Abstract
Stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the most serious diseases of wheat; therefore, exploring effective resistance-related genes is critical for breeding and studying resistance mechanisms. However, only a few stripe rust resistance genes and defence-related genes have been cloned. Moreover, transgenic wheat with enhanced stripe rust resistance has rarely been reported. Receptor-like proteins (RLPs) are known to be involved in defence and developmental pathways. In this research, a novel RLP gene TaRLP1.1 was characterized as an important stripe rust defence gene. TaRLP1.1 was screened by GeneChip and was found to be induced by Pst specifically in the resistant variety. Knock down of TaRLP1.1 in the stripe rust-resistant plants resulted in increased susceptibility to Pst, and phenolic autofluorogen accumulation at the pathogen–host interaction sites, usually correlated with the hypersensitive response, was decreased dramatically. However, when the TaRLP1.1 gene was transformed into the susceptible wheat variety Yangmai158, the transgenic plants showed highly increased resistance to Pst, and the hypersensitive response was enhanced at the infection sites. Meanwhile, the expression of pathogenesis-related genes decreased in the TaRLP1.1-silenced plants and increased in the TaRLP1.1-overexpressing plants. Thus, it was proposed that TaRLP1.1 greatly contributed to the hypersensitive response during the pathogen–host interaction. Along with the functional analysis, an evolutionary study of the TaRLP1 family was performed. Characterization of TaRLP1.1 may facilitate breeding for stripe rust resistance and better understanding of the evolution of the RLP genes in wheat.
Key words: Disease resistance, hypersensitive response, receptor-like protein, stripe rust, transgenic, virus-induced gene silencing.
Introduction
Stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the most destructive diseases of wheat in the world, and China is the largest endemic area (Kang et al., 2010). Utilization of disease-resistant varieties has proved to be the safest, most economical, and effective method to control stripe rust, and resistance gene cloning is vital for the development of resistant varieties. However, a big challenge for resistance breeding is that Pst isolates evolved very rapidly, leading to breakdown of widely used resistance genes—especially in China where most new destructive epidemics of Pst have originated. Yr10 (Laroche et al., 2000), Yr18 (Krattinger et al., 2009), and Yr36 (Fu et al., 2009) are the only stripe rust resistance genes cloned to date. Using the virus-inducing gene silencing (VIGS) approach, TaRLK-R1, -R2, and -R3 (Zhou et al., 2007), TaHsp90.2 and TaHsp90.3 (G.F. Wang et al., 2011), TaDAD2 (X.J. Wang et al., 2011), TaMCA4 (Wang et al., 2012), and TaRab7 (Liu et al., 2012) have been characterized as the genes required for stripe rust resistance. However, no stripe rust resistance-improved transgenic wheat has been developed by using these genes, so their effect in breeding remains unknown. The stripe rust resistance gene Yr26 located on chromosome 1B was introduced from Triticum turgitum cv. γ-80 (Ma et al., 2001), and has been widely used as the main gene resource in China since 2003. Yr26 is a broad-spectrum resistance gene, but, more importantly, Yr26 shows potential durable resistance. Thus cloning the genes related to stripe rust resistance from Yr26-containing wheat should help in understanding the mechanism of its resistance and in developing varieties resistant to stripe rust.
Receptor-like proteins (RLPs) are cell surface receptors which contain an extracellular lecine-rich repeat (LRR) domain, a single transmembrane domain, and a small cytoplasmic tail (Kruijt et al., 2005). Several RLP genes are known to be involved in plant–pathogen interactions. The first identified RLP gene was tomato Cf-9 that mediated resistance against Cladosporium fulvum (Jones et al., 1994), and several other Cf resistance genes have also been characterized to be RLP genes (Dixon et al., 1996, 1998). In tomato, two Ve genes involved in plant resistance against vascular wilt and two LeEix genes encoding receptors for the fungal elicitor were also reported to encode RLPs (Kawchuk et al., 2001; Ron and Avni, 2004). In apple, HcrVf2 conferred resistance to the scab fungus Venturia inaequalis (Belfanti et al., 2004). In Arabidopsis, three RLP genes were found to be involved in the resistance response, namely AtRLP52 (Ramonell et al., 2005), AtRLP30 (Wang et al., 2008), and snc2-1D (Zhang et al., 2010). Apart from defence against pathogens, RLP genes also play significant roles in plant development. In Arabidopsis, TWO MANY MOUTHS (TMM) was shown to be involved in stomatal patterning (Nadeau and Sack, 2002), while CLAVATA2 (CLV2) and its orthologue FASCINATED EAR2 in maize were shown to be involved in meristem development (Jeong et al., 1999; Taguchi-Shiobara et al., 2001). Some research was also carried out to elucidate the mechanism of RLP-mediated resistance, such as the Cf-mediated hypersensitive response (HR) in tomato (Stulemeijer et al., 2007), and the Vel-mediated signals in tomato and in Arabidopsis (Fradin et al., 2011).
Due to the availability of genome sequences, many RLP genes have been identified from model species: there are 57 predicted RLP genes distributed in 33 loci in Arabidopsis, and 90 RLP genes distributed in 38 loci in rice (Wang et al., 2008). However, despite being an important food crop, there have been few investigations of RLP genes in wheat. Identification of RLP genes is necessary for extensive study and better understanding of their biological functions in wheat. In the present study, an RLP gene, TaRLP1.1, was cloned and characterized as an important gene involved in stripe rust resistance. TaRLP1.1 has a huge potential role in future genetic engineering for development of varieties resistant to stripe rust. Along with TaRLP1.1, a large TaRLP1 family was identified, which may facilitate better understanding of the evolution of RLP genes in wheat.
Materials and methods
Plant materials, pathogen inoculation, and chemical treatments
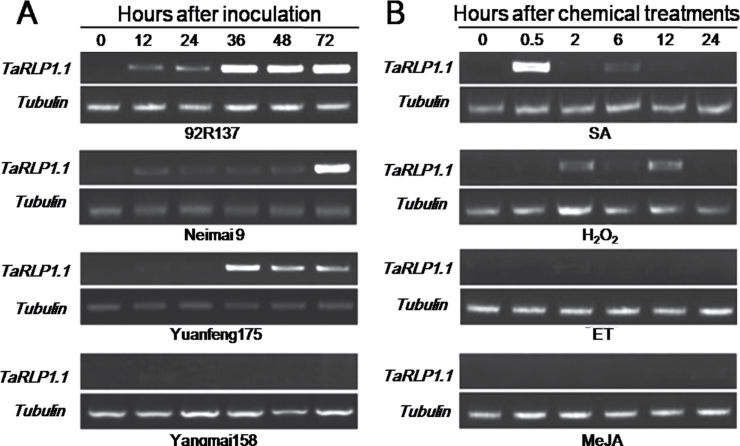
Triticum aestivum L. cv. 92R137, Yuanfeng175, Neimai9, and R236 with the stripe rust resistance gene Yr26, and cv. Yangmai158 susceptible to stripe rust were used in this study. R236 and Yangmai158 are near-isogenic lines. The seedlings were maintained and inoculated with Pst CYR32, the predominant isolate in China (X.J. Wang et al., 2011). Leaves were harvested at 0, 12, 24, 36, 48, and 72h post-inoculation (hpi) for RNA isolation. For chemical treatments, 3-week-old seedlings of cv. 92R137 were sprayed with 5mM salicylic acid (SA), 100 μΜ methyl jasmonate (MeJA), 200 μΜ ethylene (ET), or 7mM hydrogen peroxide (H2O2) in 0.05% Tween-20 solution, and with 0.05% Tween-20 used as control. Leaves were harvested at 0, 0.5, 2, 6, 12, and 24 hpi for RNA isolation. Three independent biological replications were performed.
