Abstract
Strawberries (Fragaria×ananassa) are false fruits the ripening of which follows the non-climacteric pathway. The role played by a C-type MADS-box gene [SHATTERPROOF-like (FaSHP)] in the ripening of strawberries has been studied by transiently modifying gene expression through either over-expression or RNA-interference-mediated down-regulation. The altered expression of the FaSHP gene caused a change in the time taken by the over-expressing and the down- regulated fruits to attain the pink stage, which was slightly shorter and much longer, respectively, compared to controls. In parallel with the modified ripening times, the metabolome components and the expression of ripening-related genes also appeared different in the transiently modified fruits. Differences in the response time of the analysed genes suggest that FaSHP can control the expression of ripening genes either directly or indirectly through other transcription factor-encoding genes. Because fleshy strawberries are false fruits these results indicate that C-type MADS-box genes like SHATTERPROOF may act as modulators of ripening in fleshy fruit-like structures independently of their anatomical origin. Treatment of strawberries with either auxin or abscisic acid had antagonistic impacts on both the expression of FaSHP and the expression of ripening-related genes and metabolome components.
Key words: Abscisic acid, auxin, Fragaria×ananassa, fruit metabolome, fruit ripening, MADS-box gene, non-climacteric fruit, SHATTERPROOF-like gene, strawberry.
Introduction
The evolution of seeds brought about the necessity to disperse them into the environment. A very successful tool for seed dispersal is represented by fleshy fruits, which attract frugivorous animals by means of pleasant colours and aromas, thus imposing on them the task of dispersing the seeds via their faeces. Besides being pleasant to eat, fleshy fruits also contain numerous secondary metabolites with positive effects on human health that are being unravelled by researchers around the world. However, fleshy fruits are perishable commodities due to their limited post-harvest life; therefore the idea of extending their life has led to the search for possible factors controlling the process of ripening (Seymour et al., 2012).
Since their discovery, plant hormones have been assayed for their activity on fleshy fruit development and ripening. Ethylene has striking effects on the ripening of climacteric fruits, which exhibit a respiratory climacteric peak prior to the start of ripening. Interestingly, such fruits also show a burst of ethylene production concomitant with the respiratory climacteric, whereas other fruits show neither type of climacteric rise and were thus named non-climacteric fruits (Alexander and Grierson, 2002; Klee and Giovannoni, 2011).
In spite of the economic importance of some non- climacteric fruits (e.g. grape, strawberry, citrus, etc.), ripening was mostly studied in the climacteric ones. In particular, ethylene is not the sole controller of ripening because other important checkpoints are situated upward of ethylene (Klee and Giovannoni, 2011). The latter finding was made possible by the molecular characterization of some tomato (Solanum lycopersicon) mutants, such as ripening inhibitor (rin; Vrebalov et al., 2002), non-ripening (nor; Giovannoni, 2004), and colorless non-ripening (cnr; Manning et al., 2006). Their fruits, besides being unable to produce any climacteric ethylene, are also unable to ripen when treated with the hormone. In all cases the mutation involved genes coding for transcription factors that were not expressed with their normal pattern.
In particular, the RIN gene encodes a MADS-box transcription factor (Vrebalov et al., 2002) and some of the latter transcription factors are known to be involved in the determination of flower components. Hence, as already seen for Arabidopsis dry fruit, it appears that at least some of the genes involved in flower development are recruited later on for the subsequent transformation of the ovary into a fleshy fruit. It was thus seen that MADS-box genes are involved in the development and ripening of fruits in peach (Prunus persica; Tadiello et al., 2009), tomato (Itkin et al., 2009; Vrebalov et al., 2009), banana (Musa acuminata; Elitzur et al., 2010), and oil palm (Elaeis guineensis; Tranbarger et al., 2011) but also in the non-climacteric bilberry (Vaccinium myrtillus; Jaakola et al., 2010) and strawberry (Fragaria×ananassa; Seymour et al., 2011). Although non-climacteric fruits have always been considered a group apart, the involvement of MADS-box genes in their ripening suggests that at least some aspects of this process might be shared by all fruit types.
