Abstract
Background:
To explore the possible adverse effects and search for cell phone electromagnetic field (EMF)-responsive proteins in human early reproduction, a proteomics approach was employed to investigate the changes in protein expression profile induced by cell phone EMF in human chorionic tissues of early pregnancy in vivo.
Methods:
Volunteer women about 50 days pregnant were exposed to EMF at the average absorption rate of 1.6 to 8.8 W/kg for 1 hour with the irradiation device placed 10 cm away from the umbilicus at the midline of the abdomen. The changes in protein profile were examined using 2-dimensional electrophoresis (2-DE).
Results:
Up to 15 spots have yielded significant change at least 2- to 2.5-folds up or down compared to sham-exposed group. Twelve proteins were identified— procollagen–proline, eukaryotic translation elongation factor 1 delta, chain D crystal structure of human vitamin D-binding protein, thioredoxin-like 3, capping protein, isocitrate dehydrogenase 3 alpha, calumenin, Catechol-O-methyltransferase protein, proteinase inhibitor 6 (PI-6; SerpinB6) protein, 3,2-trans-enoyl-CoA isomerase protein, chain B human erythrocyte 2,3-bisphosphoglycerate mutase, and nucleoprotein.
Conclusion:
Cell phone EMF might alter the protein profile of chorionic tissue of early pregnancy, during the most sensitive stage of the embryos. The exposure to EMF may cause adverse effects on cell proliferation and development of nervous system in early embryos. Furthermore, 2-DE coupled with mass spectrometry is a promising approach to elucidate the effects and search for new biomarkers for environmental toxic effects.
Keywords: placental villous, proteomics, cell phone EMF, radiofrequency, early development
Introduction
The wide use of cell phone networks worldwide leads to increased environmental exposure to radiofrequency electromagnetic fields (RF-EMFs). The concern of the potential health effects of the low-energy electromagnetic radiation emitted by mobile phones has been growing, especially the long-term effects and reproduction-related effects.1 Up to now, the adverse effects of RF associated with cell phones on the human reproduction are highly controversial.
Epidemiological studies of adverse pregnancy outcomes due to the exposure to RF fields have been reviewed, although the evidence on possible effects of RF fields on pregnancy outcomes is virtually limited to occupational exposures.2 The end points include spontaneous abortions, birth weight, gender ratio, and congenital malformations.1,3 Although some researchers have reported positive findings, no specific type of malformation or other adverse outcomes has been consistently reported and most of the studies have limited statistical power. The available data are not sufficient to draw any definitive conclusions. Numerous studies have clearly shown that RF fields are teratogenic at exposure levels that are high enough to cause significant increase in temperature;4 however, no consistent evidence of effects at nonthermal exposure levels was found and only a few studies have evaluated the possible effects on postnatal development using sensitive end points, such as behavioral effects.
In vitro studies reported the effects of RF radiation on DNA damage,5 chromosome aberrations,6 mutation,7 cell transformation,8 and gene expression.9 However, these experiments focused on the cell lines including immune cells, breast cancer cells, and so on.10 Recently, it is suggested that RF-electromagnetic wave (EMW) emitted from cell phones may lead to oxidative stress in human semen and keeping the cell phone in a trouser pocket in talk mode may negatively affect spermatozoa and impair male fertility.11 Our previous studies on the effects of extremely-low frequency (ELF)-EMF on preimplantation embryos also supposed that the adverse effects of ELF-EMF on preimplantation embryos might be caused by the DNA damage in the embryos in vitro.12 However, so far, as for the RF-EMF, few research on its effects on reproduction especially on early stage pregnancy in vivo has been conducted. The early stage of pregnancy is one of the most important stages of the reproduction, in which all tissues are most sensitive to the toxic effects of the environment compared to the other stages during the life span.13Consequently, the early-pregnancy villous tissues were used in the present study to explore the possible effects of RF-EMF on reproduction.
High-throughput screening techniques, such as proteomics, have been proposed to be the effective approaches to search for possible molecular targets of environmental exposure which is unable to be predicted based on current knowledge.14 The early-stage pregnancy villous is supposed to represent the fetus during the first trimester of pregnancy. In our study, the effects of RF-EMF exposure on protein expression profiles in human early-stage placental villous tissue are to clarify the possible effects of RF-EMF on the early-stage reproduction and the underlying mechanism.
