Abstract
Blastocystis, an unusual anaerobic, single-celled stramenopile, is a remarkably successful intestinal parasite of a vast array of host species including humans. Fecal Deoxyribonucleic acid (DNA) analysis by nucleic-acid based methods in particular has led to significant advances in Blastocystis diagnostics and research over the past few years enabling accurate identification of carriers and molecular characterization by high discriminatory power. Moreover, Blastocystis comprises a multitude of subtypes (STs) (arguably species) many of which have been identified only recently and molecular epidemiological studies have revealed a significant difference in the distribution of STs across host species and geographical regions. Having a cosmopolitan distribution, the parasite is a common laboratory finding in the stools of individuals with and without intestinal symptoms across the entire globe and while the parasite remains extremely difficult to eradicate and isolate in culture, appropriate molecular tools are now available to resolve important questions such as whether the clinical outcome of colonization is linked to ST and whether Blastocystis is transmitted zoonotically. This review summarizes some of the recent advances in the molecular diagnosis of Blastocystis and gives an introduction to Blastocystis STs, including a recommendation of subtyping methodology based on recent data and method comparisons. A few suggestions for future directions and research areas are given in the light of relevant technological advances and the availability of mitochondrial and nuclear genomes.
KEY WORDS: Blastocystis, deoxyribonucleic acid, diagnosis, epidemiology, polymerase chain reaction
INTRODUCTION
Over the past 10 years, screening of patients for intestinal parasites, especially protozoa, by molecular methods has started to gain a foothold in clinical microbiology laboratories. Although a variety of theoretical and practical limitations and obstacles can be identified,[1] these are counter-balanced by the many advantages of nucleic-acid based diagnostics; advantages that include higher diagnostic sensitivity, high accuracy (repeatability, reproducibility), and semi-or-fully automated processes, leading to higher cost-effectiveness and standardization.[2,3]
Conserved and variable regions within the 18S small subunit (SSU) ribosomal DNA (rDNA) constitute the basis for identifying phylogenetic relationships between species[4] and the SSU ribosomal RNA (rRNA) genes have been popular targets for diagnostic polymerase chain reaction (PCR) assays; multiple gene copy numbers ensure high diagnostic sensitivity, and SSU rDNA sequences are available for most known parasites.
Amplification of parasite-specific rDNA from genomic DNA extracted from feces (i.e., without any intermediate step such as culture or isolation of (oo) cysts) has not only opened new diagnostic avenues in the clinical microbiology laboratory but also enabled us to detect a multitude of novel ribosomal lineages, many of which are probably separate species, either in the absence of morphology data and cultures,[5,6,7,8] or in situations where morphological differentiation is not possible.[9,10]
Molecular screening assays typically include those enabling immediate detection of and differentiation between Entamoeba histolytica and Entamoeba dispar, and those that target the most common and clinically relevant parasitic genera such as Giardia and Cryptosporidium.[2,11,12] Species identification and genotyping of the latter genera have been used to clarify transmission patterns and to assist in outbreak investigations.
Much more common intestinal parasites include Blastocystis and Dientamoeba, the clinical significance of which remains to be fully understood, in part due to the absence of efficient treatment modalities.[13,14,15,16] It is clear that detection of these parasites by conventional methods is difficult for a variety of reasons, such as lack of a detectable cyst stage (Dientamoeba), fragility of vegetative stages, and general morphological inconspicuousness.[2,17,18,19]
The prevalence, intra-generic diversity and transmission modes of these two genera are only starting to be revealed following the introduction of molecular methods. For Dientamoeba, the design of conventional and real-time diagnostic PCR assays has proven relatively easy due to the fact that only one species has been found in humans (Dientamoeba fragilis), and there is relatively little genetic variation across the SSU rRNA gene.
GENETIC DIVERSITY OF BLASTOCYSTIS AND BLASTOCYSTIS SUBTYPING
The situation is quite different for Blastocystis, a remarkable, single-celled enteric parasitic stramenopile of humans and a vast array of non-human hosts. Based on SSU rDNA analysis, this genus comprises at least 17 lineages (arguably species), the so-called subtypes (STs).[20] ST1-ST4 account for probably more than 90% of human carriage,[21,22] while the remaining human carriers are colonized by isolates belonging to ST5-ST9. All of these STs, apart from ST9 have also been found in various non-human hosts. Since 2009, no less than 8 novel STs (ST10-ST17) have been detected in non-human hosts,[20] primarily by amplification of Blastocystis DNA directly from fecal DNA template.[5,7,8]
Importantly, the ST system developed in 2007 includes Blastocystis exclusively from humans, other mammals and birds.[23] Hence Blastocystis from for instance reptiles, amphibia, and insects have not been included. Incidentally, ST5 has been found in an amphibian host, and conversely, a few isolates from mammalian samples representing two of the “novel” STs, namely ST15 and ST17, are phylogenetically closer related to reptilian or arthropod isolates than to STs usually found in mammalian hosts.[20]
Genetic variation within some STs (e.g., ST3) may amount to about 3% and experience has told us that the level of genetic difference between STs is at least 4-5%.[24] In the event that a Blastocystis barcode sequence does not match those in the 18S database and has less than 97-98% similarity to sequences available at the NCBI database, it is suggested that the complete SSU rRNA gene be sequenced using a mixture of general and specific primers (and preferentially from DNA from a cultured isolate since this makes amplification easier and sequence traces often become clearer). The complete sequence should be aligned with reference sequences and the alignment should be edited and submitted to phylogenetic analyses using a variety of models, including at least distance and Maximum Likelihood analysis.[5,20] Since a standard approach to Blastocystis subtyping and nomenclature has been proposed,[23,25] it is appropriate to approach a standard methodology here also; for more detailed considerations and guidelines regarding ST analysis and nomenclature, please refer to Clark et al.[20,24]
Subtyping has given us a crucial tool for testing the current working hypothesis that the clinical outcome of Blastocystis colonization is dependent on ST.[26,27,28] Based on observations from multiple studies from various countries, no clear link between STs and clinical phenotypes can be made at present. However, at least two main confounders may play a role: Firstly, significant variation in the distribution of STs is seen across geographic ranges. A review of more than 3,000 observations from global surveys made it clear that while ST4 is almost as common as ST1 and ST3 in some countries in Europe, this ST is virtually absent in most other regions.[21] Conversely, ST6 is very rare in Europeans and Americans while human colonization by this ST appears rather common in some Asian and Middle Eastern countries, such as Egypt, Nepal, Japan and Malaysia. Although ST3 is the most common ST reported to date, a few individual surveys have seen a strong preponderance of ST1 or a “tie” between ST1 and ST3.[29,30,31,32,33] Secondly, different methodologies are used for subtyping, and differences in results from ST surveys may stem from differences in subtyping methodologies.[25] Subtyping has been approached in mainly two ways: By screening fecal samples by PCR using either (1) ST-specific primers (sequence-tagged site (STS) primers)[34] or (2) genus-specific primers with subsequent sequencing for ST identification; one such method is barcoding covering the 600 5’-most bases of the SSU rDNA.[35] These two methods, STS and barcoding have different advantages and limitations, which will be discussed in the following section.