Strawberry is not ovary-derived and is therefore a false fruit, yet many biochemical and molecular studies have demonstrated that the ripening syndrome in strawberry is similar to that of botanical fruits (Aharoni and O’Connell, 2002; Fait et al., 2008), albeit with specific differences that distinguish one fruit from another. The false strawberry fruit is comparable to the fleshy fruit-like structures produced by some Gymnosperms in which C-type MADS-box genes have been seen to be involved in development and ripening (Lovisetto et al., 2012); therefore, it appeared interesting to study the role played by C-type MADS-box genes in the ripening of strawberries. However, flower-producing plants normally have at least two different C-type MADS-box genes: one gene is included in the AGAMOUS/FARINELLI clade while the other belongs to the PLENA/SHATTERPROOF subgroup (euAGAMOUS and PLENA subgroups, respectively, according to Kramer et al., 2004).
A role in fruit development and ripening has recently been shown for the genes belonging to the PLENA/SHATTERPROOF subgroup in climacteric peaches (Tadiello et al., 2009) and tomatoes (Itkin et al., 2009; Vrebalov et al., 2009); hence it was deemed of interest to study the regulatory role played by a gene belonging to the same subgroup in the non-climacteric false fruit of the strawberry. To such a purpose the expression of a SHATTERPROOF-like gene was altered through either down-regulation or over-expression. It has thus been shown that in strawberry this gene plays a role as a modulator of the ripening syndrome. Moreover, its expression appeared to be antagonistically regulated by the hormones auxin and abscisic acid (ABA).
Materials and methods
Plant material
Strawberry plants (Fragaria×ananassa Duch. cv. Elsanta) were grown under standard conditions at 25 °C and a 16-h photoperiod in a greenhouse at the Department of Biology, University of Padua, Italy. Fruits were harvested at different developmental stages: small green (G1), large green [G2(1)], large green with enlarged achenes [G2(2)], white with green achenes (W1), white with brown achenes (W2), pink (P), and red (R). Fully expanded leaves and flowers at anthesis were also harvested. All tissues and fruit samples were frozen and stored at −80 °C.
RNA extraction and gene-expression analysis
Total RNA extraction, first-strand cDNA synthesis, and gene-expression analysis were carried out similarly to Lovisetto et al. (2012). Primer sequences for the selected genes are listed in the Supplementary Table S3.
Cloning of the FaSHP cDNA and promoter
The F.×ananassa SHATTERPROOF-like (FaSHP) full-length cDNA was isolated by screening a strawberry cDNA library (Trainotti et al., 1999). The screening was made following standard procedures using a probe of 200bp prepared by means of a reverse transcription (RT)-PCR experiment using primers [forward (FW) 5′-GGCACAGCAGCAGCAAGCAAATA-3′ and reverse (RV) 5′-AGAGGCGGAATAAATCACCAGACT-3′] designed on expressed sequence tag (EST) sequences (CO380891, EX686940, EX687722) available in public databases. The identity of the isolated cDNA was ascertained by sequencing.
The FaSHP gene promoter was obtained by means of RT-PCR carried out using primers (FW 5′-CTCCATGATAAT TTGCGAAGATGA-3′ and RV 5′-TTGCCAACCAAATGAAC GGTGTG-3′) designed on the promoter region of the Fragaria vesca SHP gene (FvSHP). The obtained amplicon was cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and was subsequently sequenced.
The sequences can be found in the GenBank database under the following accession numbers: FaSHP KC676787, FaSHP_promoter KC676788.
DNA sequencing and analysis
DNA sequencing was performed at BMR Genomics (Padua, Italy). Sequence manipulations, analyses, and alignments were performed using the LASERGENE software package (DNASTAR, Madison, WI, USA). Identification of putative cis-acting elements in the gene promoter was performed using the following software: PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html), PLACE (http://www.dna.affrc.go.jp/PLACE), and MatInspector (http://www.genomatix.de/online_help/help_matinspector/matinspector_help.html).
Construction of a MADS-Box phylogenetic tree
A set of MADS-box protein sequences downloaded from GenBank (Supplementary Table S4) plus the FaSHP sequence obtained in this work were used to construct a phylogenetic tree that was prepared similarly to that of Lovisetto et al. (2012).