GeneChip microarray analysis
The wheat GeneChip (part no. 900515, Affymetrix) was chosen to screen the Pst resistance-responsive genes in the Yr26-containing materials. Leaves of 2-week-old seedlings of resistant varieties 92R137 and R236 and susceptible Yangmai158 were collected at 0, 12, and 36 hpi with Pst CYR32, respectively. In this study, the microarray was used only to screen some genes for the downstream functional analysis, so the hybridization of each sample was not repeated. However, two resistant lines were used to increase the credibility of the screened positive genes, and the near-isogenic lines were used to reduce the interference of the genetic background. The transcript profile of each sample was obtained using genechip hybridization conducted in the National Engineering Center for Biochip in Shanghai, China, according to the Affymetrix technical manual. Six separate comparisons (categories) were performed to identify the differentially expressed genes between different samples, namely ‘R236-12h versus R236-0h’, ‘R236-36h versus R236-0h’, ‘92R137-12h versus Yangmai158-12h’, ‘R236-12h versus Yangmai158-12h’, ‘92R137-36h versus Yangmai158-36h’, and ‘R236-36h versus Yangmai158-36h’. For each category, probe sets were filtered for a fold change threshold of >2.0 and a parametric two-way analysis of variance (ANOVA) (P < 0.001) with false discovery rate (FDR) multiple testing correction at 5% was applied to identify statistically significant differentially expressed probe sets. Only when the signal ratio value was >2 was the corresponding probe regarded as up-regulated.
Semi-quantitative RT–PCR and real-time PCR analysis
A 2 μg aliquot of total RNA was used to synthesize the first-strand cDNA by the AMV reverse transcriptase. To evaluate the effectiveness of the gene silencing by VIGS, the expression of TaRLP1.1 was analysed by real-time quantitative reverse transcription–PCR (RT–PCR) with a pair of primers (TaRLP1.1-RT, Supplementary Table S1 available at JXB online) specific to TaRLP1.1, using the 18S rRNA (18rRNA-RT, Supplementary Table S1) as the internal control. The reagents were from Takara (Japan), and the reaction was performed in the ABI 7500fast (ABI, USA). The responses of the TaRLP1.1 gene to pathogen and chemical treatments, the expression of three PR genes (Supplementary Table S1) in the gene-silenced and overexpressing plants, and the expression of TaRLP1.1 in the transgenic plants were analysed by semi-quantitative RT–PCR, using the tubulin gene (Tubulin-RT, Supplementary Table S1) as the internal control. PCR was performed in 25 μl of reaction mixture. The program used was as follows: 3min at 94 °C, followed by 27 cycles at 94 °C for 30 s, 55–60 °C for 30 s, and 72° C for 40s, and then retained at 10 °C.
Isolation of cDNA and genomic sequences of TaRLP1
The fragment of TaRLP1.1, a homologue of Pst-up-regulated probe Ta.22666.2.S1_at in cv 92R137 and R236, was obtained by RT–PCR using the primers (Ta.22666.2.S1-EST, Supplementary Table S1 at JXB online) designed according to the probe. Rapid amplification of cDNA ends (RACE) was then conducted to obtain the full-length cDNA of TaRLP1.1 from cv. 92R137 inoculated for 36h using the SMART™ RACE cDNA Amplification Kit (Clontech). Primers for 5′-RACE and 3′-RACE (TaRLP-RACE, Supplementary Table S1) were designed according to the fragment of TaRLP1.1. To obtain the full-length TaRLP1.1, RT–PCR was performed with a pair of primers (TaRLP1.1-FL, Supplementary Table S1) designed based on the sequence of the 5′ and 3′ cDNA ends. The homologues of TaRLP1.1 in cv. Neimai9 and Yuanfeng175 were also amplified using the above primers. The genomic sequence of TaRLP1.1, and of its homologues in cv. 92R137 and Yangmai158, was obtained with primers (TaRLP1.1-ORF, Supplementary Table S1) designed based on the full-length cDNA of TaRLP1.1.
Sequence analysis and phylogenetic tree construction
The cDNA sequence was analysed using the software of the DNAMAN, BLAST, and ORF Finder. Protein sequences were analysed using PROSITE Scan (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_prosite.html) for prediction of the conserved domains and motifs, SignalP (http://www.cbs.dtu.dk/services/SignalP/) for prediction of the signal peptide, and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) for prediction of the transmembrane region. The MEGA 4.0 software was used for construction of the phylogenetic tree.
Subcellular localization of TaRLP1.1
The cDNA fragments containing the TaRLP1.1 open reading frame (ORF) were amplified by PCR with the specific primers containing NcoI and SacI enzyme sites (TaRLP1.1-SL, Supplementary Table S1 at JXB online), and then inserted to the 5′ end of the green fluorescent protein (GFP) coding region in the pCaMV35S:GFP vector. The plasmid DNAs of pCaMV35S:TaRLP1.1-GFP and pCaMV35S:GFP were transformed into onion epidermal cells by particle bombardment as described (Liu et al., 2012), respectively. The GFP signal was detected using a confocal microscope (Leica, Germany).
Plasmid construction and functional analysis of TaRLP1.1 by VIGS
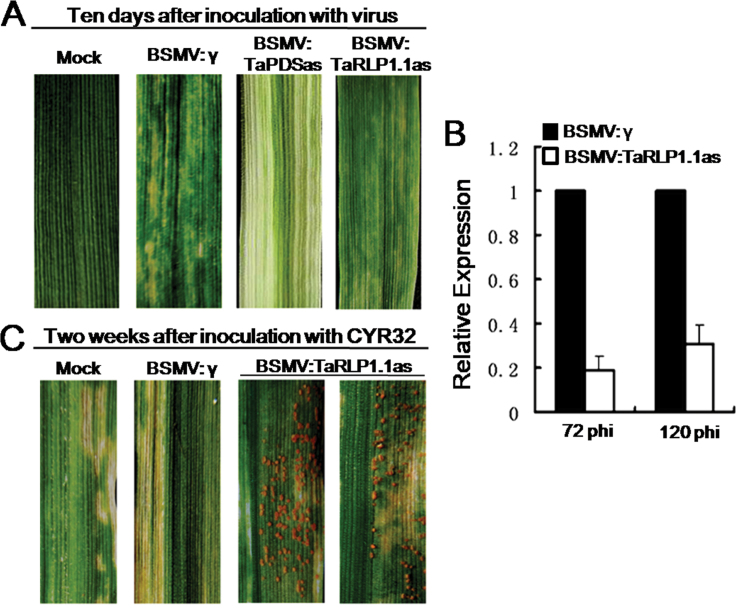
VIGS mediated by Barley stripe mosaic virus (BSMV) (Zhou et al., 2007) in 92R137 was adopted to investigate the potential involvement of TaRLP1.1 in the HR to Pst. A 186bp cDNA fragment covering both part of the coding region and part of the 3′-untranslated region (UTR) was amplified with primers (TaRLP1.1-VIGS, Supplementary Table S1 at JXB online), and then was inserted in reverse orientation into the BSMV:γ vector to form the recombinant vector BSMV:TaRLP1.1as. The second fully expanded leaves of the seedlings were infected with virus BSMV:TaRLP1.1as, using BSMV:TaPDSas and BSMV:γ as the controls. Three independent sets of inoculations were performed, with a total of 72 seedlings inoculated for each BSMV virus. For the control, 24 seedlings were mock-inoculated with 1× GPK buffer. At 10–12 d after virus inoculation, the fourth leaves were further treated with Pst CYR32. At 15 d after pathogen inoculation, when extensive fungal growth was visible in Yangmai158, the infection types of stripe rust were examined in the gene-silenced leaves of 92R137. The fourth leaves were also sampled at 72 and 120 hpi with Pst for histological observation and gene silencing efficiency analysis.