Materials and Methods
Patients
Our study was approved by the institutional review board of School of Medicine, Zhejiang University. Forty women involved in this study were recruited from the women’s hospital, School of Medicine, Zhejiang University, with a normal pregnancy and undergoing terminations of pregnancy for psychological reasons at the gestational age about 50-60 days. The fetal heart rate was detected in all pregnancy with transvaginal ultrasound. All these women were consented by the investigator and signed a consent form for EMF exposure and the use of placental villous tissue. To date, most of the animal studies conducted have demonstrated that exposure to RF-EMF does not increase the incidence or promote the development of tumor and exerts no genotoxic effects.15,16
These women were randomly divided into 2 groups, exposure group and sham exposure group, with the random number generated by Excel. There were no significant differences in age, body mass index, and days of pregnancy between these 2 groups. After the exposure for 20 minutes, induced abortion was performed. Placental villous tissues were taken through the cervix during dilatation and aspiration according to strict clinical procedures.
Exposure System
The lower abdomens of the women were exposed to EMW emitted from a commercially available cellular telephone as previously described11 with the central station of the radiation in the navel region. Briefly, the cellular telephone was in talk mode (Sony Ericsson w300; service provider China Mobile; GSM-Global System for Mobile communications network; 900 MHz frequency; maximum power < 1 W; SAR 1.46 W/kg). This phone model had an antenna placed on the top back of its handset. The distance between the phone antenna and the abdomen was kept at 2.5 cm. In PR China, the most common frequency is 850 to 900 that was used in the present study. The duration of exposure was 20 minutes. The duration was selected according to our preexperiment. We found significant changes in 2-dimensional electrophoresis (2-DE) protein maps with 20 and 40 minutes exposure; however, no significant changes were found with 5 or 10 minutes exposure. To date, the animal studies demonstrated that exposure to RF-EMF in that power and duration does not increase the incidence or promote the development of tumor or have significant toxic effects.15
The women in control group were exposed to the same exposure system with the cellular telephone in power-off state.
Peak power density was used to evaluate the reference level of exposure according to the International Commission for Non-Ionizing Radiation Protection (ICNIRP) and the Federal Communications Commission and it is a commonly used term for characterizing an RF-EMF (ICNIRP. 1998). In the present study, the power density was monitored during basal condition (no cell phone radiation) and experimental condition (cell phone in talk mode). Power density in the control condition was 0.01 to 0.1 mW/cm2 and was 1 to 40 mW/cm2 in the experimental condition.
The frequency emitted by the cell phone was confirmed with the help of a RF spectrum analyzer (Tektronix, Beaverton, Oregon).
Sample Preparation
Tissues were collected in ice-cold and sterile phosphate-buffered saline, dissected using a microscope to remove the endometrial tissues, which were finally stored at −80°C until further analysis. Six placental villous tissues randomly collected from the women with RF-EMF exposure and 6randomly from the control women were used for proteomic analysis. The protein extraction protocol is the same as described in the previous study.17 Briefly, a total of 300 mg of each placental villous sample was homogenized and solubilized in 1000 μL of lysis buffer containing 7 mol/L of urea, 2 mol/L of thiourea, 4% 3-[(3-Cholamidopropyl)dimethylammonio]-1 propanesulfonate, 1% immobilized pH gradient (IPG) buffer (pH 4-7), and 65 mmol/L dithiothreitol (DTT). Nonsoluble cellular debris was removed by centrifugation. Protein concentrations were determined using the Bradford assay. All samples were stored at −80°C prior to electrophoresis. For each sample, 3 separate experiments were conducted independently on 3 analytical 2-DE gels.
Two-Dimensional Gel Electrophoresis
The 2-DE was performed using the Amersham Bioscience 2-DE system according to the manufacturer’s instructions (Amersham Biosciences, Uppsala, Sweden). For the first dimension, 300 μg of total protein was mixed with a rehydration solution containing 7 mol/L urea, 2 mol/L thiourea, 2% CHAPS, 0.5% IPG buffer (pH 4-7), 18 mmol/L DTT. A trace of bromophenol blue to a total volume of 450 μL and analyzed for both the RF-EMF treated group and the control group and rehydrated for 12 hours. Automatically, isoelectric focusing (IEF) was conducted on the IPGphor (Amersham) platform at 20°C, and the total Vh of IEF is 65 000 to 70 000 Vh. After IEF separation, the strips were equilibrated for 15 minutes in a buffer (50 mmol/L Tris-HCl, 6 mol/L urea, 30% glycerol, 2% sodium dodecyl sulfate [SDS], and a trace of bromophenol blue) containing 1% DTT and subsequently in the same equilibration buffer containing 2.5% iodoacetamide for 15 minutes. The second separation was carried out at 15°C on 12.5% SDS slab gels using an ETTAN DALT II electrophoresis system (Amersham), with the IPG strips sealed on the top of the gels with 0.5% agarose. The SDS-polyacrylamide gel electrophoresis was run at constant power of 2.5 W/gel for 30 minutes, and then switched to 15 W/gel until the bromophenol blue marker reached the bottom of the gel.
