Abstract
Proper maintenance and duplication of the genome require accurate recombination between homologous DNA molecules. In eukaryotic cells, the Rad51 protein mediates pairing between homologous DNA molecules. This reaction is assisted by the Rad54 protein. To gain insight into how Rad54 functions, we studied the interaction of the human Rad54 (hRad54) protein with double-stranded DNA. We have recently shown that binding of hRad54 to DNA induces a change in DNA topology. To determine whether this change was caused by a protein-constrained change in twist, a protein-constrained change in writhe, or the introduction of unconstrained plectonemic supercoils, we investigated the hRad54–DNA complex by scanning force microscopy. The architecture of the observed complexes suggests that movement of the hRad54 protein complex along the DNA helix generates unconstrained plectonemic supercoils. We discuss how hRad54-induced superhelical stress in the target DNA may function to facilitate homologous DNA pairing by the hRad51 protein directly. In addition, the induction of supercoiling by hRad54 could stimulate recombination indirectly by displacing histones and/or other proteins packaging the DNA into chromatin. This function of DNA translocating motors might be of general importance in chromatin metabolism.
Recombination between homologous DNA molecules is important for maintenance and faithful duplication of the genome (1–4). Homologous recombination is a major pathway for the accurate repair of DNA double-strand breaks (DSBs) that arise from exposure to exogenous DNA-damaging agents such as ionizing radiation, which is commonly used in antitumor therapies. Furthermore, homologous recombination processes programmed DSB intermediates during meiosis. Finally, homologous recombination plays a major role in reestablishing DNA replication forks that are stalled or have collapsed because of the presence of spontaneous or induced DNA damage in one of the template strands of the DNA double helix (5–12).
Extensive genetic and biochemical experiments have revealed that DSB repair mediated by homologous recombination in yeast, chicken, and mammalian cells occurs through the close cooperation of the RAD52 group of proteins, including Rad51, Rad52, and Rad54 (13, 14). A key member of this group is the Rad51 protein. Rad51 protomers assemble a nucleoprotein filament on the single-stranded DNA tails that form at the break site. This filament pairs with homologous double-stranded DNA, resulting in a joint molecule. Joint molecules are pivotal intermediates in recombination because they allow the broken DNA to use the intact homologous double-stranded DNA as a repair template (15). The Rad52 and Rad54 proteins serve as accessory factors in Rad51-mediated joint molecule formation. The details of the molecular mechanisms through which Rad52 and Rad54 stimulate joint molecule formation are not well understood. Rad52 has been shown to increase the rate of annealing of complementary single-stranded DNA molecules, to bind to DNA ends, to stimulate homologous paring by Rad51, and to overcome the inhibitory effect of the single-stranded DNA-binding protein RPA on Rad51 nucleoprotein filament formation (16–22). Rad54 can interact with Rad51 (23–26) and has ATPase activity (27, 28). Importantly, the ATPase activity of Rad54 specifically requires the presence of double-stranded DNA. It is not active in the presence of single-stranded DNA (27, 28). This cofactor specificity is opposite to that of Rad51 (29). Because the initial substrate of Rad51 during homologous recombination is single-stranded DNA (13), it is likely that the substrate for Rad54 is the double-stranded homologous repair template.
To understand how the Rad54 protein assists Rad51 during joint molecule formation we have investigated the interaction of the human Rad54 (hRad54) protein with double-stranded DNA. We have recently demonstrated that binding of hRad54 to double-stranded DNA induces a change in the topology of the DNA (26), as is also observed for yeast Rad54 homologues (30–33). In the topological experiments, singly nicked plasmid DNA is incubated with hRad54 protein. Subsequently, the nick is closed by the addition of DNA ligase. The ligation will fix any change in linking number (ΔLk) in the DNA that is induced by protein binding. Lk describes the number of times that the two strands of the DNA double helix wind around each other. Lk is a topological parameter of double-stranded DNA that is made up of two geometrical parameters, twist (Tw) and writhe (Wr) (34). Tw and Wr give information about the shape of the DNA. The local winding of the two strands of the double helix is described by Tw, whereas Wr describes the number of times that the axis of the double helix winds around itself. The relationship between these three parameters is expressed by the equation Lk = Tw + Wr (35). Therefore, to mechanistically interpret the ΔLk induced by hRad54 binding it is necessary to determine whether protein binding changes Tw or Wr.