Plasmids construction and wheat transformation
The ORF of TaRLP1.1 was amplified using the primers containing BamHI and SacI sites (TaRLP1.1-WT, Supplementary Table S1 at JXB online), and was then inserted into the plant expression vector pBI220 under the control of the 35S promoter to form the recombinant vector pBI220:TaRLP1.1. Because there is no selective marker gene available in the vector pBI220, another plant expression vector pAHC25, containing the β-glucuronidase gene (GUS) and the herbicide tolerance gene (Bar) driven by the ubiquitin (Ubi) promoter, was used as the co-transformation vector. The pBI220:TaRLP1.1- and pAHC25-co-transformed wheat plants were produced by particle bombardment of calli cultured from immature embryos of susceptible Yangmai158 as described (Xing et al., 2008). For screening the TaRLP1.1-transformed positive transgenic plants from the 238 regenerated plants, the root tips of 20 T1 seeds from each T0 plant were first stained with X-gluc (Xing et al., 2008), and the target TaRLP1.1 was then detected by PCR using the DNA of the corresponding T0 plant whose T1 seeds showed GUS activity. The target gene was also detected in the T1 generation of the positive T0 plants. To avoid interference from the endogenous homologous genes in Yangmai158, the forward primer (35S-F, Supplementary Table S1) was located in the 35S promoter, and the reverse primer (TaRLP1.1-R, Supplementary Table S1) was located in the target TaRLP1.1 gene.
Evaluation of stripe rust resistance of transgenic plants
Stripe rust resistance of T1 transgenic plants was evaluated via inoculation with urediniospores of Pst CYR32 using cv. Yangmai158 and 92R137 as the controls at the two-leaf stage as described by X.J. Wang et al. (2011). About 30 seedlings of the T1 generation from each positive T0 line were inoculated with CYR32 and kept in a dew chamber at 10 °C for 24h in darkness, and then grown in a growth chamber at 16/8h light/dark with diurnal temperatures gradually changing from 4 °C at 02:00h to 20 °C at 14:00h. Infection type data were recorded 18 d after inoculation based on the 0–9 scale described by X.J. Wang et al. (2011). Infection types of 0–3 were considered resistant, while those >5 were considered susceptible.
Histological observations of fungal growth and host response
Wheat leaves infected with BSMV were sampled at 72 and 120 hpi with Pst and stained as described (X.J. Wang et al., 2011). Bleached leaf segments were examined with an Olympus BX-51 microscope (Olympus, Japan) for infection sites, and only those sites where substomatal vesicles had formed in the stomata were considered to be successfully penetrated. Autofluorescence of the mesophyll cells in the infected sites was measured as phenolic autofluorogen accumulation area by epifluorescence microscopy (excitation filter, 485nm; dichromic mirror, 510nm; and barrier filter, 520nm). The autofluorescence areas, hyphal length, and hyphal branches were calculated by DP-BSW software, and the final data of each index were the mean of at least 60 infection sites for each of the five randomly selected leaf segments per treatment. Standard deviations and a paired sample t-test for statistical analysis were performed with SAS software.
Results
Screening for Pst-responsive genes in Yr26-containing wheat
To screen the Pst resistance-related genes in wheat containing the stripe rust resistance gene Yr26, microarrays were used to obtain the transcriptomes of seven samples, including two resistant varieties 92R137 and R236, and one susceptible cv. Yangmai158, with or without Pst inoculation as described (X.J. Wang et al., 2011). The data of six differentially expressed probes (2-fold up- and down-regulated and P-value ≤0.001) were consequently produced. Bioinformatics analysis showed that in total 35 probes were shared by all of the above six data groups, with 17 up-regulated and 18 down-regulated; thus 35 probes showed a differentially expressed pattern not only between Pst-inoculated samples of resistant and susceptible lines, but also between Pst-inoculated and uninoculated samples of the resistant lines. Among the 17 probes, Ta.22666.2.S1 and TaAffx.55699.1.S1_at were two putative RLP genes (Supplementary Table S2 at JXB online). Although there was no homology between Ta.22666.2.S1 and TaAffx.55699.1.S1, these two probes showed similarity to the different parts of the same genes in GenBank, indicating that they represented the same RLP gene. RLP genes have been reported to be involved in the defence responses to different pathogens; however, no RLP has been identified in wheat, so the screened RLP gene was selected as of interest for investigation of its function in stripe rust resistance.
Cloning and structural analysis of the TaRLP1.1 gene from stripe rust-resistant variety 92R137
The homologue of Ta.22666.2.S1_at in cv. 92R137 inoculated with CYR32 was obtained by RT–PCR using the primers (Ta.22666.2.S1-EST, Supplementary Table S1 at JXB online) designed according to the probe Ta.22666.2.S1_at. The amplified fragment shared 99% similarity with Ta.22666.2.S1_at, so the homologue in cv. 92R137 was designated as TaRLP1.1, since it showed high homology with other RLP genes. The full-length sequence of TaRLP1.1 was further obtained by RACE from cv. 92R137, which was then verified by RT–PCR using the primers (TaRLP1.1-FL, Supplementary Table S1) designed based on the 5′- and 3′-UTR. The full length of the TaRLP1.1 was 1273bp, which contained a putative 801bp ORF coding for 266 amino acids.
The predicted primary structure of TaRLP1.1 comprised several conserved domains which have been identified in other RLPs (Supplementary Fig. S1A, B at JXB online). Domain ‘a’ is a signal peptide, a hydrophobic N-terminus found in TaRLP1.1, which may target the protein to the cytoplasmic membrane. Domain ‘b’ is an LRR region with three imperfect copies of LRR; each one is a 23–25 amino acid consensus sequence [xLxxLxLSxNxLSGxIP] associated with protein–protein interactions (Jones and Jones, 1997). The presence of a glycine within the LRR is considered as the characteristic of extracytoplasmic proteins with the ability to recognize the extracellular pathogen ligands (Jones et al., 1994). As frequently observed in membrane-spanning proteins, a hydrophobic sequence with a predicted α-helix secondary structure (domain c, containing GxxxG-type motifs) is the putative transmembrane region, following a negatively charged extracytoplasmic domain (domain d) and followed by a positively charged cytoplasmic tail. In the short cytoplasmic tail domain, there is a ‘Yxxφ’ signal sequence, where ‘φ’ is an amino acid with a hydrophobic side chain, and this motif has been shown to stimulate receptor-mediated endocytosis and degradation of mammalian cell surface receptors (Ron and Avni, 2004; Kruijt, et al., 2005).