Gel Staining, Image Analysis, Data Normalization, and Statistical Analysis
Silver staining was performed using the method described by Swain and Ross,18 except that the treatment with glutaraldehyde (a cross-linking and sensitizing agent) was omitted.19 The silver-stained 2-DE gels were scanned, and the digitized images were analyzed with PDQuest (v7.2, Bio-Rad, Hercules, California). Image analysis included the following procedures: spot detection, spot editing, background subtraction, and spot matching. Spot detection was controlled by 4 parameters, including sensitivity, operator size, noise factor, and background. The same parameters were used to detect the spots in all the gels in order to guarantee comparability between the gels. One of the gels in the control group was chosen as a reference gel before matching. The spots unmatched with the reference gel were not added to the reference gel to avoid biological variation. The amount of a protein spot was expressed as the volume of the spot, which was defined as the sum of the intensities of all the pixels that made up the spot. In order to correct for variations due to silver staining and quantify protein spots, the individual spot volume was normalized as a percentage of the total volume of all the spots detected on the gel.
Enzymatic In-Gel Digestion
Selected protein spots were excised from the gel, and in-gel digestion with trypsin was performed according to published procedures with slight modification.18,19 The gel pieces were washed twice with water and destained in freshly prepared destaining solution (30 mmol/L potassium hexacyanoferrate: 100 mmol/L sodium thiosulfate, 1:1 [v/v]) for 1 to 2 minutes until the brown color disappeared. They were then rinsed sequentially with water and 100 mmol/L ammonium bicarbonate, respectively. The gel pieces were shrunk by dehydration in acetonitrile (ACN) and dried in a vacuum centrifuge (SpeedVac, Thermo Savant, Holbrook, New York). Then, the gel pieces were swollen in a digestion buffer containing 40 mmol/L ammonium bicarbonate, 10% ACN, and 20 μg/mL of trypsin (Sigma, Germany; proteomics sequencing grade) in an ice-cold bath. After 30 minutes, the supernatant was removed and replaced with 30 μL of the same buffer but without trypsin. The gel pieces were kept wet during the enzymatic cleavage at 37°C overnight. The supernatants were collected and peptides were extracted twice by 2 changes of 5% trifluoroacetic acid (TFA)/50% ACN (15 minutes for each change) at room temperature (RT) and dried out.
MALDI-TOF Mass Spectrometry Analysis and Database Searching
The peptide extracts dissolved in 5 μL of 0.1% TFA were mixed with 5 μL of saturated a-cyano-4-hydroxycinnamic acid (HCCA, Applied Biosystems, Carlsbad, California) in 50% ACN/0.05% TFA. Aliquots of 2 μL were applied onto a target plate and allowed to air dry. Mass analysis of peptide mixtures was performed using a Voyager-DE PRO matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS; Applied Biosystems) with a 337-nm N2 ultraviolet laser. The instrument was operated in the reflector/delayed extraction mode with an accelerating voltage of 20 kV, grid voltage of 67%, and delay time of 200 nanosecond. In general, 200 shots were applied for each sample, with a mass range from 900 to 4000 Da. P14R (Mr 1532.8582) and adrenocorticotropic hormone (ACTH) fragment 18 to 29 (Mr 2464.1989) were used for external calibration. The raw spectra files were analyzed using the Data Explorer v 4.0 software (Applied Biosystems). After baseline correction, noise filter, and peak deisotoping, the raw peaks were converted into isotopic peaks. The mass list was put in the MASCOT search engine (Matrix Science, London, UK). For protein search the National Center for Biotechnology Information Homo sapiens protein database (December 2004) was used. The parameters included were as follows: using monoisotopic masses, a maximum 6 0.3 Da mass tolerance, cysteine in carboxyamidomethyl form and methionine in oxidization form, and an allowance for up to 1 missed cleavage per peptide. Proteins matching more than 4 peptides and with a MASCOT score higher than 64 were considered significant (P < .05).