One well-characterized class of proteins that has the ability to change the Lk of DNA is topoisomerases (36–38). Topoisomerases induce a ΔLk by a strand passage mechanism because these enzymes can break and rejoin DNA strands. In contrast, no strand breakage and rejoining activity has been detected for hRad54 (26). Therefore, the ΔLk measured by the assay described above could be caused by the hRad54 protein constraining either Tw or Wr. Protein-constrained ΔTw and ΔWr result from very different DNA-binding modes of the protein and therefore imply a different function for hRad54 in homologous recombination. For example, proteins induce a ΔWr by wrapping the DNA around their surface. A classical example of such a binding mode is provided by the nucleosome (39). This mode of DNA binding is not restricted to general architectural proteins, because it has also been found to occur with a protein involved in modulating DNA supercoiling (40) and DNA damage recognition (41). Proteins can induce a ΔTw by stretching the helix in a protein-stabilized filament. An example of this binding mode is provided by the Escherichia coli RecA protein, the central homologous pairing and strand exchange protein in homologous recombination (42–44). The two different binding modes are not easily distinguished by biochemical assays. However, they will result in architecturally different DNA–protein complexes, and therefore the binding modes can be distinguished if the complexes are imaged directly. A protein-constrained ΔLk caused by decreasing Tw will be evident by an extensive protein filament formed on the DNA and an increase in the contour length of the resulting complex relative to unbound DNA. A ΔLk due to wrapping of DNA around proteins will not result in an extensive region of DNA covered by protein but will cause a decrease in contour length of the DNA–protein complexes relative to naked DNA. Alternatively, the ΔLk measured in the topological assay could be due to the introduction of unconstrained supercoils in the plasmid by a protein translocation mechanism (45). In our assay, this would require the formation of specific DNA–protein complexes that create topologically separate domains. Any relevant change in the structure of the DNA–protein complexes will have to occur in the presence of ATP because ATP hydrolysis by hRad54 is required for the ΔLk induction (26). With analysis of these parameters in mind, we chose to investigate the structure of DNA–protein complexes formed between hRad54 and circular DNA molecules by scanning force microscopy (SFM).
Materials and Methods
DNA Substrates.
Substrate DNA plasmids used in this study were pTrcHisB (Invitrogen) and pDERI1. Plasmid pDERI1 was generated by deletion of the SspI–SapI fragment from plasmid pUC19, resulting in a plasmid 1,821 bp in length. Singly nicked plasmid DNA was produced in a 30-μl reaction mixture containing 0.5 μg of DNA, 20 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 360 μg/ml ethidium bromide, and 1 μg/ml DNase I at 30°C for 30 min. The reaction was stopped by the addition of 0.1 vol of 5% (wt/vol) SDS/50 mM EDTA/30 μg/ml proteinase K and subsequent incubation at 65°C for 30 min. DNA was purified by extraction with phenol and phenol/chloroform (1:1, vol/vol), precipitated with ethanol, and dissolved in 10 mM Tris⋅HCl (pH 8.0)/1 mM EDTA. Linear DNA substrates were made by digestion of plasmid pTrcHisB (Invitrogen) with EcoRV and NcoI, followed by isolation of the 732-bp and 3,672-bp DNA fragments from a 1.0% agarose gel.
Proteins and DNA-Binding Reactions.
The hRad54 protein was produced in baculovirus-infected Sf21 cells and purified as described (27). In addition, a mutant version of hRad54 was purified that contained a single amino acid substitution at position 189. The invariant lysine residue in the Walker A nucleotide-binding motif was changed to an alanine residue by using site-specific mutagenesis. This protein is referred to as hRad54K189A. hRad54K189A had no detectable DNA-dependent ATPase activity. Given the sensitivity of the ATPase assay, the ATPase activity of the mutant protein must be reduced by more than 50-fold compared with the wild-type protein (data not shown). The Ku70/80 heterodimer was produced and purified as described and was the generous gift of M. Modesti (46). E. coli RNA polymerase was purchased from Boehringer Mannheim.
Protein–DNA complexes were prepared by addition of the hRad54 and hRad54K189A protein preparations to DNA substrates. Reaction mixtures (10 μl final volume) were assembled by mixing hRad54 or hRad54K189A (up to final concentrations of 0.34 μM) and DNA (76 μM; concentration in nucleotides) in buffer containing 20 mM Hepes⋅KOH (pH 7.4), 20 mM KCl, 5 mM MgCl2, and 2 mM ATP or 2 mM adenosine 5′-[γ-thio]triphosphate (ATPγS). After incubation at 30°C for 10 min, glutaraldehyde was added to a final concentration of 0.1%, followed by additional incubation at 30°C for 10 min. Experiments done without glutaraldehyde fixation showed the same type of DNA protein complexes as observed with glutaraldehyde, but in general fewer complexes and more naked DNA were observed. The Ku70/80 heterodimer was incubated with the 732-bp linear DNA fragment in 10-μl reaction mixtures containing 75 ng of Ku 70/80 (50 nM), 270 ng of DNA substrate (81 μM), 50 mM Hepes⋅KOH (pH 8.0), 100 mM KCl, 10 mM MgCl2, and 1 mM DTT. Incubations were carried out at 37°C for 20 min. E. coli RNA polymerase was bound to the 3,672-bp DNA fragment in a 20-μl reaction mixture containing 250 ng of RNA polymerase (55 nM), 200 ng of DNA fragment (30 μM), 30 mM Hepes⋅KOH (pH 7.4), 100 mM NaCl, 10 mM MgCl2, and 0.5 mM DTT. Incubations were at 37°C for 15 min, followed by addition of glutaraldehyde to a final concentration of 0.1% and an additional incubation at 37°C for 15 min.