Barcoding appears robust for genetic characterization combining the use of a forward primer of broad, eukaryotic specificity (RD5[36]), and a genus-specific reverse primer (BhRDr[35]). Sequence traces from PCR products obtained by barcoding may be difficult to decipher in cases of mixed ST carriage. The foremost advantage is that barcode fasta files can be queried individually or by bulk submission at the “Blastocystis ST (18S) and Multi Locus Sequence Typing (MLST) Multi Locus Sequence Typing Databases”[37,38] at www.pubmlst.org/blastocystis and identified not only to ST level, but to 18S allele level; 18S allele analysis offers higher resolution than subtyping alone.[38] This is especially attractive for new colleagues in the field since this approach bypasses the need for phylogenetic analyses based on reference sequences and hence the risk of annotating sequences to STs to which they do not belong, which has been a problem previously.[24,39] It should be noted that barcoding PCR should not be used on fecal DNA template as a strictly diagnostic tool since the RD5/BhRDr primer pair typically amplifies common fungal DNA in the absence of Blastocystis with no obvious difference in PCR product size.[24]
By using the STS method, the need for sequencing PCR products is circumvented, and detection of mixed ST carriage is straight-forward. However, although highly specific, some of the primer pairs appear rather insensitive, especially those targeting ST4, for which the common 18S allele (allele 42) appears to go undetected.[25,38] Moreover, STS primer pairs are only available for ST1-ST7; hence, human carriage due to other STs remains undetected. Moreover, STS PCR products are rather long (~300-700 bp), which is a clear disadvantage since these primers are sometimes used diagnostically. Finally, there is evidence that sequence variation in the STS primer regions exists.[25]
A list of studies employing the two different methods is available in Table 1. It appears that ST4 accounts for 1% and 17% of Blastocystis carriage detected by the STS method and non-STS methods, respectively. Although potentially coincidental, it should be noted that ST4 is absent mainly in those countries where surveys have been carried out using the STS method, typically Asian and Middle Eastern countries. Therefore, the use of barcoding in these countries is especially warranted to confirm/refute the absence of ST4 in these particular regions.
Table 1.
Subtype distribution by country of sample origin (modified after[21])
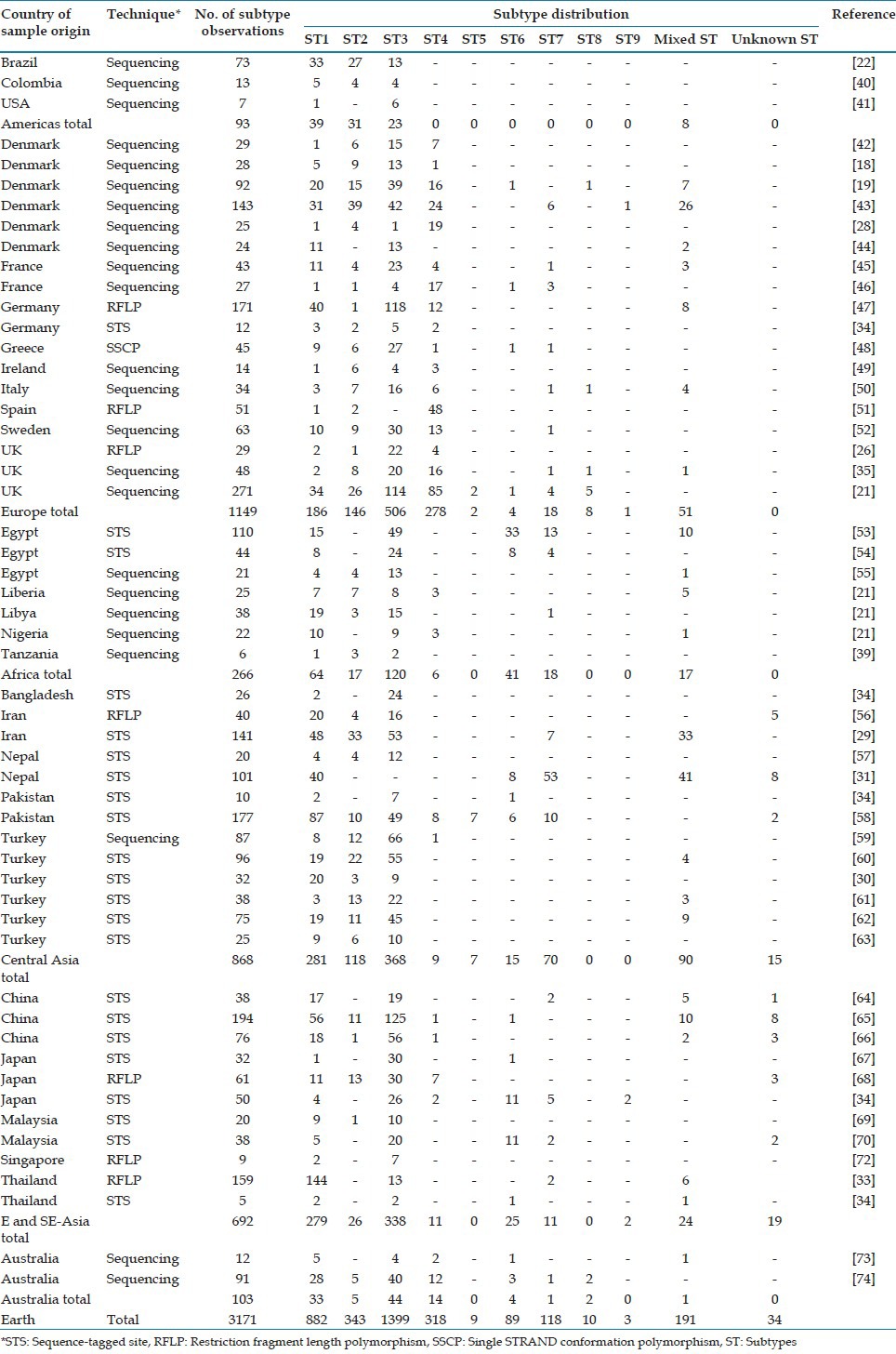
Although numerous authors have avidly pointed toward a large overlap in STs shared by humans and animals,[7,33,75,76,77,78,79] the hypothesis of zoonotic transmission of especially Blastocystis ST1 and ST3 has to a large extent been unsubstantiated by recent studies looking at intra-ST genetic diversity, such as MLST[38] and 18S allele analysis.[20,38,80] However, it is still highly likely that some STs are passed on zoonotically; for instance there are very few reports of human ST8 infections and only two 18S alleles have been identified so far. ST8 is probably mainly a ST of non-human primates (NHP), and human carriage may very well be a result of exposure to NHP manure.[35,38,80,81]