Western Blot Analysis
For western blot analysis, protein samples from the tissue lysate of 20 pairs of placental villous both from the RF-EMF-treated group and the control group were separated on a 8% SDS polyacrylamide gel and subsequently blotted onto a protran nitrocellulose transfer membrane (Schleicher & Schuell Bioscience, Germany). The membranes were blocked for 2 hours at RT in 5% milk powder in Tris-buffered saline, containing 0.1% Tween 20, incubated overnight at 4°C with the primary antibodies, Goat anti-catechol-O-methyltransferase (COMT; internal, Sigma), CAPZB polyclonal antibody (Abnova, Taiwan), and monoclonal anti-TXNL-2 antibody (Sigma). This was followed by incubating with horseradish peroxidase (HRP)-coupled secondary antibodies and the proteins were detected by developing with ECL using the Chemiluminescence Detection Kit for HRP (Biological Industries, Israel). Equal protein loading was confirmed by exposing the membranes to the anti-b-actin antibody.
Statistical Analysis
Data from 2-DE image and Western blot analysis were exported into Microsoft Excel 2003. Statistical analysis was performed using the paired t test. A P value <.05 was considered significant. Only proteins that demonstrated significant and consistent changes (increased or decreased) were used for further proteomic analysis. Data were expressed as mean ± standard deviation.
Results
Differential Analysis of 2-DE Protein Maps of Exposure and Control Groups
Three pairs of gels from the exposure and the control groups were analyzed for quantitative spot comparisons with image analysis software from PDQuest to investigate the difference in gene expression between RF-exposure villi and control villi. Approximately, 2000 spots were detected both in the exposure group and in the sham-exposed group. The representative 2-DE images are shown in Figure 1. A total of 15 protein spots were decreased or increased 2-fold in the exposure group compared to the control. Among them, 9 proteins were downregulated and 6 proteins were upregulated significantly in the exposure group, determined by spot volume (P < .05; Figure 1).
Figure 1.
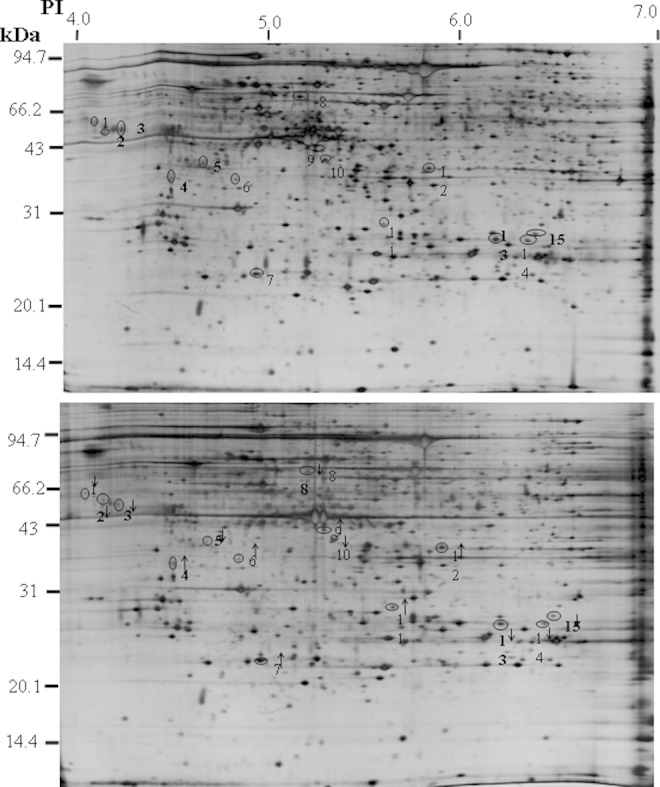
Representative 2-dimensional electrophoresis (2-DE) protein maps of radiofrequency electromagnetic field (RF-EMF)-exposed group and control groups. Two representative 2-DE maps indicating protein spots that changed in volume in placental villous samples from the control (A) and RF-EMF-exposed group (B). The 15 proteins found to be significantly (P < .05) decreased or increased in placental villous tissues from the RF-EMF-exposed group compared to the control, which are marked with numbers and downward or upward arrows.
Proteins With Altered Expression Identified by MALDI-TOF MS and Database Searching
The 15 protein spots with altered intensity in the RF-exposed group were excised from the gels, digested with trypsin, and subsequently analyzed by MALDI-TOF MS. Differentially expressed proteins were identified by the resultant spectra. Of the 15 spots excised from the gels, 12 spots were successfully identified (Table 1). The search results were evaluated on the basis of accepted standards that take account of the number of peptides matched to the candidate protein, the difference in the number of matched peptides between the candidate protein and the next best fit, the coverage of the candidate proteins sequence by the matching peptides. The remaining 3 protein spots were not identified mainly because there was no satisfactory spectrum available.
Table 1.