Identification and chromosome location of different members of the TaRLP1 family in resistant cv. 92R137 and susceptible cv. Yangmai158
To determine whether TaRLP1.1 belonged to a gene family and whether there was a difference in TaRLP1.1 between the resistant varity 92R137 and susceptible cv. Yangmai158, PCR was conducted using primers (TaRLP1.1-ORF, Supplementary Table S1 at JXB online) to identify the homologues of TaRLP1.1 in the DNA of both materials. In cv. 92R137, in addition to TaRLP1.1, another nine TaRLP1 genes of different sizes were identified; and in cv. Yangmai158, eight TaRLP1 genes of different sizes were identified (Supplementary Table S3). Sequence analysis showed that TaRLP1 738, TaRLP1 800-1, TaRLP1 801-2, TaRLP1 804-1, and TaRLP1 813 existed in both cv. 92R137 and Yangmai158; however, other genes were identified to be specific to each cultivar.
The specific primers were designed based on the sequence of the above genes (Supplementary Table S1 at JXB online) and were used for gene chromosome location in cv. Chinese Spring-derived nulli-tetrasomic lines. TaRLP1.1 was located to chromosome 3D, TaRLP1 804-1 to 3B, and TaRLP1 813 to 3A (Supplementary Fig. S2). The result indicated that the TaRLP1 gene family may be located in the homoeologous group 3 chromosomes.
Phylogenetic analysis of TaRLP1 family members
The result of the multisequence alignment and phylogenetic analysis of the RLP genes from common wheat suggested that TaRLP1 is a rapidly evolving gene family due to single nucleotide variation, deletion or insertion, and recombination (Supplementary Fig. S3 at JXB online) (Dixon et al., 1998). For example, compared with TaRLP1 801, TaRLP1 800 was produced by one missing nucleotide, TaRLP1 804 was the outcome of a three-nucleotide (TCA) insertion into TaRLP1 801, and TaRLP1 813 was the product of a nine-nucleotide insertion (CCTATCC and TAA) into TaRLP1 804. Gene duplication occurred on TaRLP1 801 and TaRLP1 804, resulting in more members of both types of genes in wheat (Supplementary Fig. S4).
Recombination between the homologous regions of different members of one gene cluster is a common event that produces new genes. In this study, TaRLP1 800-2 was the product of recombination between TaRLP1 738 and TaRLP1 804-4: the first half of TaRLP1 800-2 was identical to TaRLP1 738, and the second half was identical to TaRLP1 804-4.
Polymorphism of the DNA sequence leads directly to the variation in the putative protein sequences (Supplementary Fig. S5 at JXB online). Compared with TaRLP1.1, TaRLP1800-1 and TaRLP1800-2 are two truncated proteins due to one nucleotide missing and pre-stop in the reading frame; while in TaRLP1738 there were 22 amino acids missing. Although many variations were detected, the effect on gene function needs to be elucidated.
As to the homology of TaRLP1.1 with other putative RLPs, TaRLP1.1 showed the highest similarity (>70% identity) to its homologues from other monocots (Supplementary Fig. S6A at JXB online); however, it showed the lowest sequence similarity to Arabidopsis AtRLP44 (38% identity) (Supplementary Fig. S6B).
Expression analysis of the TaRLP1 family in the resistant cv. 92R137 and susceptible cv. Yangmai158
To determine how many TaRLP1 genes were expressed in both the resistant and susceptible plants, the primers used to amplify the homologous gene from 92R137 and Yangmai158 were also used to amplify the expressed genes from the cDNA of the cv. 92R137 and Yangmai158. There were 22 positive clones from the cv. 92R137 sequenced, and three TaRLP1 genes were found to be expressed: TaRLP1.1 (19 of 22), TaRLP1800-1 (two of 22), and TaRLP1813 (one of 22). Twenty positive clones from cv. Yangmai158 were sequenced, and two RLP genes, TaRLP1 800-1 (19 of 20) and TaRLP1 801-2 (one of 20), were detected as expressed. In two other resistant cultivars, Neimai 9 and Yuanfeng 175, TaRLP1.1 was also detected as the predominant transcript as in cv. 92R137. Sequence analysis showed that TaRLP1 800-1 produced a truncated 57 amino acid protein due to a missing nucleotide compared with TaRLP1.1. TaRLP1 801-2 also produced a 266 amino acid protein; however, the similarity with TaRLP1.1 was only 88%. TaRLP1 813 produced a 270 amino acid protein, and similarity with TaRLP1.1 was 81% (Supplementary Fig. S5 at JXB online).
TaRLP1.1 was the dominant expressed gene specifically in the Yr26-containing cv. 92R137, Neimai 9, and Yuanfeng 175; thus, it was proposed that TaRLP1.1 may play an important role in the Yr26-mediated resistance pathway.
Responses of TaRLP1.1 to Pst infection and chemical stress treatments
Because TaRLP1.1 was the dominant transcript in cv. 92R137 and another two Yr26-containing materials based on expression analysis, to examine further whether the expression of TaRLP1.1 was really induced by Pst, semi-quantitative RT–PCR analysis was conducted using primers (TaRLP1.1-RT, Supplementary Table S1 at JXB online) specific to TaRLP1.1. It was found that TaRLP1.1 was significantly up-regulated in cv. 92R137 at 12 hpi with Pst, and reached a peak at 36 hpi (Fig. 1A). In another two Yr26-containing cultivars, Neimai 9 and Yuanfeng175, TaRLP1.1 was also induced by CYR32 and reached a peak at 72 and 36 hpi, respectively. However, no transcript was detected in susceptible Yangmai158. Therefore, TaRLP1.1 may play an important role in resistance to stripe rust fungus, and perhaps take part in the signal transduction mediated by Yr26.
Fig. 1.
Evaluation of expression changes of TaRLP1.1 in response to biotic (stripe rust infection), and chemical treatments (SA, ET, MeJA, and H2O2) stresses in wheat seedlings by semi-quantitative RT–PCR. The tubulin gene was used as the internal control. (A) TaRLP1.1 was up-regulated by Pst CYR32 in resistant wheat (cv. 92R137, Neimai 9, and Yuanfeng 175), while it was not induced in susceptible cv. Yangmai158. (B) TaRLP1.1 was induced by SA and H2O2, but not by ET and MeJA.
Seedlings of cv. 92R137 were treated with exogenous chemicals—SA (5mM), MeJA (100 μΜ), ET (200 μΜ), and H2O2 (7mM)—for analysis of the responses of TaRLP1.1 by semi-quantitative RT–PCR. The TaRLP1.1 was substantially up-regulated by both SA and H2O2 treatments; however, there was no response to the MeJA and ET (Fig. 1B).
Subcellular localization of TaRLP1.1–GFP fusion protein
The subcellular localization of TaRLP1.1 was studied using the constructed fused protein TaRLP1.1–GFP driven by the CaMV 35S promoter by the transcient expression system in onion epidermal cells. The result showed that the green signal produced by GFP protein alone was uniformly distributed throughout the cell; however, the green signal produced by the fused protein TaRLP1.1–GFP was clearly localized only to the cytomembrane (Supplementary Fig. S7 at JXB online). The subcellular localization indicated that TaRLP1.1 was localized to the membrane as predicted by its transmembrane ‘GxxxG’ motif and TaRLP1.1 may act as a receptor to receive the stimuli from outside of the cell in the resistance pathway.