Intra-ST diversity appears to vary dramatically from ST to ST. ST4 SSU rDNA sequences from humans are strikingly homogenous compared to those stemming from ST3.[38] A total of 132 ST3 and ST4 samples from human and non-human primates were studied by multilocus sequence typing, and while 58 sequence types were found among the 81 ST3 samples, only five sequence types were found among 50 ST4s. Intra-ST diversity of ST1 and ST2 are currently being undertaken by MLST analysis.
Molecular screening tools for human Blastocystis carriage
The extensive genetic diversity of Blastocystis was clearly and independently substantiated by Clark and Böhm-Gloning et al., in 1997.[26,47] Pairwise genetic distances among some of the STs amount to at least 14.8%,[5] and this situation has made the design of genus-specific primers applicable to fecal DNA template amplification much more challenging than for most other parasites encountered in the clinical microbiology laboratory.
The first diagnostic PCR for detection of Blastocystis was introduced in 2006,[42] i.e. before the introduction of the consensus terminology. This conventional PCR was based on sequences present in GenBank at that time and targets ~ 300 bp toward the 3′-end of the SSU rRNA gene, but later observations suggest that it may exhibit preferential amplification of ST3 over others due to ST-specific polymorphisms in both primer annealing regions (unpublished observations).
Only two “pan-Blastocystis” real-time PCR assays have been published so far: A SYBR-green-based real-time PCR assay allowing subtyping by direct sequencing of SSU rRNA gene PCR products was introduced in 2011.[46] Despite the fact that the assay was designed to be applicable to fecal DNA, no inhibition control was incorporated. Moreover, in silico amplification shows that PCR products as large as 345 may be amplified from Blastocystis-specific template in human fecal samples (unpublished observations); this is a relatively large product for a real-time PCR assay, potentially impairing test sensitivity. The assay had a specificity of only 95%, which despite the high prevalence of Blastocystis gives a positive predictive value only around 91%, given a prevalence of 35%.
It may initially seem appropriate to evaluate the PCR assay specificity against other intestinal parasites such as Entamoeba, Giardia, Cryptosporidium, and helminths;[46] however, Blastocystis is genetically not even remotely related to any of these organisms. Neither are fungi, but specificity testing of diagnostic Blastocystis PCR assays ought include species of fungal genera such as Candida, Geotrichum and Saccharomyces,[82] all of which are common components of human fecal samples, and potentially quite abundant in samples that have been on their way to the laboratory for a couple of days at ambient temperature.
The other real-time PCR method was published by Stensvold et al.,[82] and was based on TaqMan technology including a specific probe and an internal process control to identify cases of PCR inhibition. The assay was evaluated against a large panel of fungal and bacterial DNAs and proved 100% specific. ST data were generated for positive samples from the test panel, and interestingly, samples positive for ST3 had lower cycle threshold (Ct) values than samples positive for ST1, indicating that ST1 colonization is generally lighter than ST3 colonization (P = 0.022). So far, this method has not been used for screening/surveys, but among the potentially interesting results using this method are whether Ct value ranges are associated with ST and whether the subjective experience of symptoms is related to infection intensity as measured by Ct-values.
