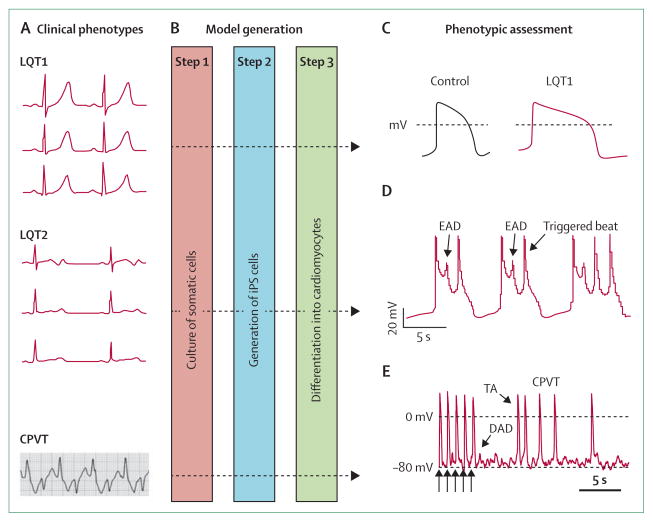
Genetically modified mouse models have provided one research mainstay to link human genetic variants to arrhythmogenic phenotypes.7,11–13 Advances in cell biology now allow the opportunity to study electro-physiology in human cardiomyocytes derived from iPS cells.8 These cells are difficult to obtain reliably and can have a fairly immature phenotype, but they do have sarcomeric structures, ion-channel profiles, calcium handling, and drug responses that broadly conform to observations made clinically and in conventional cells.20,21,43,44 Models reported so far include those of some subtypes of long QT syndrome43,44 and CPVT (figure 5).20
Figure 5. Induced pluripotent stem (iPS)-cell models of ion-channel diseases.
(A) iPS-cell models of long QT syndromes and catecholaminergic polymorphic ventricular tachycardia (CPVT) have been established (figure shows representative electrocardiograms of each).20,43,44 (B) Steps 1–2: fibroblasts and other somatic cells can be reprogrammed to become iPS cells after transduction with retroviral vectors that express four transcription factors (POU5F1 [also known as OCT4], SOX2, KLF4, and MYC).8 Step 3: directed differentiation into cardiomyocytes provides cellular models for physiological assessment. Cardiomyocytes from patients with long QT syndrome 1 (LQT1) that harbour an Arg190Gln variant in KCNQ3 recapitulate features of clinical phenotypes (C), with prolonged action potentials, defective potassium-channel characteristics, and inducible arrhythmias attenuated by β-adrenergic blockade.43 Similar characteristics are seen with cells from LQT2 patients—the Ala614Val variant in KCNH3 (D) shows early afterdepolarisations (EADs) and triggered activity.44 Typical action potentials from a ventricular myocyte obtained from a patient with CPVT that carried a Ser406Leu variant in RYR3 (E) show 5 paced beats (arrows) followed by delayed afterdepolarisations (DADs) and triggered activity (TA).20 (C) is reproduced from reference 43, by permission of the Massachusetts Medical Society; (D) is reproduced from reference 44, by permission of Nature Publishing Group; (E) is reproduced from reference 20, by permission of John Wiley and Sons.
Developing new treatments
An important aim of the development of disease models is to allow responses to therapeutic interventions to be assessed.9 New models, combined with other features of a systems approach, might well identify molecules the targeting of which could be antiarrhythmic. These insights might come from family or population genetic approaches, or from laboratory studies of proarrhythmic signalling pathways.5 The potential relevance of individual targets could then be assessed in wild-type or genetically modified organisms99 or in iPS cell-derived cardiomyocytes.20,43,44
Encouragingly, mouse models have already been used to provide evidence for drug repurposing. For example, flecainide (a nominal class Ic sodium-channel blocker) has been shown to directly block ryanodine-receptor mediated calcium release.21 Flecainide was then reported to be effective in arrhythmia suppression in both calsequestrin bi-allelic knockout (Casq2−/−) mouse models of CPVT21 and in a subsequent clinical study into CPVT,100 which has provided clinicians with a much needed therapeutic option. In similar Casq2−/− mutant mice, carvedilol and its engineered analogues also suppressed both spontaneous and induced arrhythmia.22 Additionally, in human iPS-cell models, well established interventions (eg, propranolol43 and nifedipine44) rescued arrhythmia phenotypes in long QT syndrome 1 and 2, respectively.43,44 Similarly, dantrolene rescued the phenotype of CPVT (figure 5).20 The use of iPS cell technology to screen non-cardiac drugs to address potential toxicity is also appealing.8,56 In addition to experimental approaches, computational models have been used to describe drug–tissue interactions, which allow for predictions of efficacy and risk.26
Conclusions
Systematic approaches to human disease are at an early stage, but they offer the possibility of refined diagnosis, risk prediction, and targeted treatment decisions (figure 1).18 We believe that cardiac arrhythmias are especially amenable to a systems biology approach, partly because of the range of measureable determinants.6,9 Next-generation sequencing,52 exploi tation of stem cell-based models,8 and new uses of omics technologies87 are starting to provide tangible evidence of what can be achieved. The use of these experimental technologies in combination with holistic, network-based approaches6,10,18 can be used to find rational solutions to many of the unresolved questions in arrhythmia management.