Silencing of TaRLP1.1 compromised resistance to Pst and suppressed expression of PR genes in cv. 92R137
To evaluate the function of TaRLP1.1, a BSMV-based VIGS approach (Zhou et al., 2007) was used to silence the genes in the stripe rust resistance cv. 92R137. Fifteen days after BSMV:TaPDSas inoculatation, leaf bleaching was observed due to PDS gene silencing, indicating that the VIGS system for gene functional analysis was successfully silencing the target gene (Fig. 2A). The efficiency of VIGS was analysed by quantitative RT–PCR using the fourth leaves of wheat seedlings, and expression levels of TaRLP1.1 were reduced by 81% and 69% in leaves challenged with BSMV:TaRLP1.1as at 72 and 120 hpi, respectively, compared with the control inoculated with BSMV:γ (Fig. 2B). At 2 weeks after Pst isolate CYR32 inoculation, a robust HR was observed on leaves infected with BSMV:γ, or mock-inoculated only with GPK buffer. However, fungal sporulation emerged only on fourth leaves previously infected with BSMV:TaRLP1.1as (Fig. 2C).
Fig. 2.
Functional characterization of TaRLP1.1 by the BSMV-based VIGS approach. (A) Successful virus infection. Mild chlorotic mosaic symptoms were observed on the fourth leaves inoculated with BSMV:γ or BSMV:TaRLP1.1as at 10 days post-inoculation (dpi). Photobleaching displayed on leaves infected with BSMV:TaPDSas at 15 dpi. Typical photographs were taken at 15 d after virus inoculation. (B) The expression level of TaRLP1.1 was decreased dramatically at both 72h and 120h after CYR32 inoculation in the BSMV:TaRLP1.1as-infected plants. (C) The resistance to Pst CYR32 was compromised in the BSMV:TaRLP1.1as-infected plants, and the rust sori were observed in the TaRLP1.1-silenced leaves. The photographs were taken 2 weeks after fungal inoculation.
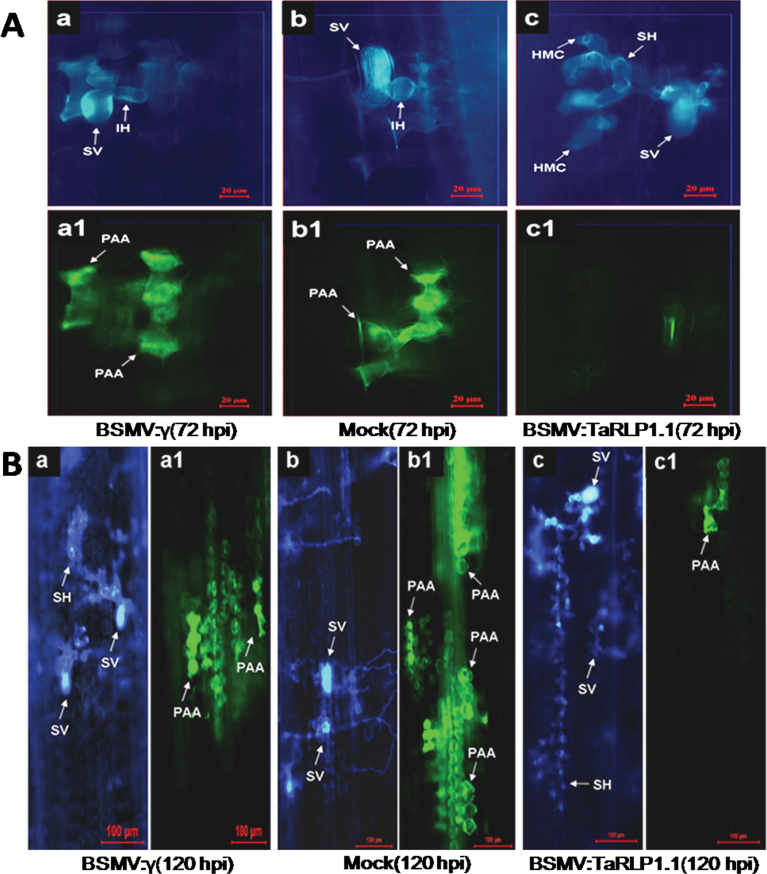
In the incompatible host–pathogen interaction, autofluorescence due to the accumulation of phenylpropanoid compounds is common, and usually correlated with the HR. Because the incompatible interaction of 92R137 and Pst CYR32 was accompanied by a sharp HR, an investigation was carried out to determine what would happen to the HR or phenolic autofluorogen accumulation when the compatible interaction formed in the TaRLP1.1-silenced 92R137. Detailed histological changes were also observed at 72h and 120h after Pst CYR32 inoculation of the BSMV:TaRLP1.1as-, BSMV:γ-, and mock-infected plants, and four parameters were measured: proportion of the infection sites producing phenolic autofluorogen accumulation, autofluorescence area, hyphal length, and hyphal branches (Fig. 3). In the control plants infected with BSMV:γ or mock infected, a large proportion of infection sites producing phenolic autofluorogen accumulation could be detected (Fig. 3A), and a large autofluorescence area could be observed at 72h [Fig. 4A (a1, b1)] or 120h [Fig. 4B (a1, b1)] after Pst CYR32 inoculation (Fig. 3B). In test plants infected with BSMV:TaRLP1.1as, the proportion of infection sites producing phenolic autofluorogen accumulation and the autofluorescence area were significantly (P < 0.001) lower than for control leaves at 72h [Figs 3A, B, 4A (c1)] and 120h [Figs 3A, 4B, B (c1)] after CYR32 inoculation, and hyphal length was significantly (P < 0.001) longer than for the control [Figs 3C, 4A (c), 4B (c)]. However, there was no significant difference (P < 0.05) in hyphal branching between control and test plants (Fig. 3D). These results indicated that knocking down TaRLP1.1 decreased the autofluorescence areas generated at infection sites, and the HR was compromised.
Fig. 3.
Quantitative comparisons of the percentage of the infection sites producing phenolic autofluorogen accumulation, average area of autofluorescence, and average linear length of hyphae at 72 and 120 hpi after CYR32 inoculation on leaves pre-infected with BSMV:TaRLP1.1as or BSMV:γ, or mock infected. The data from each treatment were calculated from at least 60 infection sites on each seedling. Statistical significance was determined using ANOVA and t-test. Mean values followed by different letters are significantly different from each other (b* indicates P < 0.001 and b indicates P < 0.05). (A) Percentage of infection sites producing phenolic autofluorogen accumulation. (B) Average area of autofluorescence in the mesophyll cells. (C) Average hyphal length from the base of substomatal vesicles to hyphal tips. (D) Average number of hyphal branches.
Fig. 4.