It was previously shown that conventional PCR was not significantly more sensitive than short-term xenic in-vitro culture (XIVC);[18] however, XIVC had a sensitivity of only 79% and 52%, respectively, compared to the real-time PCRs developed.[46,82] An estimate of the number of copies of rDNA in one Blastocystis cell is not available, although, it may lie somewhere between 10 and 100 based on currently available data.[82,83]
It is already clear that the prevalence of Blastocystis even in Western countries is much higher than expected only few years ago, when prevalence figures were generated using traditional parasitological methods. Accurate identification of carriers and non-carriers by real-time PCR is key to understanding fundamental questions about transmission, treatment efficacy, longevity of colonization, and thereby overall clinical significance.
Mixed subtypes infection
It was recently argued that sequencing-based subtyping methods likely underestimate mixed ST infections.[84] However, a review of the data collected from various surveys across the globe (especially those using the STS primers that enable direct detection of mixed ST infections) strongly indicates that the level of mixed ST infections is rather low compared to the prevalence of the parasite, probably <10% of all cases. Roughly, the extent of mixed ST infection detected by the STS method corresponds to that seen by PCR and sequencing (e.g., barcoding).[34]
The impact of sampling and sample processing on Blastocystis detection and subtyping
Methodological issues other than those pertaining to the choice of subtyping method may influence our ability to accurately identify the distribution of Blastocystis STs in any given population. For instance, sometimes Blastocystis is subtyped directly from fecal DNA template, at other times from DNA template prepared from cultured isolates; there is evidence that XIVC may favor one ST over another in cases of mixed ST colonization,[75] and it is also possible that different strains (e.g. some animal vs. human strains) may have different requirements in terms of growth conditions, including temperature and nutrients; however, in some labs, short-term XVIC appears not to lead to differential growth of STs.[20]
Storage conditions before sample processing may be another variable potentially influencing our ability to detect Blastocystis. Samples are often collected in locations far from the lab and may be kept under varying conditions such as on cold/freeze storage, at ambient temperature, or in preservatives such as lysis buffer or ethanol.[22,52]
DNA extraction can be performed in many ways and differences in quality may obviously impact our ability to amplify and sequence Blastocystis; here, the size of the PCR product is also important since short PCR products may be amplifiable even from low quality DNA template while longer reads (eg. 600 bp used for barcoding) may be more difficult to obtain (unpublished observations). Furthermore, the amount of competing DNA template in DNA extracted directly from feces may influence PCR results.
Although clearly advantageous, it may prove much more difficult to standardize sampling and sample processing than standardizing subtyping methods. Suffice to say that direct screening of fecal DNA templates by PCR (preferably real-time PCR or a conventional PCR amplifying a “short” PCR product) with subsequent barcoding of positive samples may be considered “state-of-the-art” due to high diagnostic sensitivity and the added benefit of automatically assigning sequences to a ST number and even 18S allele when using www.pubmlst.org/Blastocystis, and this should be feasible in most laboratories.
New technologies and perspectives
There is no doubt that the future use of relevant technologies will facilitate and improve epidemiological surveys plus molecular analyses and detection of Blastocystis. So far, TaqMan array technology has been used for instance to generate a “one-size-fits-all” array for the detection of viral, bacterial, and parasitic enteropathogens.[85] It is not unlikely that this technology could prove useful in terms of facilitating the simultaneous detection and differentiation of Blastocystis STs. Another alternative that could potentially be looked into is the design and applicability of a real-time PCR assay using molecular beacons or scorpion probes for ST/allele analysis by single nucleotide polymorphism (SNP) detection; use of molecular beacons has proven useful for instance in screening for hotspot resistance mutations in Aspergillus fumigatus.[86] ST-specific SNP analysis by pyro sequencing has been developed, but never applied to large-scale studies.[87]
In this era of “omics,” it is possible that Blastocystis data can be extracted from pools of data already available and obtained for instance by metagenomic approaches. For instance, we are currently identifying and analyzing Blastocystis-specific mitochondrion-like organelle sequences (ST1-ST4) retrieved from metagenomic species originally generated in a study of the intestinal microbiomes of 124 Danish and Spanish individuals conducted by the Meta HIT Consortium.[88] After comparing the respective prevalence figures of STs identified by this method with data obtained, e.g. by barcoding in Denmark and Spain, this approach appears valid and useful for extracting ST data on Blastocystis. Moreover, it enables a characterization of the bacterial flora that accompanies Blastocystis colonization (unpublished observations).