Search Results of Peptide for the Altered Expression Proteins With Relatively High Confidence in Placental Villi of RF-Exposed Group.
| Spot no. | Mass | Calculated PI | Expression Regulationa | Score | Masses Matched | Coverage (%) | Protein Name |
|---|---|---|---|---|---|---|---|
| 2 | 37 050 | 4.47 | − | 115 | 10 (20) | 39 | Calumenin |
| 4 | 57 081 | 4.76 | + | 64 | 7 (18) | 18 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), β-polypeptide |
| 6 | 20 032 | 5.36 | + | 105 | 11 (21) | 63 | COMT protein |
| 7 | 31 103 | 4.90 | + | 106 | 9 (22) | 43 | Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| 8 | 42 562 | 5.18 | − | 88 | 9 (19) | 33 | SERPINB6 protein |
| 9 | 51 210 | 5.22 | + | 95 | 10 (19) | 35 | Chain D, crystal structure of human vitamin D-binding protein in complex with skeletal actin |
| 10 | 37 408 | 5.31 | − | 112 | 9 (16) | 34 | Thioredoxin-like 2 |
| 11 | 31 331 | 5.36 | + | 96 | 10 (32) | 36 | Capping protein (actin filament) muscle Z-line, beta |
| 12 | 39 566 | 6.47 | + | 149 | 12 (18) | 32 | Isocitrate dehydrogenase 3 (NAD+) alpha |
| 13 | 26 965 | 6.32 | − | 104 | 8 (18) | 50 | DCI protein |
| 14 | 31 051 | 6.28 | − | 135 | 10 (19) | 49 | Chain B, human erythrocyte 2,3-bisphosphoglycerate mutase |
| 15 | 32 097 | 6.45 | − | 113 | 10 (26) | 53 | Nucleoprotein |
Abbreviations: COMT, catechol-O-methyltransferase; NAD, nicotinamide adenine dinucleotide; PI, proteinase inhibitor; RF, radiofrequency.
a +, upregulation; −, downregulation.
Characteristics of the Identified Proteins
By comparing RF-exposed and control villi, 15 protein spots were shown to be expressed differentially and 12 protein were successfully identified, from which 6 were upregulated, including procollagen–proline, eukaryotic translation elongation factor 1 δ, chain D crystal structure of human vitamin D-binding protein, thioredoxin-like 3, capping protein (CP), isocitrate dehydrogenase 3 (nicotinamide adenine dinucleotide [NAD+]) alpha, and rest 6 were downregulated, including calumenin, COMT protein, SERPINB6 protein, DCI protein, chain B human erythrocyte 2,3-bisphosphoglycerate mutase, nucleoprotein. According to annotations from UniProt knowledgebase (Swiss-Prot/TrEMBL) and Gene Ontology Database, the identified proteins are related to a wide range of cell functions, including antioxidant enzymes relevant to oxidative stress, such as TXNL-2, eukaryotic translation elongation factor involved in transcription; protein involved in vitamin A metabolism.
Validation of Differentially Expressed Proteins by Western Blot
To further confirm the alterations of protein expression in RF-exposed group, 3 of the differentially expressed proteins were validated. As shown in Figure 2, COMT and Capzb showed consistently increased expression in RF-exposed groups. In contrast, TXNL-2 showed significantly decreased expression. The changes in the expression of these proteins were consistent with the 2-DE and silver-staining results.
Figure 2.