Histological observation in wheat cv. 92R137 at 72 hpi (A) and 120 hpi (B) after Pst CYR32 inoculation in the BSMV:γ- (a, a1), mock- (b, b1), and BSMV:TaRLP1.1as- (c, c1) pre-infected leaves. The same infection site was observed by white fluorescence microscopy for fungal growth (a, b, c) and epifluorescence microscopy for hypersensitive cell death (a1, b1, c1). (A) The spreading of secondary hyphae was suppressed (a, b), and mesophyll cells at the infection sites showed strong phenolic autofluorogen accumulation (a1, b1). However, secondary hyphae were formed (c) and autofluorescence was not obvious (c1) in the BSMV:TaRLP1.1as-infected leaves. Bars=20 μm. (B) Secondary hyphae were longer in the BSMV:TaRLP1.1as-infected leaves (c) than in the BSMV:γ- (a) and mock- (b) infected leaves. However, the area of autofluorescence was smaller in the BSMV:TaRLP1.1as-infected leaves (c1) than in the BSMV:γ- (a1) and mock- (b1) infected leaves. Bars=100 μm. SV, substomatal vesicle; IH, infection hypha; HMC, haustorial mother cell; SH, secondary hyphae; PAA, phenolic autofluorogen accumulation.
The loss of resistance usually correlated with the decreased expression of some defence-related genes. Some pathogenesis-related genes were tested in the TaRLP1.1-silenced plants and controls. Expression levels of TaPR1, TaPR2, and TaPR5 were decreased in TaRLP1.1 knocked-down leaves at 72 or 120 hpi, in contrast to BSMV:γ-infected control (Supplementary Fig. S8 at JXB online). PR1, PR2, and PR5 are usually considered to be marker genes of the SA pathway, so their decreased expression in TaRLP1.1-silenced plants implied some damage to the SA pathway.
Overexpression of TaRLP1.1 enhanced the stripe rust resistance of susceptible Yangmai158
The function of TaRLP1.1 was further studied by developing TaRLP1.1-overexpressing transgenic plants. Susceptible cv. Yangmai158 is a near-isogenic line of Yr26-containing resistant cv. R236, and cv. Yangmai158 lacks TaRLP1.1. Additionally, cv. Yangmai158 is a good variety for tissue culture, and so TaRLP1.1 was transformed into cv. Yangmai158 by particle bombardment in this study. The pBI220:TaRLP1.1 was co-transformed with the GUS gene-containing vector pAHC-25. GUS staining using the root tips of the T1 generation seeds and target gene detection in the T0 generation was used to identify positive transgenic plants—22 T0 plants were determined to be positive transgenic plants (Supplementary Fig. S9A, B at JXB online). Expression analysis in the T1 generation showed that TaRLP1.1 was successfully overexpressed in the transgenic plants (Supplementary Fig. S9C).
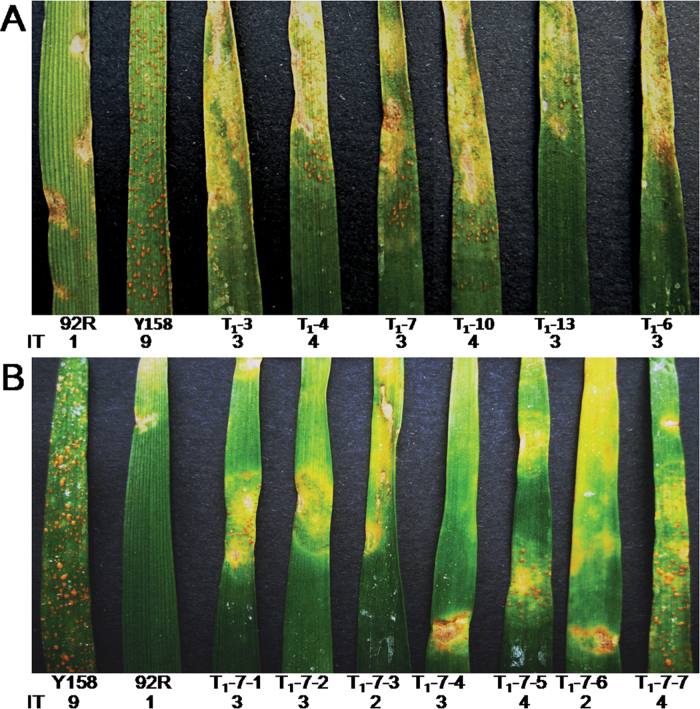
Evaluation of the resistance to Pst CYR32 was performed using the T1 plants of each positive T0 plant. The second leaves of plants were inoculated with Pst isolate CYR32, and 2 weeks later robust hypersensitive cell death was observed—the transgenic plants showed high resistance to CYR32 (Fig. 5A). Because of heterozygosity of TaRLP1.1 in the T0 generation, the TaRLP1.1 was segregated in T1 generation seedlings, and only those plants transformed with TaRLP1.1 showed increased resistance with resistance of grade 2–4, while the recipient Yangmai158 showed the highest level of susceptibility of grade 9 (Fig. 5B; Supplementary Fig. S10) at JXB online.
Fig. 5.
Stripe rust resistance evaluation of TaRLP1.1-transformed plants. (A) T1 transgenic plants from different T0 plants showed a varied level of resistance to Pst CYR32. (B) T1 transgenic plants from the T0-7 showed a high level of resistance to Pst CYR32. IT, infection type; 92R, cv. 92R137; Y158, cv. Yangmai158.
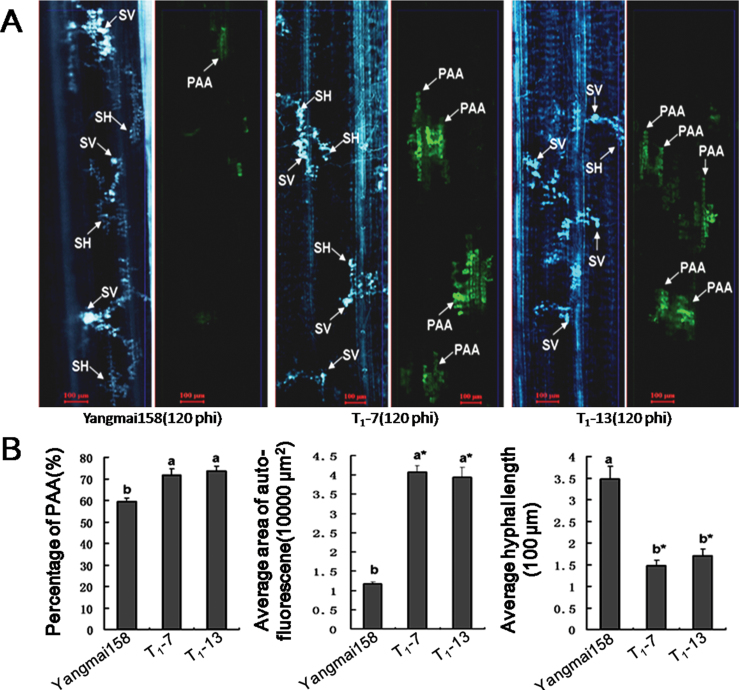
To determine whether the HR contributed to increased resistance in the TaRLP1.1 transgenic plants, the proportion of infection sites producing phenolic autofluorogen accumulation, the autofluorescence area, and the hyphal length at the infection site were investigated by histological observations at 120h after CYR32 inoculation. A higher percentage of infection sites producing phenolic autofluorogen accumulation (73.8%), a larger area of autofluorescence (40 798.8 μm2), and shorter hyphae (147.6 μm) at infection sites were observed in transgenic plants compared with corresponding values of 59.5%, 11 754.2 μm2, and 348.2 μm in Yangmai158 (Fig. 6A, B). It is proposed that the increased resistance of transgenic plants was closely related to the HR.