While efforts to generate data on the extent (prevalence) and nature (ST) of Blastocystis carriage in various cohorts (mainly patients with intestinal disease versus healthy individuals) go on, it is also important to identify the methods relevant for investigating the function of Blastocystis genes.[89] The genome of Blastocystis ST7 was obtained by traditional Sanger sequencing and published in 2011,[83] and more genomes from other, more common, STs may be published as early as 2013-2014, thanks to faster and less expensive new generation sequencing methods. Comparative genomic and transcriptomic analysis should assist us in identifying effector proteins of potential interest that can be detected, monitored and correlated with health and disease phenotypes. Finally, analyzing Blastocystis and other common micro-eukaryotes in an ecological context is highly warranted for a complete understanding of their function, life cycle, and clinical significance.[89,90]
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Stensvold CR, Lebbad M, Verweij JJ. The impact of genetic diversity in protozoa on molecular diagnostics. Trends Parasitol. 2011;27:53–8. doi: 10.1016/j.pt.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Stensvold CR, Nielsen HV. Comparison of microscopy and PCR for detection of intestinal parasites in Danish patients supports an incentive for molecular screening platforms. J Clin Microbiol. 2012;50:540–1. doi: 10.1128/JCM.06012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Lieshout L, Verweij JJ. Newer diagnostic approaches to intestinal protozoa. Curr Opin Infect Dis. 2010;23:488–93. doi: 10.1097/QCO.0b013e32833de0eb. [DOI] [PubMed] [Google Scholar]
- 4.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, et al. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol. 2009;39:473–9. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Stensvold CR, Lebbad M, Victory EL, Verweij JJ, Tannich E, Alfellani M, et al. Increased sampling reveals novel lineages of Entamoeba: Consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist. 2011;162:525–41. doi: 10.1016/j.protis.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, et al. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol. 2010;169:8–17. doi: 10.1016/j.vetpar.2009.12.032. [DOI] [PubMed] [Google Scholar]
- 8.Fayer R, Santin M, Macarisin D. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol Res. 2012;111:1349–55. doi: 10.1007/s00436-012-2971-1. [DOI] [PubMed] [Google Scholar]
- 9.Royer TL, Gilchrist C, Kabir M, Arju T, Ralston KS, Haque R, et al. Entamoeba bangladeshi nov. sp., Bangladesh. Emerg Infect Dis. 2012;18:1543–5. doi: 10.3201/eid1809.120122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stensvold CR, Lebbad M, Clark CG. Last of the human protists: The phylogeny and genetic diversity of Iodamoeba. Mol Biol Evol. 2012;29:39–42. doi: 10.1093/molbev/msr238. [DOI] [PubMed] [Google Scholar]
- 11.Bruijnesteijn van Coppenraet LE, Wallinga JA, Ruijs GJ, Bruins MJ, Verweij JJ. Parasitological diagnosis combining an internally controlled real-time PCR assay for the detection of four protozoa in stool samples with a testing algorithm for microscopy. Clin Microbiol Infect. 2009;15:869–74. doi: 10.1111/j.1469-0691.2009.02894.x. [DOI] [PubMed] [Google Scholar]
- 12.Verweij JJ, Blangé RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, et al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–3. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engsbro AL, Stensvold CR. Blastocystis: To treat or not to treat… but how? Clin Infect Dis. 2012;55:1431–2. doi: 10.1093/cid/cis699. [DOI] [PubMed] [Google Scholar]
- 14.Stensvold CR, Smith HV, Nagel R, Olsen KE, Traub RJ. Eradication of Blastocystis carriage with antimicrobials: Reality or delusion? J Clin Gastroenterol. 2010;44:85–90. doi: 10.1097/MCG.0b013e3181bb86ba. [DOI] [PubMed] [Google Scholar]
- 15.Vogelberg C, Stensvold CR, Monecke S, Ditzen A, Stopsack K, Heinrich-Gräfe U, et al. Blastocystis sp. subtype 2 detection during recurrence of gastrointestinal and urticarial symptoms. Parasitol Int. 2010;59:469–71. doi: 10.1016/j.parint.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. Treatment of Dientamoeba fragilis in patients with irritable bowel syndrome. Am J Trop Med Hyg. 2012;87:1046–52. doi: 10.4269/ajtmh.2012.11-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stensvold CR, Arendrup MC, Mølbak K, Nielsen HV. The prevalence of Dientamoeba fragilis in patients with suspected enteroparasitic disease in a metropolitan area in Denmark. Clin Microbiol Infect. 2007;13:839–42. doi: 10.1111/j.1469-0691.2007.01760.x. [DOI] [PubMed] [Google Scholar]
- 18.Stensvold CR, Arendrup MC, Jespersgaard C, Mølbak K, Nielsen HV. Detecting Blastocystis using parasitologic and DNA-based methods: A comparative study. Diagn Microbiol Infect Dis. 2007;59:303–7. doi: 10.1016/j.diagmicrobio.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Rene BA, Stensvold CR, Badsberg JH, Nielsen HV. Subtype analysis of Blastocystis isolates from Blastocystis cyst excreting patients. Am J Trop Med Hyg. 2009;80:588–92. [PubMed] [Google Scholar]
- 20.Alfellani MA, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR, et al. Genetic diversity of Blastocystis in livestock and zoo animals. Protist. doi: 10.1016/j.protis.2013.05.003. In Press. [DOI] [PubMed] [Google Scholar]