SFM.
Reaction mixtures were diluted 15- to 30-fold in deposition buffer, consisting of 5 mM Hepes⋅KOH (pH 7.5) and 5 mM MgCl2. Within 15 sec a 10- to 15-μl drop was placed onto freshly cleaved mica. After 30 sec the mica surface was washed with H2O (glass-distilled, Sigma), followed by drying with a stream of filtered air. For the simultaneous deposition of protein–DNA complexes from different binding reactions, the separate reaction mixtures were combined at the dilution step. The nucleoprotein complexes were imaged in air at room temperature and humidity by using a NanoScope IIIa (Digital Instruments, Santa Barbara, CA) operating in the tapping mode with a type E scanner. Silicon tips (Nanoprobes) were obtained from Digital Instruments. DNA length and the size of protein complexes on DNA were measured from NanoScope images imported into IMAGE SXM 1.62 (National Institutes of Health IMAGE version modified by Steve Barrett, Surface Science Research Centre, Univ. of Liverpool, Liverpool, U.K.). DNA contours were manually traced. In the case of DNA–protein complexes, contour length was traced as the shortest possible DNA path through the bound protein. The volume of DNA-bound protein complexes was determined as described (47). Briefly, the object was manually traced and its area and average height were measured, and a background volume of the same traced area at an adjacent position including DNA was subtracted. Volume measurements are given in arbitrary units because they are used only to compare the relative sizes of objects.
Results
Visualization of hRad54–DNA Complexes by SFM.
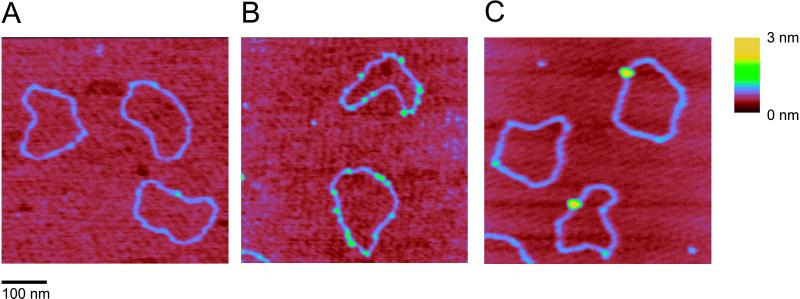
Complexes between DNA and hRad54 were formed by incubating the protein with a singly nicked, circular DNA substrate. Reaction mixtures were deposited on mica and imaged by SFM. In the absence of protein, the majority (80–85%) of DNA molecules looked like a relaxed circle with uniform height (Fig. 1A). The remaining circles had some DNA crossings or small regions where two double-stranded regions were next to each other, which is typical of DNA prepared in this manner for SFM (C.W., unpublished observation). In the absence of ATP, hRad54 bound to DNA as small complexes with several on each DNA circle (Fig. 1B). Addition of ATP to the binding reaction resulted in a dramatic change both in frequency and in size of hRad54–DNA complexes. The DNA-bound protein now formed much larger complexes with only one or sometimes two on each DNA circle (Fig. 1C). In neither case were protein-coated filaments of any kind observed. Thus, it appears likely that hRad54 does not alter DNA topology by inducing a ΔTw through constraining the DNA double helix in a protein filament.
Figure 1.
SFM images of nicked circular DNA molecules in the absence or presence of hRad54. The images were processed only by flattening to remove background slope and are presented as top views. All images show an area of 500 nm × 500 nm, zoomed in from 2-μm × 2-μm scans. The z dimension is indicated by color as shown on the bar at the right. (A) DNA molecules without hRad54. (B) hRad54–DNA complexes formed in the absence ATP. (C) hRad54–DNA complexes formed in the presence of ATP.
hRad54 Binding Does Not Induce DNA Wrapping.