Fig. 6.
Histological observation of the hyphal development and phenolic autofluorogen accumulation at the infection sites between Pst CYR32 and TaRLP1.1-overexpressing transgenic plants at 120 hpi using the susceptible cv. Yangmai158 as the control. (A) In cv. Yangmai158, the hyphae elongated normally, and there was almost no autofluorescence at the infection site. However, in transgenic plants, the fungal growth was slower, and autofluorescence was obvious at the infection sites. The same infection site was observed by white fluorescence or epifluorescence microscopy. Bars=100 μm. SV, substomatal vesicle; SH, secondary hyphae; PAA, phenolic autofluorogen accumulation. (B) Quantitative comparisons of the percentage of infection sites producing phenolic autofluorogen accumulation, the average area of autofluorescence, and average linear length of hyphae emerging on the leaves of infection sites. Means (± SE) were calculated using the measurements from six seedlings, and at least 30 infection sites for each seedling. Significance was determined according to the paired sample t-test method (b* indicates P < 0.001 and b indicates P < 0.05).
In addition, TaPR1, TaPR2, and TaPR5 were induced in transgenic wheat leaves challenged by CYR32 at 72 and 120 hpi (Supplementary Fig. S11 at JXB online). These results suggest that TaRLP1.1 played a positive role in plant defence reactions and enhanced resistance to the Pst fungus.
Discussion
Plants are exposed to diverse pathogen infection, and various responses are activated to deal with such attacks. Among the complex defence networks, the signal pathways mediated by RLP genes have recently been found to be required in the response to pathogen attacks in Arabidopsis and tomato. However, no RLP genes have been studied in monocots. Wheat is one of the world’s most important crops and is adversely affected by Pst. In this study, a TaRLP1.1 gene was isolated from a stripe rust-resistant variety. The putative protein structure of TaRLP1.1 indicated that TaRLP1.1 was a typical RLP, and strong evidence suggested that TaRLP1.1, a member of the RLP gene family, was involved in the defence response to stripe rust.
The role of TaRLP1.1 in stripe rust resistance of wheat
TaRLP1.1 was induced in all four tested and genetically diverse Yr26-containing resistant wheat varieties after Pst inoculation, but not in susceptible cv. Yangmai158. Furthermore, a gene silencing experiment indicated that TaRLP1.1 was required for the full resistance to Pst, while the complementary experiment showed that overexpression of TaRLP1.1 could dramatically improve resistance. Histological analysis implied that it had important roles in HR regulation and activation of PR genes. The above results indicate that TaRLP1.1 was involved in the stripe rust resistance.
SA, JA, and ET are signalling molecules to regulate complex defence responses by inducing PR genes, and the SA signal pathway is generally correlated with biotrophic pathogen defences and HR (Spoel et al., 2008). In the present study, strong HR could be induced, and the PR genes (PR1, PR2, and PR5—the marker genes of SA) were up-regulated in the Yr26-containing wheat after the biotrophic Pst infection; thus the SA pathway probably participated in defence. TaRLP1.1 expression was dominantly induced by exogenous SA; meanwhile, the extent of HR and the expression of PR1, PR2, and PR5 changed along with the expression abundance of TaRLP1.1. This suggests that TaRLP1.1 participated in the SA-mediated signalling pathway in the wheat–stripe rust interactions.
The Cf type RLP genes were characterized as disease resistance genes (Jones et al., 1994; Dixon et al., 1996, 1998). In the present study, the silencing of TaRLP1.1 resulted in decreased resistance of cv. 92R137; however, the level of susceptibility was not as serious as that of the susceptible cv. Yangmai158, Conversely, overexpression of TaRLP1.1 increased the resistance of cv. Yangmai158, but not to a level as high as that of cv. 92R137. Thus, gene expression analysis and functional evaluation indicated that TaRLP1.1 was a potential downstream factor involved in the Yr26-mediated signal pathway. In addition to the signal pathway containing TaRLP1.1, there are probably other branches of signal pathways contributing to full resistance to Pst mediated by Yr26.
CYR32 is the most frequently identified isolate of Pst, accounting for a frequency of 29.6% throughout China. More importantly, with broad virulence and high fitness, CYR32 will probably cause widespread epidemics of stripe rust in the near future (Kang et al., 2010). The identification of TaRLP1.1 as a positive regulator of the HR and an enhancer of resistance to Pst CYR32 may provide molecular targets for the development of varieties resistant to stripe rust. The spectrum of resistance will be determined in the higher generations of TaRLP1.1 transgenic plants using more Pst isolates.
Protein–protein interaction cascades are crucial for signalling pathways. Like other resistance proteins, the LRR domain of RLPs is widely involved in disease resistance by binding to pathogen elicitors or other proteins in the plants to activate downstream signals (Kruijt et al., 2005). The ‘GxxxG’ conserved motif in the transmembrane regions of RLP genes is likely to be involved in protein–protein interactions, as found in the study of the Cf-RLP genes, and this protein interaction plays a critical role in RLP signal transduction from the extracellular to the cytoplasmic space (Stulemeijer et al., 2007).The binding protein and the mechanism of the resistance mediated by TaRLP1.1 remain to be elucidated. Thus, transgenic plants of TaRLP1.1 fused with a Myc-tag in the C-terminus are being produced to facilitate the search for the interacting proteins of TaRLP1.1.
The evolution of TaRLP1 genes in wheat
The disease resistance genes are usually distributed on chromosomes as clusters, and this has been widely shown concerning the NBS-LRR and some RLK resistance genes. For RLP resistance genes, a similar manner of gene organization was identified in several cases; for example, the Cf-2 gene cluster in Lycopersicon (now Solanum) pimpinellifolium included three Hcr2 genes (Dixon et al., 1996), and the Cf-5 gene cluster in Lycopersicon esculentum (now Solanum lycopersicum) var. cerasiforme included four members (Dixon et al., 1998). In Arabidopsis, 57 RLP genes were distributed on 33 loci, and in rice 90 RLP genes were distributed on 38 loci (Wang et al., 2008). In the present study, 10 members of TaRLP1 in resistant cv. 92R137 and eight members in susceptible cv. Yangmai158 were identified, and it was proposed that the TaRLP1 family clustered on the homoeologous group 3 chromosomes.
To adapt to the quick evolution of the pathogen, disease resistance genes often undergo dramatic evolution, such as duplication, deletion/insertion, and recombination, to produce new members to recognize the new pathogens. The gene structure and function analysis of Cf genes showed that gene duplication and recombination played an important role in the evolution of RLP genes. In some cases, the duplicated gene preserved the function of the original genes (Dixon et al., 1996). In L. pimpinellifolium, Hcr9 is the product of recombination between Hcr9-9D and Cf-9 (Kruijt et al., 2004). In the TaRLP1 family, there were identical members shared by the two materials; however, there were also some members specific to the individual materials. Gene duplication, deletion/insertion, and recombination were identified in the gene evolution of the TaRLP1 family.