- 21.Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ES, Fagbenro-Beyioku AF, Clark CG. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–8. doi: 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Malheiros AF, Stensvold CR, Clark CG, Braga GB, Shaw JJ. Short report: Molecular characterization of Blastocystis obtained from members of the indigenous Tapirapé ethnic group from the Brazilian Amazon region, Brazil. Am J Trop Med Hyg. 2011;85:1050–3. doi: 10.4269/ajtmh.2011.11-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stensvold CR, Suresh GK, Tan KS, Thompson RC, Traub RJ, Viscogliosi E, et al. Terminology for Blastocystis subtypes: A consensus. Trends Parasitol. 2007;23:93–6. doi: 10.1016/j.pt.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Clark CG, van der Giezen M, Alfellani M, Stensvold CR. Recent developments in Blastocystis research. Adv Parasitol Adv Parasitol. 2013;82:1–32. doi: 10.1016/B978-0-12-407706-5.00001-0. [DOI] [PubMed] [Google Scholar]
- 25.Stensvold CR. Comparison of sequencing (barcode region) and sequence-tagged-site PCR for Blastocystis subtyping. J Clin Microbiol. 2013;51:190–4. doi: 10.1128/JCM.02541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark CG. Extensive genetic diversity in Blastocystis hominis. Mol Biochem Parasitol. 1997;87:79–83. doi: 10.1016/s0166-6851(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 27.Stensvold CR, Nielsen HV, Mølbak K, Smith HV. Pursuing the clinical significance of Blastocystis: Diagnostic limitations. Trends Parasitol. 2009;25:23–9. doi: 10.1016/j.pt.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Stensvold CR, Christiansen DB, Olsen KE, Nielsen HV. Blastocystis sp. subtype 4 is common in Danish Blastocystis-positive patients presenting with acute diarrhea. Am J Trop Med Hyg. 2011;84:883–5. doi: 10.4269/ajtmh.2011.11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moosavi A, Haghighi A, Mojarad EN, Zayeri F, Alebouyeh M, Khazan H, et al. Genetic variability of Blastocystis sp. isolated from symptomatic and asymptomatic individuals in Iran. Parasitol Res. 2012;111:2311–5. doi: 10.1007/s00436-012-3085-5. [DOI] [PubMed] [Google Scholar]
- 30.Eroglu F, Genc A, Elgun G, Koltas IS. Identification of Blastocystis hominis isolates from asymptomatic and symptomatic patients by PCR. Parasitol Res. 2009;105:1589–92. doi: 10.1007/s00436-009-1595-6. [DOI] [PubMed] [Google Scholar]
- 31.Lee IL, Tan TC, Tan PC, Nanthiney DR, Biraj MK, Surendra KM, et al. Predominance of Blastocystis sp. subtype 4 in rural communities, Nepal. Parasitol Res. 2012;110:1553–62. doi: 10.1007/s00436-011-2665-0. [DOI] [PubMed] [Google Scholar]
- 32.Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S, Islam M, et al. Blastocystis hominis and Dientamoeba fragilis in patients fulfilling irritable bowel syndrome criteria. Parasitol Res. 2010;107:679–84. doi: 10.1007/s00436-010-1918-7. [DOI] [PubMed] [Google Scholar]
- 33.Thathaisong U, Worapong J, Mungthin M, Tan-Ariya P, Viputtigul K, Sudatis A, et al. Blastocystis isolates from a pig and a horse are closely related to Blastocystis hominis. J Clin Microbiol. 2003;41:967–75. doi: 10.1128/JCM.41.3.967-975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa H, Wu Z, Kimata I, Iseki M, Ali IK, Hossain MB, et al. Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol Res. 2004;92:22–9. doi: 10.1007/s00436-003-0995-2. [DOI] [PubMed] [Google Scholar]
- 35.Scicluna SM, Tawari B, Clark CG. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Clark CG, Diamond LS. Intraspecific variation and phylogenetic relationships in the genus Entamoeba as revealed by riboprinting. J Eukaryot Microbiol. 1997;44:142–54. doi: 10.1111/j.1550-7408.1997.tb05951.x. [DOI] [PubMed] [Google Scholar]
- 37.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12:263–73. doi: 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Petrášová J, Uzlíková M, Kostka M, Petrželková KJ, Huffman MA, Modrý D. Diversity and host specificity of Blastocystis in syntopic primates on Rubondo Island, Tanzania. Int J Parasitol. 2011;41:1113–20. doi: 10.1016/j.ijpara.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Santín M, Gómez-Muñoz MT, Solano-Aguilar G, Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol Res. 2011;109:205–12. doi: 10.1007/s00436-010-2244-9. [DOI] [PubMed] [Google Scholar]
- 41.Jones MS, Whipps CM, Ganac RD, Hudson NR, Boorom K. Association of Blastocystis subtype 3 and 1 with patients from an Oregon community presenting with chronic gastrointestinal illness. Parasitol Res. 2009;104:341–5. doi: 10.1007/s00436-008-1198-7. [DOI] [PubMed] [Google Scholar]
- 42.Stensvold R, Brillowska-Dabrowska A, Nielsen HV, Arendrup MC. Detection of Blastocystis hominis in unpreserved stool specimens by using polymerase chain reaction. J Parasitol. 2006;92:1081–7. doi: 10.1645/GE-840R.1. [DOI] [PubMed] [Google Scholar]
- 43.Stensvold CR, Lewis HC, Hammerum AM, Porsbo LJ, Nielsen SS, Olsen KE, et al. Blastocystis: Unravelling potential risk factors and clinical significance of a common but neglected parasite. Epidemiol Infect. 2009;137:1655–63. doi: 10.1017/S0950268809002672. [DOI] [PubMed] [Google Scholar]
- 44.Stensvold CR, Nielsen SD, Badsberg JH, Engberg J, Friis-Møller N, Nielsen SS, et al. The prevalence and clinical significance of intestinal parasites in HIV-infected patients in Denmark. Scand J Infect Dis. 2011;43:129–35. doi: 10.3109/00365548.2010.524659. [DOI] [PubMed] [Google Scholar]
- 45.Souppart L, Sanciu G, Cian A, Wawrzyniak I, Delbac F, Capron M, et al. Molecular epidemiology of human Blastocystis isolates in France. Parasitol Res. 2009;105:413–21. doi: 10.1007/s00436-009-1398-9. [DOI] [PubMed] [Google Scholar]