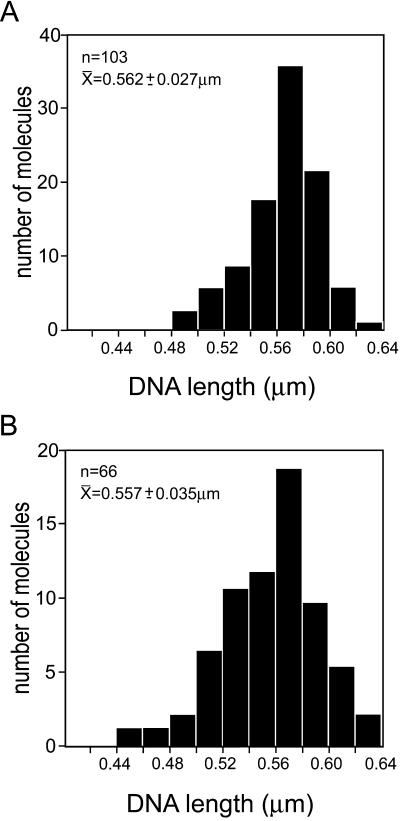
To determine whether hRad54 binding resulted in ΔWr by wrapping of DNA around the large protein complexes formed in the presence of ATP, we measured the contour length of the DNA. Histograms of contour length for DNA alone and DNA bound by large protein complexes are shown in Fig. 2 A and B, respectively. On the basis of the size of hRad54 and the stiffness of DNA, we would expect a minimum wrap of about 60 bp of DNA or loss of about 20 nm in length per complex to introduce each ΔLk of −1 or +1, depending on the handedness of the wrapping. Thus, according to this model significant length changes should occur to account for the large ΔLk seen in the topological assays in which topoisomers with a ΔLk of up to −23 have been resolved (26). However, the data presented in Fig. 2 show that there is no difference in either the mean DNA length or the distribution of DNA lengths in the presence or absence of bound large hRad54 complexes. Thus, it appears that hRad54 does not alter DNA topology by introducing ΔWr through DNA wrapping.
Figure 2.
Histograms of DNA contour length measured from molecules with or without bound hRad54. hRad54–DNA complexes were formed in the presence of ATP. All DNA length measurements were from images collected from one deposition of molecules from one reaction mixture. The top left of each panel shows the number of DNA molecules measured, their average contour length, and standard deviation. (A) Contour length of DNA molecules without bound protein. (B) Contour length of DNA molecules with bound hRad54.
Supercoiled Domains Anchored by the hRad54 Protein.
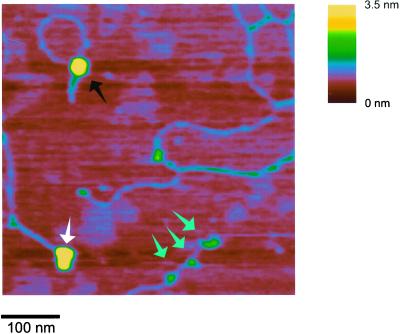
The SFM experiments did reveal some intriguing complexes formed between hRad54 and DNA in the presence of ATP. Occasionally we observed a large hRad54 complex anchoring the junction between relaxed and apparently plectonemically supercoiled domains of the plasmid (Fig. 3). To determine whether the structures with hRad54 anchoring a supercoiled domain were a relevant representation of hRad54 activity on DNA, we correlated the occurrence of these structures with the ability to hydrolyze ATP. In similar SFM experiments either ATP was replaced by the slowly hydrolyzable ATP analogue ATPγS or the wild-type hRad54 protein was replaced by hRad54K189A, a mutant defective in ATP hydrolysis (27) (data not shown). The number of DNA–protein complexes in which the protein anchors a supercoiled domain was counted and calculated as a percentage of the total number of protein-bound DNA molecules observed (Table 1). While the structures indicative of domain anchoring were 10.5% of the total protein-bound DNA for wild-type hRad54 in the presence of ATP, they were only 3% when ATP was replaced with ATPγS. It should be noted that wild-type hRad54 has some residual activity in topological assays in the presence of ATPγS (26). The difference was even more dramatic when hRad54K189A was used. Both the frequency of DNA-bound protein and the size of the complexes formed by this mutant were the same in the presence and absence of ATP. Most of the DNA-bound complexes appeared to be the same size as those formed by the wild-type protein in the presence of ATP. Although compared with wild-type protein significantly more of the total DNA was bound by protein (about 90% of DNA molecules compared with 15–20% for wild-type hRad54 in the presence of either ATP or ATPγS), the protein was never observed to anchor a supercoiled DNA domain. Thus, our results show a direct correlation between ATP hydrolysis, the ability to induce a ΔLk, and the appearance of protein complexes anchoring a plectonemically supercoiled domain.
Figure 3.
SFM image of hRad54–DNA complexes formed in the presence of ATP. The image is presented as line plot at a 60° viewing angle to emphasize topography. Height is indicated by color as shown on the bar at the right. One plasmid has a hRad54 complex bound at the junction of relaxed and supercoiled domains. The other plasmid has two hRad54 complexes bound.
Table 1.