Search strategy and selection criteria.
We started with a thorough review of our personal collections of articles and reviews accumulated over several years. We then searched PubMed, Google Scholar, Scopus, and other sources to ensure that we included the most important ideas and articles. We have tended to cite recent original research papers and supplemented these with some reviews and commentaries that provide an overview of older material.
Acknowledgments
AAG is supported by the British Heart Foundation, the UK Medical Research Council, the Wellcome Trust, and the UK Biotechnology and Biological Sciences Research Council. DMR is supported by grants from the US Public Health Service, including grant numbers HL49989 and HL65962.
Footnotes
Contributors
AAG and DMR both contributed to the drafting and revision of the report.
Conflicts of interest
AAG is a consultant for Xention. DMR has received patent royalties from Clinical Data (Forest Laboratories).
Contributor Information
Andrew A Grace, Department of Biochemistry, University of Cambridge, Cambridge, UK; Papworth Hospital, Cambridge, UK.
Prof. Dan M Roden, School of Medicine, Vanderbilt University, Nashville, TN, USA.
References
- 1.Lee G, Sanders P, Kalman JM. Catheter ablation of cardiac arrhythmias: state of the art. Lancet. 2012;380:1509–19. doi: 10.1016/S0140-6736(12)61463-9. [DOI] [PubMed] [Google Scholar]
- 2.John RM, Tedrow UB, Koplan BA, et al. Ventricular arrhythmias and sudden cardiac death. Lancet. 2012;380:1520–29. doi: 10.1016/S0140-6736(12)61413-5. [DOI] [PubMed] [Google Scholar]
- 3.Delacretaz E. Supraventricular tachycardia. N Engl J Med. 2006;354:1039–51. doi: 10.1056/NEJMcp051145. [DOI] [PubMed] [Google Scholar]
- 4.Chugh SS. Early identification of risk factors for sudden cardiac death. Nat Rev Cardiol. 2010;7:318–26. doi: 10.1038/nrcardio.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrev D, Carlsson L, Nattel S. Novel molecular targets for atrial fibrillation therapy. Nat Rev Drug Discov. 2012;11:275–91. doi: 10.1038/nrd3682. [DOI] [PubMed] [Google Scholar]
- 6.Rudy Y, Ackerman MJ, Bers DM, et al. Systems approach to understanding electromechanical activity in the human heart: a national heart, lung, and blood institute workshop summary. Circulation. 2008;118:1202–11. doi: 10.1161/CIRCULATIONAHA.108.772715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dow LE, Lowe SW. Life in the fast lane: mammalian disease models in the genomics era. Cell. 2012;148:1099–109. doi: 10.1016/j.cell.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherry ABC, Daley GQ. Reprogramming cellular identity for regenerative medicine. Cell. 2012;148:1110–22. doi: 10.1016/j.cell.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noble D. Biophysics and systems biology. Philos Transact A Math Phys Eng Sci. 2010;368:1125–39. doi: 10.1098/rsta.2009.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss JN, Nivala M, Garfinkel A, Qu Z. Alternans and arrhythmias: from cell to heart. Circ Res. 2011;108:98–112. doi: 10.1161/CIRCRESAHA.110.223586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papadatos GA, Wallerstein PM, Head CE, et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci USA. 2002;99:6210–15. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe H, Yang T, Stroud DM, et al. Striking In vivo phenotype of a disease-associated human SCN5A mutation producing minimal changes in vitro. Circulation. 2011;124:1001–11. doi: 10.1161/CIRCULATIONAHA.110.987248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeyaraj D, Haldar SM, Wan X, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483:96–99. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnani JW, Rienstra M, Lin H, et al. Atrial fibrillation: current knowledge and future directions in epidemiology and genomics. Circulation. 2011;124:1982–93. doi: 10.1161/CIRCULATIONAHA.111.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rienstra M, Lubitz SA, Mahida S, et al. Symptoms and functional status of patients with atrial fibrillation: state of the art and future research opportunities. Circulation. 2012;125:2933–43. doi: 10.1161/CIRCULATIONAHA.111.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloch Thomsen PE, Jons C, Raatikainen MJ, et al. for the CARISMA Study Group. Long-term recording of cardiac arrhythmias with an implantable cardiac monitor in patients with reduced ejection fraction after acute myocardial infarction: the cardiac arrhythmias and risk stratification after acute myocardial infarction (CARISMA) study. Circulation. 2010;122:1258–64. doi: 10.1161/CIRCULATIONAHA.109.902148. [DOI] [PubMed] [Google Scholar]