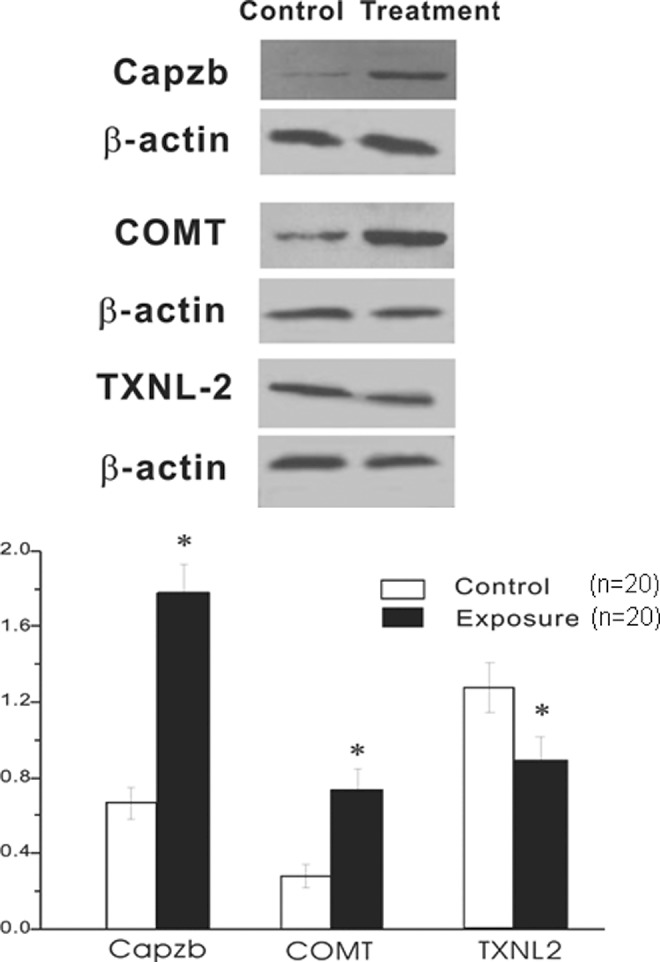
Western blot analysis of deferentially expressed proteins by radiofrequency electromagnetic field (RF-EMF) exposure. COMT, Capzb, and TXNL-2 were detected in RF-EMF exposed and control placental villous tissues. The representative autoradiographs are shown (A). Intensities of protein loading were confirmed by exposing the membranes to the anti-b-actin antibody. Data were normalized by using the b-actin signals (mean ± standard deviation [SD], n = 20; B). *Value significantly differs from the control (P < .05).
Discussion
A comparison of the 2-DE patterns from control and exposure groups in the present study revealed that RF-EMF exposure changes the protein expression profile of early-stage placental villous tissue. The results indicated that 15 of the proteins with significant and reproducible changes were identified. Among the 15 proteins detected either increased or decreased in the placental villi from the RF-EMF-exposed group, 12 proteins were identified by peptide mass fingerprinting. Western blotting analysis on 3 proteins (COMT, TXNL-2, and Capzb) validated that the differential expressions of proteins obtained from proteomic analysis were convincing. The remaining 3 proteins were not identified successfully, as the confidence of database search through peptide mass fingerprint was not sufficient to yield unambiguous results. Our database search results illustrated that these differential proteins relate with a wide range of cell functions including the development of cytoskeleton, oxidation–reduction reaction, cellular metabolism, and so on.
Proteomics combining 2-DE and MS is the large-scale study of gene expression at the protein level in contrast to conventional biochemical approaches that detect one or a few specific proteins at a time.20 In the present study, the differential protein patterns between RF-EMF-exposed placental villous tissues and the sham-exposed ones were analyzed using the 2-DE, and differences on the specific protein differences were assessed by MS.
The trophoblastic cells in early pregnancy are extremely sensitive to oxidative stress because of extensive cell divisions and the concomitant exposure of their DNA. In vivo study supports the concept of physiologically low O2 environment inside the human gestational sac during most of the first trimester of pregnancy.21 The TXNL-2 is one of the most important antioxidant enzymes. In the present study, placental villous tissues were found to possess lower concentrations of antioxidant enzyme.22 The TXNL-2 at early-stage pregnancy with RF-EMF exposure was compared to without exposure, suggesting that the depletion of placental antioxidant defenses might exist as a key mechanism of the biological effects of RF-EMF on early-stage pregnancy. In these circumstances, the damage to proteins and nucleic acid might severely impair normal trophoblastic functions and may even lead to cell death.
Alteration of other functional protein was also evident. The altered expression of COMT protein was found by proteomics analysis in the present study. The enzyme COMT, identified in the 1950s, is involved in catabolism of monoamines that are influenced by psychotropic medications, including neuroleptics and antidepressants.23 Consequently, COMT has been one of the most studied genes for psychosis.24 Recent study indicated that decreased COMT expression may have a role in preeclampsia.25,26 The alteration in expression of COMT protein in early-pregnancy villous tissue with RF-EMF exposure in our study indicated that the RF-EMF exposure may exert adverse effects on the placenta function and even the developing brain function especially the psychic dysfunction consequently. It is widely recognized that high strength EMF might exert adverse effects on brain functions. The alteration of COMT proteins in placenta villi with RF-EMF exposure might partly explain that phenomenon.27
Unexpectedly, we found the levels of Capzb, one isoform of actin CP, which is an F-actin binding protein and regulates F-actin assembly in a variety of cell types, were increased in the RF-EMF-exposed early-pregnancy villous tissues. The CP functions as an a/b heterodimer.28 The vertebrates have conserved isoforms of each subunit. It is reported by one previous study that Capzb2, the neuronal CPb subunit, is necessary for the regulation of F-actin assembly in neuron and inhibiting microtubule polymerization.29 Capzb in muscle cells were found to attach actin filaments to the Z-line, which organizes the actin at the intercalated discs.30 The increased expression of Capzb in villous tissue might indicate that RF-EMF has adverse effects on cell skeleton, leading the actin to bind abnormally and even to cell death. This suggests another way in which RF-EMF radiation may be harmful to early-stage reproduction. In conclusion, we performed a proteomic approach on the human early pregnant placental villi derived from RF-EMF exposure with a combination of 2-DE and MALDI-TOF/TOF MS. By the application of this strict selection criteria for the determination of potentially important proteins, 15 proteins were identified in the exposed group with significantly altered abundance in comparison with the sham-exposed group. These proteins seem to be related to various cellular functions. Our results offer insights into the underlying molecular mechanisms of RF-EMF radiation, and provide possible reasons for the adverse effects of RF-EMF on human reproduction.