Correlation of hRad54 ATP hydrolysis activity and observation of hRad54-anchored supercoiled domains
| Protein | Nucleotide | No. of protein–DNA complexes | No. of protein complexes anchoring DNA domains | Percentage of complexes anchoring DNA domains |
|---|---|---|---|---|
| hRad54 | ATP | 496 | 52 | 10.5 |
| hRad54 | ATPγS | 100 | 3 | 3 |
| hRad54K189A | ATP | 217 | 0 | <0.5 |
Multimeric State of hRad54 Bound to DNA.
We believe that the hRad54 complexes bound to DNA in the presence of ATP are the functional form(s) of this protein (see Discussion). These complexes were much larger than those bound to DNA in the absence of ATP. We wished to estimate the size of the presumptive functional form of hRad54 from our SFM images. Because of the well-known distortions in the absolute dimensions of biomolecules imaged by SFM (48, 49), it is necessary to measure the volume of the protein of interest, as well as proteins of known size from the same deposition with the same tip. The approximate size of the unknown complex is then determined by comparison to the standards (47). In this way, we can estimate the size of the protein complexes bound to DNA, even if there is more than one complex per DNA molecule, a measurement that would not be possible with biochemical methods. Three separate protein–DNA complexes, E. coli RNA polymerase (450 kDa) bound to a long linear DNA (3,672 bp), Ku70/80 heterodimer (155 kDa) bound to a short linear DNA (732 bp), and hRad54 (87.8-kDa monomers) bound to singly nicked circular DNA (1,821 bp) in the presence of ATP, were prepared and mixed together for a single deposition. Fig. 4 shows a field containing these three protein–DNA complexes; it is obvious that the hRad54 complex is much larger than the Ku heterodimer and nearly the same size as RNA polymerase. An average volume for the different DNA-bound proteins was determined from over 100 individual complexes of each kind. In one experiment the average volume of the hRad54 complexes was close to that of RNA polymerase, indicative of a molecular mass of about 450 kDa or slightly above five 87.8-kDa hRad54 monomers. In two other experiments the average volume of the hRad54 complexes was between that of Ku70/80 and RNA polymerase at a value corresponding to approximately three hRad54 monomers. There is large variation in these volume measurements, partially because of the inaccuracies in SFM dimensions of biomolecules. For the hRad54 complexes the variation between experiments is possibly caused by variation in the population of complexes with different sizes. We have no unbiased way of sorting the DNA-bound complexes that are not anchoring a supercoiled domain. The Rad54 complexes anchoring a supercoiled domain do appear larger than the isolated complexes (see Fig. 3). However, there were not enough of them in the experiments with size standards to determine a significant average volume. Although this is only an estimation of molecular mass, the active form of hRad54 bound to DNA is at least a trimer and possibly as large as a hexamer.
Figure 4.
Comparison of the size of large hRad54 complexes bound to DNA in the presence of ATP and proteins of known size. SFM image of a mixture of hRad54, RNA polymerase, and Ku70/80 bound to the different DNA substrates. Upper left, circular DNA–hRad54 complex (hRad54 monomer is 87.8 kDa); lower left, RNA polymerase (450 kDa) bound to a long linear DNA (partially shown); lower middle, three Ku70/80 heterodimers (155 kDa) bound to a short linear DNA.
Discussion
Rad54 belongs to a superfamily of proteins that includes known helicases but could more generally be described as DNA-translocating motors (50). The detailed mechanism(s) by which proteins use the energy of ATP hydrolysis to track along the DNA helix and accomplish work such as strand separation, supercoiling, and possibly chromatin remodeling is not well understood. Rad54 belongs to the Swi2/Snf2 subfamily on the basis of its amino acid sequence. Rad54 has double-stranded DNA-dependent ATPase activity but, like other members of this family, it has not been demonstrated to have any strand displacement activity (27, 28). We have recently demonstrated that hRad54 can use the energy of ATP hydrolysis to change the topology of DNA (26). As classical topoisomerase activity, involving breakage, passage, and rejoining of DNA strands, could not be demonstrated for hRad54, we hypothesized that the observed changes in DNA topology might result from hRad54 constrained changes in DNA Tw or Wr. We used SFM to distinguish between the protein–DNA structures that would have induced changes in DNA Tw and those that would have induced changes in DNA Wr. We observed neither protein filaments on DNA that would have constrained Tw nor protein-induced DNA length changes that would have accompanied changes in Tw or Wr. Instead, in conditions where DNA topology is altered, we observed single large hRad54 protein complexes anchoring supercoiled DNA domains. We believe these structures result from interaction between two DNA-bound hRad54 complexes and movement of one of them. We will now discuss how these structures may arise and, in light of other information on the function of Rad54, how this activity fits into a recombination reaction.