- 46.Poirier P, Wawrzyniak I, Albert A, El Alaoui H, Delbac F, Livrelli V. Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: Prospective study of patients with hematological malignancies. J Clin Microbiol. 2011;49:975–83. doi: 10.1128/JCM.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Böhm-Gloning B, Knobloch J, Walderich B. Five subgroups of Blastocystis hominis from symptomatic and asymptomatic patients revealed by restriction site analysis of PCR-amplified 16S-like rDNA. Trop Med Int Health. 1997;2:771–8. doi: 10.1046/j.1365-3156.1997.d01-383.x. [DOI] [PubMed] [Google Scholar]
- 48.Menounos PG, Spanakos G, Tegos N, Vassalos CM, Papadopoulou C, Vakalis NC. Direct detection of Blastocystis sp. in human faecal samples and subtype assignment using single strand conformational polymorphism and sequencing. Mol Cell Probes. 2008;22:24–9. doi: 10.1016/j.mcp.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: Qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183–93. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 50.Meloni D, Sanciu G, Poirier P, El Alaoui H, Chabé M, Delhaes L, et al. Molecular subtyping of Blastocystis sp. isolates from symptomatic patients in Italy. Parasitol Res. 2011;109:613–9. doi: 10.1007/s00436-011-2294-7. [DOI] [PubMed] [Google Scholar]
- 51.Domínguez-Márquez MV, Guna R, Muñoz C, Gómez-Muñoz MT, Borrás R. High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain) Parasitol Res. 2009;105:949–55. doi: 10.1007/s00436-009-1485-y. [DOI] [PubMed] [Google Scholar]
- 52.Forsell J, Granlund M, Stensvold CR, Clark CG, Evengård B. Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis. 2012;31:1689–96. doi: 10.1007/s10096-011-1416-6. [DOI] [PubMed] [Google Scholar]
- 53.Fouad SA, Basyoni MM, Fahmy RA, Kobaisi MH. The pathogenic role of different Blastocystis hominis genotypes isolated from patients with irritable bowel syndrome. Arab J Gastroenterol. 2011;12:194–200. doi: 10.1016/j.ajg.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Hussein EM, Hussein AM, Eida MM, Atwa MM. Pathophysiological variability of different genotypes of human Blastocystis hominis Egyptian isolates in experimentally infected rats. Parasitol Res. 2008;102:853–60. doi: 10.1007/s00436-007-0833-z. [DOI] [PubMed] [Google Scholar]
- 55.Souppart L, Moussa H, Cian A, Sanciu G, Poirier P, El Alaoui H, et al. Subtype analysis of Blastocystis isolates from symptomatic patients in Egypt. Parasitol Res. 2010;106:505–11. doi: 10.1007/s00436-009-1693-5. [DOI] [PubMed] [Google Scholar]
- 56.Motazedian H, Ghasemi H, Sadjjadi SM. Genomic diversity of Blastocystis hominis from patients in southern Iran. Ann Trop Med Parasitol. 2008;102:85–8. doi: 10.1179/136485908X252197. [DOI] [PubMed] [Google Scholar]
- 57.Yoshikawa H, Wu Z, Pandey K, Pandey BD, Sherchand JB, Yanagi T, et al. Molecular characterization of Blastocystis isolates from children and rhesus monkeys in Kathmandu, Nepal. Vet Parasitol. 2009;160:295–300. doi: 10.1016/j.vetpar.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 58.Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S, Islam M, et al. Irritable bowel syndrome: Is it associated with genotypes of Blastocystis hominis. Parasitol Res. 2010;106:1033–8. doi: 10.1007/s00436-010-1761-x. [DOI] [PubMed] [Google Scholar]
- 59.Ozyurt M, Kurt O, Mølbak K, Nielsen HV, Haznedaroglu T, Stensvold CR. Molecular epidemiology of Blastocystis infections in Turkey. Parasitol Int. 2008;57:300–6. doi: 10.1016/j.parint.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 60.Dogruman-Al F, Dagci H, Yoshikawa H, Kurt O, Demirel M. A possible link between subtype 2 and asymptomatic infections of Blastocystis hominis. Parasitol Res. 2008;103:685–9. doi: 10.1007/s00436-008-1031-3. [DOI] [PubMed] [Google Scholar]
- 61.Dogruman-Al F, Kustimur S, Yoshikawa H, Tuncer C, Simsek Z, Tanyuksel M, et al. Blastocystis subtypes in irritable bowel syndrome and inflammatory bowel disease in Ankara, Turkey. Mem Inst Oswaldo Cruz. 2009;104:724–7. doi: 10.1590/s0074-02762009000500011. [DOI] [PubMed] [Google Scholar]
- 62.Dogruman-Al F, Yoshikawa H, Kustimur S, Balaban N. PCR-based subtyping of Blastocystis isolates from symptomatic and asymptomatic individuals in a major hospital in Ankara, Turkey. Parasitol Res. 2009;106:263–8. doi: 10.1007/s00436-009-1658-8. [DOI] [PubMed] [Google Scholar]
- 63.Eroglu F, Koltas IS. Evaluation of the transmission mode of B. hominis by using PCR method. Parasitol Res. 2010;107:841–5. doi: 10.1007/s00436-010-1937-4. [DOI] [PubMed] [Google Scholar]
- 64.Yan Y, Su S, Lai R, Liao H, Ye J, Li X, et al. Genetic variability of Blastocystis hominis isolates in China. Parasitol Res. 2006;99:597–601. doi: 10.1007/s00436-006-0186-z. [DOI] [PubMed] [Google Scholar]
- 65.Li LH, Zhang XP, Lv S, Zhang L, Yoshikawa H, Wu Z, et al. Cross-sectional surveys and subtype classification of human Blastocystis isolates from four epidemiological settings in China. Parasitol Res. 2007;102:83–90. doi: 10.1007/s00436-007-0727-0. [DOI] [PubMed] [Google Scholar]
- 66.Li LH, Zhou XN, Du ZW, Wang XZ, Wang LB, Jiang JY, et al. Molecular epidemiology of human Blastocystis in a village in Yunnan province, China. Parasitol Int. 2007;56:281–6. doi: 10.1016/j.parint.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Yoshikawa H, Abe N, Iwasawa M, Kitano S, Nagano I, Wu Z, et al. Genomic analysis of Blastocystis hominis strains isolated from two long-term health care facilities. J Clin Microbiol. 2000;38:1324–30. doi: 10.1128/jcm.38.4.1324-1330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaneda Y, Horiki N, Cheng XJ, Fujita Y, Maruyama M, Tachibana H. Ribodemes of Blastocystis hominis isolated in Japan. Am J Trop Med Hyg. 2001;65:393–6. doi: 10.4269/ajtmh.2001.65.393. [DOI] [PubMed] [Google Scholar]