- 17.Nurse P, Hayles J. The cell in an era of systems biology. Cell. 2011;144:850–54. doi: 10.1016/j.cell.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 18.Chan SY, Loscalzo J. The emerging paradigm of network medicine in the study of human disease. Circ Res. 2012;111:359–74. doi: 10.1161/CIRCRESAHA.111.258541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–57. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung CB, Moretti A, Mederos YSM, et al. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4:180–91. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe H, Chopra N, Laver D, et al. Flecainide prevents catecholaminergic polymorphic ventricular tachycardia in mice and humans. Nat Med. 2009;15:380–83. doi: 10.1038/nm.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Q, Xiao J, Jiang D, et al. Carvedilol and its new analogs suppress arrhythmogenic store overload-induced Ca2+ release. Nat Med. 2011;17:1003–09. doi: 10.1038/nm.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Hara T, Virag L, Varro A, Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol. 2011;7:e1002061. doi: 10.1371/journal.pcbi.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandi E, Pandit SV, Voigt N, et al. Human atrial action potential and Ca2+ model: sinus rhythm and chronic atrial fibrillation. Circ Res. 2011;109:1055–66. doi: 10.1161/CIRCRESAHA.111.253955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thul R, Coombes S, Roderick HL, Bootman MD. Subcellular calcium dynamics in a whole-cell model of an atrial myocyte. Proc Natl Acad Sci USA. 2012;109:2150–55. doi: 10.1073/pnas.1115855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno JD, Zhu ZI, Yang PC, et al. A computational model to predict the effects of class I anti-arrhythmic drugs on ventricular rhythms. Sci Transl Med. 2011;3:98ra83. doi: 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefansdottir H, Aspelund T, Gudnason V, Arnar DO. Trends in the incidence and prevalence of atrial fibrillation in Iceland and future projections. Europace. 2011;13:1110–17. doi: 10.1093/europace/eur132. [DOI] [PubMed] [Google Scholar]
- 28.Miyasaka Y, Barnes ME, Gersh BJ, et al. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation. 2006;114:119–25. doi: 10.1161/CIRCULATIONAHA.105.595140. [DOI] [PubMed] [Google Scholar]
- 29.Wang TJ, Parise H, Levy D, et al. Obesity and the risk of new-onset atrial fibrillation. JAMA. 2004;292:2471–77. doi: 10.1001/jama.292.20.2471. [DOI] [PubMed] [Google Scholar]
- 30.Nichols GA, Reinier K, Chugh SS. Independent contribution of diabetes to increased prevalence and incidence of atrial fibrillation. Diabetes Care. 2009;32:1851–56. doi: 10.2337/dc09-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noseworthy PA, Newton-Cheh C. Genetic determinants of sudden cardiac death. Circulation. 2008;118:1854–63. doi: 10.1161/CIRCULATIONAHA.108.783654. [DOI] [PubMed] [Google Scholar]
- 32.Go AS, Hylek EM, Phillips KA, et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the anticoagulation and risk factors in atrial fibrillation (ATRIA) study. JAMA. 2001;285:2370–75. doi: 10.1001/jama.285.18.2370. [DOI] [PubMed] [Google Scholar]
- 33.Tedrow UB, Conen D, Ridker PM, et al. The long- and short-term impact of elevated body mass index on the risk of new atrial fibrillation the WHS (women’s health study) J Am Coll Cardiol. 2010;55:2319–27. doi: 10.1016/j.jacc.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong MH, Fonarow GC, Peterson ED, et al. Systematic review of the incidence of sudden cardiac death in the United States. J Am Coll Cardiol. 2011;57:794–801. doi: 10.1016/j.jacc.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jouven X, Lemaitre RN, Rea TD, Sotoodehnia N, Empana JP, Siscovick DS. Diabetes, glucose level, and risk of sudden cardiac death. Eur Heart J. 2005;26:2142–47. doi: 10.1093/eurheartj/ehi376. [DOI] [PubMed] [Google Scholar]