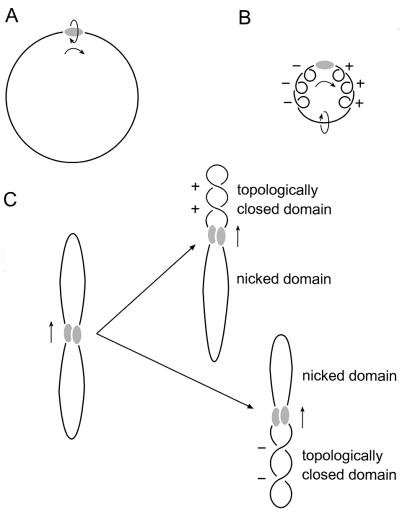
Proteins that move along DNA by tracking the helix can introduce supercoiling if certain conditions are met (45). There has to be significant effective frictional torque to prevent the protein from freely rotating around the DNA in order for movement alone to cause supercoiling, positive ahead of movement and negative behind (Fig. 5). It has recently been suggested that yeast Rad54 movement induces unconstrained supercoils in DNA (31, 32). However, the Rad54 protein alone tracking along the helix, even as part of the large complexes observed here in the presence of ATP, would not cause DNA supercoiling because the protein would be free to rotate around the DNA double helix axis as it moved. In contrast, if two DNA-bound hRad54 complexes interact and one of them moves along the helix, rotation of the protein relative to the DNA is prevented and supercoils will accumulate both ahead and behind the movement. The observation that the hRad54 complexes anchoring supercoiled DNA domains appear larger than those bound simply to other positions on the plasmid (Fig. 3) implies that proteins bound at two sites initially interact to form these structures. In our case such an association of DNA-bound hRad54 complexes would divide the plasmid into two domains. Because the DNA is singly nicked, one domain will contain a nick, whereas the other will contain two covalently closed DNA strands. When the protein complex translocates along the helix, the nicked domain will not accumulate any supercoiling because the strands are free to rotate around each other. On the other hand, the domain containing the covalently closed strands will become supercoiled (Fig. 5C). This is how we interpret the origin of the type of complex shown in Fig. 3. This mechanism predicts that both negative and positive supercoils would be produced, depending on the random occurrence of the nick in the domain either ahead or behind the moving proteins. Indeed the topological experiment, also performed on singly nicked plasmids, did show that hRad54 introduced both negative and positive supercoils into plasmids (26). Thus, if free rotation of DNA and protein is prevented, hRad54 can use the energy of ATP hydrolysis to supercoil DNA domains.
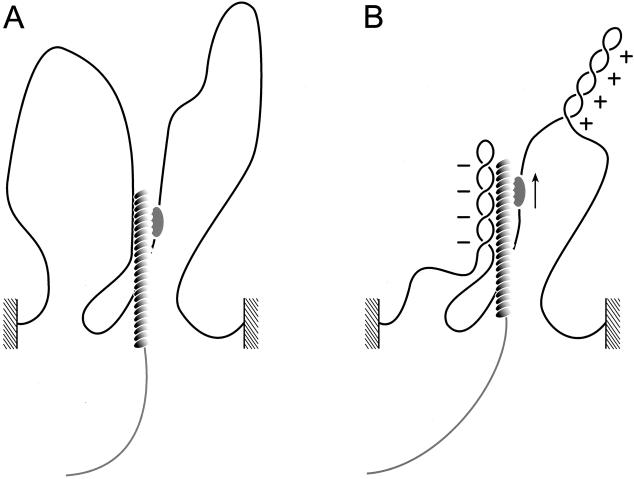
Figure 5.
Model for generation of supercoiling by hRad54 translocation along DNA. The hRad54 complex and plasmid DNA are indicated by the shaded oval and black line, respectively. (A) Movement of the hRad54 complex by tracking along the helical path of DNA is indicated by the arrows. When the complex is free to rotate around the DNA, no change in supercoiling will be induced in the plasmid DNA. (B) When the hRad54 complex tracks along the helix, while being prevented from rotating around the DNA, positive supercoils will arise ahead of the protein complex and negative supercoils behind it. These supercoils can freely distribute along the plasmid and therefore they will cancel each other out. (C) The interaction of two hRad54 complexes on a plasmid will divide the plasmid into two domains. Because the plasmid is singly nicked, one domain will contain a nick, whereas the other contains two covalently closed DNA strands. Depending on the position of the nick relative to the movement of the protein complex along the DNA, topoisomers containing either negative or positive supercoils will result after ligation of the nick.