Clearly, more work is needed to understand the different strength of EMF on the early pregnancy tissues. Moreover, for ethical reasons, we cannot verify the altered expressed proteins on the human fetus of babies. The relationship of the outcome of fetus developing as well as baby born and the alteration of the proteins by EMF exposure should be explored in animal models. Our further research will focus on this issue.
Acknowledgments
We are grateful to Jing Shen, Department of Pathology And Pathophysiology, Center For Environmental Genomics, Zhejiang University School of Medicine for technical support during MALDI-TOF MS measurement and to Jun Hu, Huilan Sun, and Xuxia Zhu for their help in sample collection.
Footnotes
Authors’ Note: Qiong Luo and Ying Jiang contributed equally to this study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: National Basic Research Program of China (NO. 2012CB944900), National Natural Science Foundation of China (NO. 30901616).
References
- 1. Feychting M, Ahlbom A, Kheifets L. EMF and health. Annu Rev Public Health. 2005;26:165–189. [DOI] [PubMed] [Google Scholar]
- 2. Breckenkamp J, Berg-Beckhoff G, Münster E, et al. Feasibility of a cohort study on health risks caused by occupational exposure to radiofrequency electromagnetic fields. Environ Health. 2009;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feychting M. Non-cancer EMF effects related to children. Bioelectromagnetics. 2005;(suppl 7):S69–S74. [DOI] [PubMed] [Google Scholar]
- 4. D'Andrea JA, Ziriax JM, Adair ER. Radio frequency electromagnetic fields: mild hyperthermia and safety standards. Prog Brain Res. 2007;162:107–135. [DOI] [PubMed] [Google Scholar]
- 5. Lai H, Singh NP. Selective cancer cell cytotoxicity from exposure to dihydroartemisinin and holotransferrin. Cancer Lett. 1995;91(1):41–46. [DOI] [PubMed] [Google Scholar]
- 6. Vijayalaxmi Bisht KS, Pickard WF, Meltz ML, Roti Roti JL, Moros EG. Chromosome damage and micronucleus formation in human blood lymphocytes exposed in vitro to radiofrequency radiation at a cellular telephone frequency (847.74 MHz, CDMA). Radiat Res. 2001;156(4):430–432. [DOI] [PubMed] [Google Scholar]
- 7. Gos P, Eicher B, Kohli J, Heyer WD. No mutagenic or recombinogenic effects of mobile phone fields at 900 MHz detected in the yeast Saccharomyces cerevisiae. Bioelectromagnetics. 2000;21(7):515–523. [DOI] [PubMed] [Google Scholar]
- 8. Roti JL, Malyapa RS, Bisht KS, et al. Neoplastic transformation in C3H 10T(1/2) cells after exposure to 835.62 MHz FDMA and 847.74 MHz CDMA radiations. Radiat Res. 2001;155:239–247. [DOI] [PubMed] [Google Scholar]
- 9. Kwee S, Raskmark P, Velizarov S. Resonant interactions of charged lipid vesicles with microwave electromagnetic field. Biofizika. 2001;44:1078–1082. [PubMed] [Google Scholar]
- 10. Takashima Y, Hirose H, Koyama S, Suzuki Y, Taki M, Miyakoshi J. Effects of continuous and intermittent exposure to RF fields with a wide range of SARs on cell growth, survival, and cell cycle distribution. Bioelectromagnetics. 2006;27(5):392–400. [DOI] [PubMed] [Google Scholar]
- 11. Agarwal A, Desai NR, Makker K, et al. Effects of radiofrequency electromagnetic waves (RF-EMW) from cellular phones on human ejaculated semen: an in vitro pilot study. Fertil Steril. 2009;92(4):1318–1325. [DOI] [PubMed] [Google Scholar]
- 12. Luo Q, Yang J, Zeng QL, Zhu XM, Qian YL, Huang HF. 50-Hertz electromagnetic fields induce gammaH2AX foci formation in mouse preimplantation embryos in vitro. Biol Reprod. 2006;75(5):673–680. [DOI] [PubMed] [Google Scholar]