- 36.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104:569–80. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 37.Ackerman MJ, Mohler PJ. Defining a new paradigm for human arrhythmia syndromes: phenotypic manifestations of gene mutations in ion channel- and transporter-associated proteins. Circ Res. 2010;107:457–65. doi: 10.1161/CIRCRESAHA.110.224592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Shen J, Splawski I, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–11. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz PJ, Crotti L, Insolia R. Long-QT syndrome: from genetics to management. Circ Arrhythm Electrophysiol. 2012;5:868–77. doi: 10.1161/CIRCEP.111.962019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 42.Vandenberg JI, Perry MD, Perrin MJ, Mann SA, Ke Y, Hill AP. hERG K+ channels: structure, function, and clinical significance. Physiol Rev. 2012;92:1393–478. doi: 10.1152/physrev.00036.2011. [DOI] [PubMed] [Google Scholar]
- 43.Moretti A, Bellin M, Welling A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 44.Itzhaki I, Maizels L, Huber I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–29. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 45.Brink PA, Crotti L, Corfield V, et al. Phenotypic variability and unusual clinical severity of congenital long-QT syndrome in a founder population. Circulation. 2005;112:2602–10. doi: 10.1161/CIRCULATIONAHA.105.572453. [DOI] [PubMed] [Google Scholar]
- 46.Golbus JR, Puckelwartz MJ, Fahrenbach JP, Dellefave-Castillo LM, Wolfgeher D, McNally EM. Population-based variation in cardiomyopathy genes. Circ Cardiovasc Genet. 2012;5:391–99. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Q, Kirsch GE, Zhang D, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–96. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 48.Cerrone M, Napolitano C, Priori SG. Catecholaminergic polymorphic ventricular tachycardia: a paradigm to understand mechanisms of arrhythmias associated to impaired Ca(2+) regulation. Heart Rhythm. 2009;6:1652–59. doi: 10.1016/j.hrthm.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 49.Haissaguerre M, Derval N, Sacher F, et al. Sudden cardiac arrest associated with early repolarization. N Engl J Med. 2008;358:2016–23. doi: 10.1056/NEJMoa071968. [DOI] [PubMed] [Google Scholar]
- 50.Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res. 2010;107:700–14. doi: 10.1161/CIRCRESAHA.110.223412. [DOI] [PubMed] [Google Scholar]
- 51.Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathies. N Engl J Med. 2011;364:1643–56. doi: 10.1056/NEJMra0902923. [DOI] [PubMed] [Google Scholar]
- 52.Dewey FE, Pan S, Wheeler MT, Quake SR, Ashley EA. DNA sequencing: clinical applications of new DNA sequencing technologies. Circulation. 2012;125:931–44. doi: 10.1161/CIRCULATIONAHA.110.972828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holm H, Gudbjartsson DF, Sulem P, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–20. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jons C, O-Uchi J, Moss AJ, et al. Use of mutant-specific ion channel characteristics for risk stratification of long QT syndrome patients. Sci Transl Med. 2011;3:76ra28. doi: 10.1126/scitranslmed.3001551. [DOI] [PubMed] [Google Scholar]
- 55.Barsheshet A, Goldenberg I, O-Uchi J, et al. Mutations in cytoplasmic loops of the KCNQ1 channel and the risk of life-threatening events: implications for mutation-specific response to beta-blocker therapy in type-1 long-QT syndrome. Circulation. 2012;125:1988–96. doi: 10.1161/CIRCULATIONAHA.111.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kannankeril P, Roden DM, Darbar D. Drug-induced long QT syndrome. Pharmacol Rev. 2010;62:760–81. doi: 10.1124/pr.110.003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kääb S, Crawford DC, Sinner MF, et al. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circ Cardiovasc Genet. 2012;5:91–99. doi: 10.1161/CIRCGENETICS.111.960930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–96. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 59.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–75. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sotoodehnia N, Isaacs A, de Bakker PI, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–76. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mommersteeg MT, Brown NA, Prall OW, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–09. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 62.Holm H, Gudbjartsson DF, Arnar DO, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–22. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 63.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–59. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chambers JC, Zhao J, Terracciano CM, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–52. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 65.Noseworthy PA, Havulinna AS, Porthan K, et al. Common genetic variants, QT interval, and sudden cardiac death in a Finnish population-based study. Circ Cardiovasc Genet. 2011;4:305–11. doi: 10.1161/CIRCGENETICS.110.959049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kao WH, Arking DE, Post W, et al. Genetic variations in nitric oxide synthase 1 adaptor protein are associated with sudden cardiac death in US white community-based populations. Circulation. 2009;119:940–51. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng S, Keyes MJ, Larson MG, et al. Long-term outcomes in individuals with prolonged PR interval or first-degree atrioventricular block. JAMA. 2009;301:2571–77. doi: 10.1001/jama.2009.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–21. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritchie MD, Rowan S, Kucera G, et al. Chromosome 4q25 variants are genetic modifiers of rare ion channel mutations associated with familial atrial fibrillation. J Am Coll Cardiol. 2012;60:1173–81. doi: 10.1016/j.jacc.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darbar D, Kannankeril PJ, Donahue BS, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–35. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–57. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 72.Chinchilla A, Daimi H, Lozano-Velasco E, et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–79. doi: 10.1161/CIRCGENETICS.110.958116. [DOI] [PubMed] [Google Scholar]
- 73.Kirchhof P, Kahr PC, Kaese S, et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–33. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 74.Jouven X, Desnos M, Guerot C, Ducimetiere P. Predicting sudden death in the population: the Paris prospective study I. Circulation. 1999;99:1978–83. doi: 10.1161/01.cir.99.15.1978. [DOI] [PubMed] [Google Scholar]
- 75.Bezzina CR, Pazoki R, Bardai A, et al. Genome-wide association study identifies a susceptibility locus at 21q21 for ventricular fibrillation in acute myocardial infarction. Nat Genet. 2010;42:688–91. doi: 10.1038/ng.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Albert CM, MacRae CA, Chasman DI, et al. Common variants in cardiac ion channel genes are associated with sudden cardiac death. Circ Arrhythm Electrophysiol. 2010;3:222–29. doi: 10.1161/CIRCEP.110.944934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westaway SK, Reinier K, Huertas-Vazquez A, et al. Common variants in CASQ2, GPD1L, and NOS1AP are significantly associated with risk of sudden death in patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4:397–402. doi: 10.1161/CIRCGENETICS.111.959916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newton-Cheh C, Cook NR, VanDenburgh M, Rimm EB, Ridker PM, Albert CM. A common variant at 9p21 is associated with sudden and arrhythmic cardiac death. Circulation. 2009;120:2062–68. doi: 10.1161/CIRCULATIONAHA.109.879049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murray SS, Smith EN, Villarasa N, et al. Genome-wide association of implantable cardioverter-defibrillator activation with life-threatening arrhythmias. PLoS One. 2012;7:e25387. doi: 10.1371/journal.pone.0025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mann SA, Otway R, Guo G, et al. Epistatic effects of potassium channel variation on cardiac repolarization and atrial fibrillation risk. J Am Coll Cardiol. 2012;59:1017–25. doi: 10.1016/j.jacc.2011.11.039. [DOI] [PubMed] [Google Scholar]
- 81.Narayan SM, Krummen DE, Shivkumar K, Clopton P, Rappel WJ, Miller JM. Treatment of atrial fibrillation by the ablation of localized sources: CONFIRM (conventional ablation for atrial fibrillation with or without focal impulse and rotor modulation) trial. J Am Coll Cardiol. 2012;60:628–36. doi: 10.1016/j.jacc.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maurits CEF, Wijffels MC, Kirchhof CJ, Dorland R, Allessie MA. Atrial fibrillation begets atrial fibrillation. A study in awake chronically instrumented goats. Circulation. 1995;92:1954–68. doi: 10.1161/01.cir.92.7.1954. [DOI] [PubMed] [Google Scholar]