In our experiments hRad54 is most likely prevented from rotating around the helix as it moves because of interaction with another hRad54 complex on the same plasmid. This is probably not how this protein works in vivo. On the small plasmid we used hRad54 complexes tethered by chance to the same DNA molecule are in very high local concentration, possibly allowing even nonspecific interactions. The activity of Rad54 relevant to homologous recombination is as an accessory factor, along with Rad52, in promoting Rad51-mediated joint molecule formation. Specific interaction between hRad54 and hRad51 has been demonstrated biochemically (23, 26). More interestingly, it has recently been demonstrated that the ATPase activity of yeast Rad54 and its activity in altering DNA topology are both stimulated by the Rad51–single-stranded DNA filament (31, 33). Physical association with and activation of Rad54 by the Rad51–single-stranded DNA filament would have two very favorable consequences for the mechanism of homologous recombination. The filament would favor interaction with homologous double-stranded DNA, thereby targeting Rad54 to the correct chromosomal location for activity. In addition, Rad54 attached to a Rad51 filament that is itself part of a broken chromosome would provide more than sufficient frictional torque to prevent rotation of Rad54 along the target DNA helix and thus create differentially supercoiled domains ahead and behind the moving Rad54 (Fig. 6). The negative supercoiling created behind the protein could promote joint molecule formation. This is because negative supercoiling will promote unpairing of the target DNA (31, 34) and thereby facilitate the hybridization of the incoming single-stranded DNA in the filament. Furthermore, one could also envision Rad54 at the end of a Rad51–single-stranded DNA filament effectively threading the filament into the target duplex by dragging it along as it tracks the helix.
Figure 6.
Model for stimulation of Rad51-mediated joint molecule formation by Rad54 translocation. A chromosomal domain is indicated by the black line connecting the hatched areas. The Rad54 protein complex is represented by the shaded oval, and it is shown to interact with the Rad51 nucleoprotein filament that is assembled on the broken DNA indicated in gray (A, before hRad54 translocation). This interaction will provide the frictional torque that prevents the Rad54 complex from rotating around the DNA as it tracks along the helix. In this way, movement of the Rad54 complex along the DNA (B, after hRad54 translocation) will generate negative and positive supercoils into the domains divided by the Rad54 complex. See text for details on how this process might stimulate Rad51-mediated joint molecule formation.
In addition, Rad54-induced superhelical stress in the target DNA may function to displace histones and/or other proteins packaging the DNA into chromatin and inhibiting recombination. This function of DNA-translocating motors may be of general importance in chromatin metabolism. The archetypal chromatin remodeling factor, yeast SWI/SNF, as well as others, has recently been shown to induce superhelical stress in DNA (51). This ability to change DNA topology has also been shown to be required for SWI/SNF chromatin remodeling activity (52). The ability of hRad54 to induce topological stress in the target chromosome may also disrupt chromatin structure, making it accessible for Rad51 nucleoprotein filament invasion.
It would be very interesting to know more about the structure of these protein complexes that use the energy of ATP hydrolysis to translocate along DNA. Structural details that would reveal how these protein machines grasp DNA strands and move are currently very limited. The available information indicates there is a subclass of helicases that are hexameric rings, some of which are known to encircle DNA (50). Our SFM images cannot resolve the path of the DNA as either going through a protein structure or simply covered by a protein on top of it. However, the images do show a clear difference in the size of hRad54 complexes bound to DNA in the presence and absence of ATP. Our estimate of the size of these large complexes is that they are at least trimers and may be as large as hexamers. Although hexamers might be expected, either trimers or hexamers could produce similar functional structures. For example, the DNA polymerase processivity factors from prokaryotes and eukaryotes form nearly identical six-lobed rings around DNA while one is a trimer of two-lobed subunits and the other dimer of three-lobed subunits (53). Clearly an accurate answer to the question of the multimeric state of active hRad54 requires additional experiments such as negative stain electron microscopy and image reconstruction. We have at least identified the type of protein complexes formed on DNA that should be the subjects for further analysis.
Using SFM, we were able to detect hRad54 in complex structures with DNA. The images simultaneously revealed that, in the presence of ATP, the protein complexes were large and could anchor a supercoiled DNA domain. These two pieces of information together suggested a mechanism for hRad54 DNA supercoiling that we had not initially considered. It now seems likely the role of hRad54 in recombination is to induce superhelical torsion in the target DNA. This superhelical torsion may result in melting of the target duplex (31), removal of nucleosomes from the target, or both of these. We anticipate that such direct imaging combined with biochemistry will continue to provide interesting insights into the mechanism of action of the hRad54 in the context of a hRad51–single-stranded DNA filament and interactions of this structure with target DNA and chromatin.
Acknowledgments
This work was supported by grants from the Dutch Cancer Society (NKB/KWF) and the Netherlands Organization for Scientific Research (NWO). We thank Mauro Modesti for purified Ku70/80.