Gene clusters are considered to be the outcome of co-evolution of host and pathogen, and different members of a gene cluster may be aimed at different virulent races (Bhullar et al., 2009). In a gene cluster, some members confer resistance to available pathogen races, while others have no identified resistance function and represent a reservoir to deal with new emerging races or producing new genes (Kruijt et al., 2004). For example, among the four members in the Cf-5 family of cherry tomato, Hcr2-5C is the only determinant of Avr5 recognition (Dixon et al., 1998). In this study, a number of TaRLP1 members were identified in wheat, but only two and three were expressed in cv. Yangmai158 and 92R137 under the tested conditions, respectively. It will be interesting to study the expression pattern of the other TaRLP1 genes, and their response to stimuli.
In conclusion, strong evidence in the present study suggested that TaRLP1.1 plays an important role in the stripe rust defence response in the monocot wheat. Furthermore, the rapid evolution of the TaRLP1 family was revealed. This work supplied a good gene resource for genetic engineering in wheat breeding.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The primary structure and conserved domains of TaRLP1.1.
Figure S2. Chromosomal location of TaRLP1.1, TaRLP 1804-1, and TaRLP1813 with specific primers using the nulli-tetrasomic (NT) lines derived from cv. Chinese Spring.
Figure S3. Phylogenetic analysis of the DNA sequence of TaRLP1 genes from common wheat using MEGA 4.0.
Figure S4. The multisequence alignment of the identified RLP genes from the stripe rust-resistant wheat cv. 92R137 and susceptible cv. Yangmai158.
Figure S5. The multisequence alignment of the deduced proteins of the identified RLP genes from the stripe rust-resistant wheat cv. 92R137 and susceptible cv. Yangmai158.
Figure S6. Sequence alignment and phylogenetic analysis of TaRLP1.1 and other putative RLP proteins using DNAMAN.
Figure S7. Subcellular localization of TaRLP1.1 protein.
Figure S8. Expression analysis by semi-quantitative RT–PCR of three pathogenesis-related genes, TaPR1, TaPR2, and TaPR5, at both 72 and 120h after CYR32 inoculation in the BSMV:TaRLP1.1as-infected plants and the BSMV-infected control plants.
Figure S9. Identification of positive transgenic plants by GUS staining, gene detection, and gene expression analysis.
Figure S10. Positive TaRLP1.1 transgenic plants showed robust hypersensitive response to Pst 15 d after inoculation.
Figure S11. Expression analysis by semi-quantitative RT–PCR of three pathogenesis-related genes TaPR1, TaPR2, and TaPR5, at both 72h and 120h after CYR32 inoculation in TaRLP1.1-overexpressing transgenic plants, the Yr26-containing cv. 92R137 and the susceptible recipient Yangmai158.
Table S1. Designed primers.
Table S2. The fold up-regulation of Ta.22666.2.S1_at and TaAffx.55699.1.S1_at between the different samples of tests and the controls.
Table S3. The genes identified in resistant cv. 92R137 and susceptible cv. Yangmai158.
Acknowledgements
We thank Professor Wenbiao Shen for critically reviewing the manuscript. This research was supported by grants from the Important National Science & Technology Specific Projects of Transgenic Research (2009ZX08009-051B), the Program for New Century Excellent Talents in University (NCET-10–0496), the National Basic Research Program of China (2009CB118304), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Belfanti E, Silfverberg-Dilworth E, Tartarini S, et al. 2004. The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proceedings of the National Academy of Sciences, USA 101, 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B. 2009. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proceedings of the National Academy Sciences, USA 106, 9519–9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG. 1998. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. The Plant Cell 10, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD. 1996. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459. [DOI] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP. 2011. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiology 156, 2255–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DL, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J. 2009. A kinase START gene confers temperature dependent resistance to wheat stripe rust. Science 323, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. 1999. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. The Plant Cell 11, 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Jones JD. 1997. The role of leucine-rich repeat proteins in plant defences. Advances in Botanical Research 24, 89–167. [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD. 1994. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Kang ZS, Zhao J, Han DJ, Zhang HC, Wang XJ, Wang CF, Han QM, Guo J, Huang LL. 2010. Status of wheat rust research and control in China. St Petersburg, BGRI Technical Workshop
- Kawchuk LM, Hachey J, Lynch DR, et al. 2001. Tomato Ve disease resistance genes encode cell surface-like receptors. Proceedings of the National Academy of Sciences, USA 98, 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. 2009. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Kruijt M, Brandwagt BF, de Wit PJ. 2004. Rearrangements in the Cf-9 disease resistance gene cluster of wild tomato have resulted in three genes that mediate Avr9 responsiveness. Genetics 168, 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijt M, De Kock MJ, de Wit PJ. 2005. Receptor-like proteins involved in plant disease resistance. Molecular Plant Pathology 16, 85–97. [DOI] [PubMed] [Google Scholar]
- Laroche A, Frick MM, Huel R, Nykiforuk C, Conner B, Kuzyk A. 2000. Molecular identification of the wheat stripe rust resistance gene Yr10, the first full-length leucine zipper-nucleotide binding site-leucine-rich-repeat resistance gene in cereals. http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?Db=nucleotide&val=11990496
- Liu FR, Guo J, Bai PF, Duan YH, Wang XD, Cheng YL, Feng F, Huang LL, Kang ZS. 2012. Wheat TaRab7 GTPase is part of the signaling pathway in responses to stripe rust and abiotic stimuli. PLoS One 7, e37146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JX, Zhou RH, Dong YS, Wang LF, Wang XM, Jia JZ. 2001. Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica 120, 219–226. [Google Scholar]
- Nadeau JA, Sack FD. 2002. Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan JR, Edwards H, Stacey G, Somerville S. 2005. Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiology 138, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Avni A. 2004. The receptor for the fungal elicitor ethylene inducing xylanase is a member of a resistance-like gene family in tomato. The Plant Cell 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. 2008. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3, 348–351. [DOI] [PubMed] [Google Scholar]
- Stulemeijer IJ, Stratmann JW, Joosten MH. 2007. Tomato mitogen activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiology 144, 1481–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D. 2001. The fascinated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes and Development 15, 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, Ellendorff U, Kemp B, et al. 2008. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiology 147, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Wei XN, Fan RH, et al. 2011. Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytologist 191, 418–431. [DOI] [PubMed] [Google Scholar]
- Wang XD, Wang XJ, Feng H, Tang CL, Bai PF, Wei GR, Huang LL, Kang ZS. 2012. TaMCA4, a novel wheat metacaspase gene functions in programmed cell death induced by the fungal pathogen Puccinia striiformis f. sp. Tritici . Molecular Plant-Microbe Interactions 25, 755–764. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Tang CL, Zhang H, Xu JR, Liu B, Lv J, Han DJ, Huang LL, Kang ZS. 2011. TaDAD2, a negative regulator of programmed cell death, is important for the interaction between wheat and the stripe rust fungus. Molecular Plant-Microbe Interactions 24, 79–90. [DOI] [PubMed] [Google Scholar]
- Xing LP, Wang HZ, Jiang ZN, Ni JS, Cao AZ, Yu L, Chen PD. 2008. Transformation of wheat thaumatin-like protein gene and diseases resistance analysis of the transgenic plants. Acta Agronomic Sinica 34, 349–354. [Google Scholar]
- Zhang YX, Yang YN, Fang B, Gannon P, Ding PT, Li X, Zhang YL. 2010. Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. The Plant Cell 22, 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HB, Li SF, Deng ZY, et al. 2007. Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. The Plant Journal 52, 420–434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.