- 69.Tan TC, Suresh KG, Smith HV. Phenotypic and genotypic characterisation of Blastocystis hominis isolates implicates subtype 3 as a subtype with pathogenic potential. Parasitol Res. 2008;104:85–93. doi: 10.1007/s00436-008-1163-5. [DOI] [PubMed] [Google Scholar]
- 70.Tan TC, Ong SC, Suresh KG. Genetic variability of Blastocystis sp. isolates obtained from cancer and HIV/AIDS patients. Parasitol Res. 2009;105:1283–6. doi: 10.1007/s00436-009-1551-5. [DOI] [PubMed] [Google Scholar]
- 71.Rivera WL, Tan MA. Molecular characterization of Blastocystis isolates in the Philippines by riboprinting. Parasitol Res. 2005;96:253–7. doi: 10.1007/s00436-005-1344-4. [DOI] [PubMed] [Google Scholar]
- 72.Wong KH, Ng GC, Lin RT, Yoshikawa H, Taylor MB, Tan KS. Predominance of subtype 3 among Blastocystis isolates from a major hospital in Singapore. Parasitol Res. 2008;102:663–70. doi: 10.1007/s00436-007-0808-0. [DOI] [PubMed] [Google Scholar]
- 73.Nagel R, Cuttell L, Stensvold CR, Mills PC, Bielefeldt-Ohmann H, Traub RJ. Blastocystis subtypes in symptomatic and asymptomatic family members and pets and response to therapy. Intern Med J. 2012;42:1187–95. doi: 10.1111/j.1445-5994.2011.02626.x. [DOI] [PubMed] [Google Scholar]
- 74.Roberts T, Stark D, Harkness J, Ellis J. Subtype distribution of Blastocystis isolates identified in a Sydney population and pathogenic potential of Blastocystis. Eur J Clin Microbiol Infect Dis. 2013;32:335–43. doi: 10.1007/s10096-012-1746-z. [DOI] [PubMed] [Google Scholar]
- 75.Parkar U, Traub RJ, Kumar S, Mungthin M, Vitali S, Leelayoova S, et al. Direct characterization of Blastocystis from faeces by PCR and evidence of zoonotic potential. Parasitology. 2007;134:359–67. doi: 10.1017/S0031182006001582. [DOI] [PubMed] [Google Scholar]
- 76.Noël C, Dufernez F, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Ho LC, et al. Molecular phylogenies of Blastocystis isolates from different hosts: Implications for genetic diversity, identification of species, and zoonosis. J Clin Microbiol. 2005;43:348–55. doi: 10.1128/JCM.43.1.348-355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abe N. Molecular and phylogenetic analysis of Blastocystis isolates from various hosts. Vet Parasitol. 2004;120:235–42. doi: 10.1016/j.vetpar.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Abe N, Wu Z, Yoshikawa H. Molecular characterization of Blastocystis isolates from primates. Vet Parasitol. 2003;113:321–5. doi: 10.1016/s0304-4017(03)00081-5. [DOI] [PubMed] [Google Scholar]
- 79.Abe N, Nagoshi M, Takami K, Sawano Y, Yoshikawa H. A survey of Blastocystis sp. in livestock, pets, and zoo animals in Japan. Vet Parasitol. 2002;106:203–12. doi: 10.1016/s0304-4017(02)00050-x. [DOI] [PubMed] [Google Scholar]
- 80.Alfellani MA, Jacob AS, Perea NO, Krecek RC, Taner-Mulla D, Verweij JJ, et al. Blastocystis subtypes in non-human primates. Parasitology. 2013 doi: 10.1017/S0031182013000255. In Press. [DOI] [PubMed] [Google Scholar]
- 81.Stensvold CR, Arendrup MC, Nielsen HV, Bada A, Thorsen S. Symptomatic infection with Blastocystis sp. subtype 8 successfully treated with trimethoprim-sulfamethoxazole. Ann Trop Med Parasitol. 2008;102:271–4. doi: 10.1179/136485908X278847. [DOI] [PubMed] [Google Scholar]
- 82.Stensvold CR, Ahmed UN, Andersen LO, Nielsen HV. Development and evaluation of a genus-specific, probe-based, internal-process-controlled real-time PCR assay for sensitive and specific detection of Blastocystis spp. J Clin Microbiol. 2012;50:1847–51. doi: 10.1128/JCM.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Denoeud F, Roussel M, Noel B, Wawrzyniak I, Da Silva C, Diogon M, et al. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 2011;12:R29. doi: 10.1186/gb-2011-12-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meloni D, Poirier P, Mantini C, Noël C, Gantois N, Wawrzyniak I, et al. Mixed human intra- and inter-subtype infections with the parasite Blastocystis sp. Parasitol Int. 2012;61:719–22. doi: 10.1016/j.parint.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 85.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, et al. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51:472–80. doi: 10.1128/JCM.02658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Balashov SV, Gardiner R, Park S, Perlin DS. Rapid, high- throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J Clin Microbiol. 2005;43:214–22. doi: 10.1128/JCM.43.1.214-222.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stensvold CR, Traub RJ, von Samson-Himmelstjerna G, Jespersgaard C, Nielsen HV, Thompson RC. Blastocystis: Subtyping isolates using pyrosequencing technology. Exp Parasitol. 2007;116:111–9. doi: 10.1016/j.exppara.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 88.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stensvold CR. Thinking Blastocystis out of the box. Trends Parasitol. 2012;28:305. doi: 10.1016/j.pt.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 90.Andersen LO, Vedel Nielsen H, Stensvold CR. Waiting for the human intestinal Eukaryotome. ISME J. 2013 doi: 10.1038/ismej.2013.21. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]