Abbreviations
- ΔLk
change in linking number
- SFM
scanning force microscopy
- Tw
twist
- Wr
writhe
- ATPγS
adenosine 5′-[γ-thio]triphosphate
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Haber J E. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 2.Morrison C, Takeda S. Int J Biochem Cell Biol. 2000;32:817–831. doi: 10.1016/s1357-2725(00)00033-9. [DOI] [PubMed] [Google Scholar]
- 3.Thompson L H, Schild D. Biochimie. 1999;81:87–105. doi: 10.1016/s0300-9084(99)80042-x. [DOI] [PubMed] [Google Scholar]
- 4.Karran P. Curr Opin Genet Dev. 2000;10:144–150. doi: 10.1016/s0959-437x(00)00069-1. [DOI] [PubMed] [Google Scholar]
- 5.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuzminov A. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 7.Marians K J. Curr Opin Genet Dev. 2000;10:151–156. doi: 10.1016/s0959-437x(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 8.Kowalczykowski S C. Trends Biochem Sci. 2000;25:165–173. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 9.Rothstein R, Michel B, Gangloff S. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 10.Haber J E. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 11.Flores-Rozas H, Kolodner R D. Trends Biochem Sci. 2000;25:200–204. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 13.Baumann P, West S C. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 14.Kanaar R, Hoeijmakers J H, van Gent D C. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 15.Pâques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung P. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 17.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Nature (London) 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 18.Benson F E, Baumann P, West S C. Nature (London) 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 19.Van Dyck E, Stasiak A Z, Stasiak A, West S C. Nature (London) 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 20.Shinohara A, Ogawa T. Nature (London) 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 21.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy G, Golub E I, Radding C M. Mutat Res. 1997;377:53–59. doi: 10.1016/s0027-5107(97)00057-2. [DOI] [PubMed] [Google Scholar]
- 23.Golub E I, Kovalenko O V, Gupta R C, Ward D C, Radding C M. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer W-D. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen U H, Rothstein R, Kodadek T. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 26.Tan T L, Essers J, Citterio E, Swagemakers S M, de Wit J, Benson F E, Hoeijmakers J H, Kanaar R. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 27.Swagemakers S M, Essers J, de Wit J, Hoeijmakers J H, Kanaar R. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 28.Petukhova G, Stratton S, Sung P. Nature (London) 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 29.Baumann P, Benson F E, West S C. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 30.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 31.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 32.Petukhova G, Sung P, Klein H. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazin A V, Bornarth C J, Solinger J A, Heyer W D, Kowalczykowski S C. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 34.Cozzarelli N R, Boles T C, White J H. In: DNA Topology and Its Biological Effects. Cozzarelli N R, Wang J C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 139–215. [Google Scholar]
- 35.White J H. Am J Math. 1969;91:693–728. [Google Scholar]
- 36.Champoux J J. Adv Pharmacol. 1994;29A:71–82. doi: 10.1016/s1054-3589(08)60540-2. [DOI] [PubMed] [Google Scholar]
- 37.Cozzarelli N R. Science. 1980;207:953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- 38.Berger J M, Wang J C. Curr Opin Struct Biol. 1996;6:84–90. doi: 10.1016/s0959-440x(96)80099-6. [DOI] [PubMed] [Google Scholar]
- 39.Travers A, Klug A. Nature (London) 1987;327:280–281. doi: 10.1038/327280a0. [DOI] [PubMed] [Google Scholar]
- 40.Kirkegaard K, Wang J C. Cell. 1981;23:721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- 41.Verhoeven E E A, Wyman C, Moolenaar G F, Hoeijmakers J H J, Goosen N. EMBO J. 2001;20:601–611. doi: 10.1093/emboj/20.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stasiak A, Di Capua E, Koller T. J Mol Biol. 1981;151:557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- 43.Conley E C, West S C. J Biol Chem. 1990;265:10156–10163. [PubMed] [Google Scholar]
- 44.Cunningham R P, Shibata T, DasGupta C, Radding C M. Nature (London) 1979;281:191–195. doi: 10.1038/281191a0. [DOI] [PubMed] [Google Scholar]
- 45.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono M, Tucker P W, Capra J D. Nucleic Acids Res. 1994;22:3918–3924. doi: 10.1093/nar/22.19.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 48.Bustamante C, Keller D, Yang G. Curr Opin Struct Biol. 1993;3:363–372. [Google Scholar]
- 49.Van Noort S J T, van der Werf K O, de Grooth B G, van Hulst N F, Greve J. Ultramicroscopy. 1997;64:117–127. [Google Scholar]
- 50.West S C. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 51.Havas K, Flaus A, Phelan M, Kingston R, Wade P A, Lilley D M J, Owen-Hughes T. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 52.Gavin I, Horn P J, Peterson C L. Mol Cell. 2001;7:97–104. doi: 10.1016/s1097-2765(01)00158-7. [DOI] [PubMed] [Google Scholar]
- 53.Wyman C, Botchan M. Curr Biol. 1995;5:334–337. doi: 10.1016/s0960-9822(95)00065-0. [DOI] [PubMed] [Google Scholar]