- 83.Wakili R, Voigt N, Kaab S, Dobrev D, Nattel S. Recent advances in the molecular pathophysiology of atrial fibrillation. J Clin Invest. 2011;121:2955–68. doi: 10.1172/JCI46315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schotten U, Verheule S, Kirchhof P, Goette A. Pathophysiological mechanisms of atrial fibrillation: a translational appraisal. Physiol Rev. 2011;91:265–325. doi: 10.1152/physrev.00031.2009. [DOI] [PubMed] [Google Scholar]
- 85.Xie Y, Sato D, Garfinkel A, Qu Z, Weiss JN. So little source, so much sink: requirements for afterdepolarizations to propagate in tissue. Biophys J. 2010;99:1408–15. doi: 10.1016/j.bpj.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herron TJ, Lee P, Jalife J. Optical imaging of voltage and calcium in cardiac cells & tissues. Circ Res. 2012;110:609–23. doi: 10.1161/CIRCRESAHA.111.247494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Voigt N, Li N, Wang Q, et al. Enhanced sarcoplasmic reticulum Ca2+ leak and increased Na+-Ca2+ exchanger function underlie delayed afterdepolarizations in patients with chronic atrial fibrillation. Circulation. 2012;125:2059–70. doi: 10.1161/CIRCULATIONAHA.111.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rentschler S, Harris BS, Kuznekoff L, et al. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest. 2011;121:525–33. doi: 10.1172/JCI44470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vaquero M, Calvo D, Jalife J. Cardiac fibrillation: from ion channels to rotors in the human heart. Heart Rhythm. 2008;5:872–79. doi: 10.1016/j.hrthm.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Narayan SM, Franz MR, Clopton P, Pruvot EJ, Krummen DE. Repolarization alternans reveals vulnerability to human atrial fibrillation. Circulation. 2011;123:2922–30. doi: 10.1161/CIRCULATIONAHA.110.977827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Atienza F, Almendral J, Moreno J, et al. Activation of inward rectifier potassium channels accelerates atrial fibrillation in humans: evidence for a reentrant mechanism. Circulation. 2006;114:2434–42. doi: 10.1161/CIRCULATIONAHA.106.633735. [DOI] [PubMed] [Google Scholar]
- 93.Jahangir A, Lee V, Friedman PA, et al. Long-term progression and outcomes with aging in patients with lone atrial fibrillation: a 30-year follow-up study. Circulation. 2007;115:3050–56. doi: 10.1161/CIRCULATIONAHA.106.644484. [DOI] [PubMed] [Google Scholar]
- 94.Lalani GG, Schricker A, Gibson M, Rostamian A, Krummen DE, Narayan SM. Atrial conduction slows immediately before the onset of human atrial fibrillation: a bi-atrial contact mapping study of transitions to atrial fibrillation. J Am Coll Cardiol. 2012;59:595–606. doi: 10.1016/j.jacc.2011.10.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tanaka K, Zlochiver S, Vikstrom KL, et al. Spatial distribution of fibrosis governs fibrillation wave dynamics in the posterior left atrium during heart failure. Circ Res. 2007;101:839–47. doi: 10.1161/CIRCRESAHA.107.153858. [DOI] [PubMed] [Google Scholar]
- 96.Voigt N, Trausch A, Knaut M, et al. Left-to-right atrial inward rectifier potassium current gradients in patients with paroxysmal versus chronic atrial fibrillation. Circ Arrhythm Electrophysiol. 2010;3:472–80. doi: 10.1161/CIRCEP.110.954636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peterson RT, Macrae CA. Systematic approaches to toxicology in the zebrafish. Annu Rev Pharmacol Toxicol. 2012;52:433–53. doi: 10.1146/annurev-pharmtox-010611-134751. [DOI] [PubMed] [Google Scholar]
- 98.Cunha SR, Hund TJ, Hashemi S, et al. Defects in ankyrin-based membrane protein targeting pathways underlie atrial fibrillation. Circulation. 2011;124:1212–22. doi: 10.1161/CIRCULATIONAHA.111.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He BJ, Joiner ML, Singh MV, et al. Oxidation of CaMKII determines the cardiotoxic effects of aldosterone. Nat Med. 2011;17:1610–18. doi: 10.1038/nm.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Werf C, Kannankeril PJ, Sacher F, et al. Flecainide therapy reduces exercise-induced ventricular arrhythmias in patients with catecholaminergic polymorphic ventricular tachycardia. J Am Coll Cardiol. 2011;57:2244–54. doi: 10.1016/j.jacc.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]