- 13. Glasser SR, Julian J, Munir MI, Soares MJ. Biological markers during early pregnancy: trophoblastic signals of the peri-implantation period. Environ Health Perspect. 1987;74:129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoang VM, Foulk R, Clauser K, Burlingame A, Gibson BW, Fisher SJ. Functional proteomics: examining the effects of hypoxia on the cytotrophoblast protein repertoire. Biochemistry. 2001;40(13):4077–4086. [DOI] [PubMed] [Google Scholar]
- 15. Heynick LN, Johnston SA, Mason PA. Guidelines for limiting exposure to time-varying electric, magnetic, and electromagnetic fields (up to 300 GHz). Health Phys. 2003;74(4):494–522. [PubMed] [Google Scholar]
- 16. Vijayalaxmi Obe G. Controversial cytogenetic observations in mammalian somatic cells exposed to radiofrequency radiation. Radiat Res. 2004;162(5):481–496. [DOI] [PubMed] [Google Scholar]
- 17. Liu AX, Jin F, Zhang WW, et al. Proteomic analysis on the alteration of protein expression in the placental villous tissue of early pregnancy loss. Biol Reprod. 2006;75(3):414–420. [DOI] [PubMed] [Google Scholar]
- 18. Swain M, Ross NW. A silver stain protocol for proteins yielding high resolution and transparent background in sodium dodecyl sulfatepolyacrylamide gels. Electrophoresis. 1995;16(6):948–951. [DOI] [PubMed] [Google Scholar]
- 19. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. [DOI] [PubMed] [Google Scholar]
- 20. Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence database. Proc Natl Acad Sci U S A. 1993;90(11):5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jauniaux E, Watson AL, Hempstock J, Bao YP, Skepper JN, Burton GJ. Onset of maternal arterial bloodflow and placental oxidative stress; a possible factor in human early pregnancy failure. Am J Pathol. 2000;157(6):2111–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Godoy JR, Funke M, Ackermann W, et al. Immunohistochemical detection of glutaredoxin-, peroxiredoxin-, and thioredoxin-family proteins in various tissues of the laboratory mouse. Biochim Biophys Acta. 2011;1810(1):2–92. [DOI] [PubMed] [Google Scholar]
- 23. Kambur O, Männistö PT. Catechol-O-methyltransferase and pain. Int Rev Neurobiol. 2010;95:227–279. [DOI] [PubMed] [Google Scholar]
- 24. Sagud M, Mück-Seler D, Mihaljević-Peles A, et al. Catechol-O-methyl transferase and schizophrenia. Psychiatr Danub. 2010;22:270–274. [PubMed] [Google Scholar]
- 25. Roten LT, Fenstad MH, Forsmo S, et al. A low COMT activity haplotype is associated with recurrent preeclampsia in a Norwegian population cohort (HUNT2). Mol Hum Reprod. 2011;17(7):439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee SB, Wong AP, Kanasaki K, et al. Preeclampsia: 2-methoxyestradiol induces cytotrophoblast invasion and vascular development specifically under hypoxic conditions. Am J Pathol. 2010;176(2):710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bak M, Dudarewicz A, Zmyślony M, Sliwinska-Kowalska M. Effects of GSM signals during exposure to event related potentials (ERPs). Int J Occup Med Environ Health. 2010;23(2):191–199. [DOI] [PubMed] [Google Scholar]
- 28. Barron-Casella EA, Torres MA, Scherer SW, Heng HH, Tsui LC, Casella JF. Sequence analysis and chromosomal localization of human Cap Z. Conserved residues within the actin-binding domain may link Cap Z to gelsolin/severin and profilin protein families. J Biol Chem. 1995;270(37):21472–21479. [DOI] [PubMed] [Google Scholar]
- 29. Davis DA, Wilson MH, Giraud J, et al. Capzb2 interacts with beta-tubulin to regulate growth cone morphology and neurite outgrowth. PLoS Biol. 2009;7(10):e1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pyle WG, Hart MC, Cooper JA, Sumandea MP, de Tombe PP, Solaro RJ. Actin capping protein: an essential element in protein kinase signaling to the myofilaments. Circ Res. 2002;90(12):1299–1306. [DOI] [PubMed] [Google Scholar]