Generation of FaSHP over-expression and RNA interference constructs
For the gene over-expression experiments the FaSHP coding sequence was cloned in the binary expression vector pBin-AR (Liu et al., 1990). The obtained construct was named pBinAR_FaSHP. The construct for FaSHP RNA interference was instead prepared using the pKANNIBAL plasmid (CSIRO, Highett, Australia). A 350-bp fragment corresponding to nucleotides 687–1037 of the FaSHP gene was amplified by PCR using forward and reverse primers (FW 5′-AAATCTAGAGAATTCGTCCGCGTATGAGCAACCAAT-3′ and RV 5′-AAAAAGCTTGGTACCCAAGGAGTGCCTGGCTAG TTC-3′). The FaSHP fragment was then inserted into pKANNIBAL vector in inverted repeat configuration according to Wesley et al. (2001). Subsequently, the fragment of pKANNIBAL containing the gene-silencing hairpin was subcloned into the binary vector pBINplus (Van Engelen et al., 1995), and the resulting construct was named pBINplus_FaSHPi. A construct to be used as a negative control in the transfection experiments was prepared by cloning the empty pKANNIBAL hairpin into the binary vector pBINplus; this plasmid was named pBINplus_intron (it had a construct carrying an intron placed downstream of the cauliflower mosaic virus 35S promoter). All the prepared binary vectors were introduced into Agrobacterium tumefaciens strain AGL0.
Transfection of strawberry fruits by agro-infiltration
Flowers with similar positions on comparable inflorescences of several plants were tagged at anthesis and after 15 days the transfection of fruits that had reached the G2(2) stage was carried out according to Hoffmann et al. (2006). These fruits were left attached to the plant and were harvested after reaching the P stage. A total of 30 agro-injected fruits from about 20 plants per each construct were inoculated and analysed.
Auxin and ABA treatments
Three pools, each consisting of 60 strawberry fruits at the W2 stage, were randomly selected on the basis of comparable size and absence of physical damage. The first group was sprayed with a synthetic auxin [2mM 1-naphthalene acetic acid (NAA) in 0.5% (v/v) Tween 20 (Trainotti et al., 1999)], the second group was sprayed with a solution of 100mM ABA in 0.5% (v/v) Tween 20 (Chen et al., 2011), and the third group, the control one, was sprayed only with Tween 20 (0.5% v/v). Ten fruits for each group were collected after 0, 2, 4, 6, 12, and 18h from the beginning of the treatment. Throughout the experiment all the fruits were kept in a growth chamber at 22 °C.
Metabolomics analysis
The powdered frozen fruits were extracted, prepared, and analysed by high-performance liquid chromatography-electrospray ionization-mass spectrometry (HPLC-ESI-MS) as described in Supplementary File S1.
To identify the metabolites under FaSHP control, an orthogonal projections to latent structures (OPLS) analysis (Simca P+, version 12.0.0.0, Umetrix, Umeå, Sweden) was performed using the 80 HPLC-ESI-MS putatively identified and quantified metabolites as x variables and the FaSHP mean normalized expression data as y variables. In this analysis both untreated and agro-infiltrated fruits were used as controls. The control versus FaSHP-down-regulated fruits OPLS model was fully cross-validated with a permutation test, using 200 permutations, and by analysis of variance, while no valid models were obtained using control versus FaSHP-up-regulated fruits.
Results
Isolation and characterization of a strawberry SHP cDNA
A publicly available cDNA fragment coding for a strawberry SHATTERPROOF-like gene was used as a probe to screen a strawberry cDNA library present in our laboratory. The result of this screening was a cDNA of 1134 nucleotides that when sequenced showed that it encoded a full-length SHP transcript with untranslated regions at both ends (not shown).
The cDNA was named F.×ananassa SHATTERPROOF-like (FaSHP) and the sequence was used to construct a phylogenetic tree (Fig. 1A) together with other known C-type sequences belonging to both the euAGAMOUS and the PLENA clades (Kramer et al., 2004). As expected, FaSHP was situated in the PLENA clade while the other strawberry C-class sequence (STAG1; Aharoni et al., 1999) was located in the euAGAMOUS clade. In both cases the strawberry sequences showed a higher similarity with the corresponding sequences of other rosaceous species (Fig. 1A).
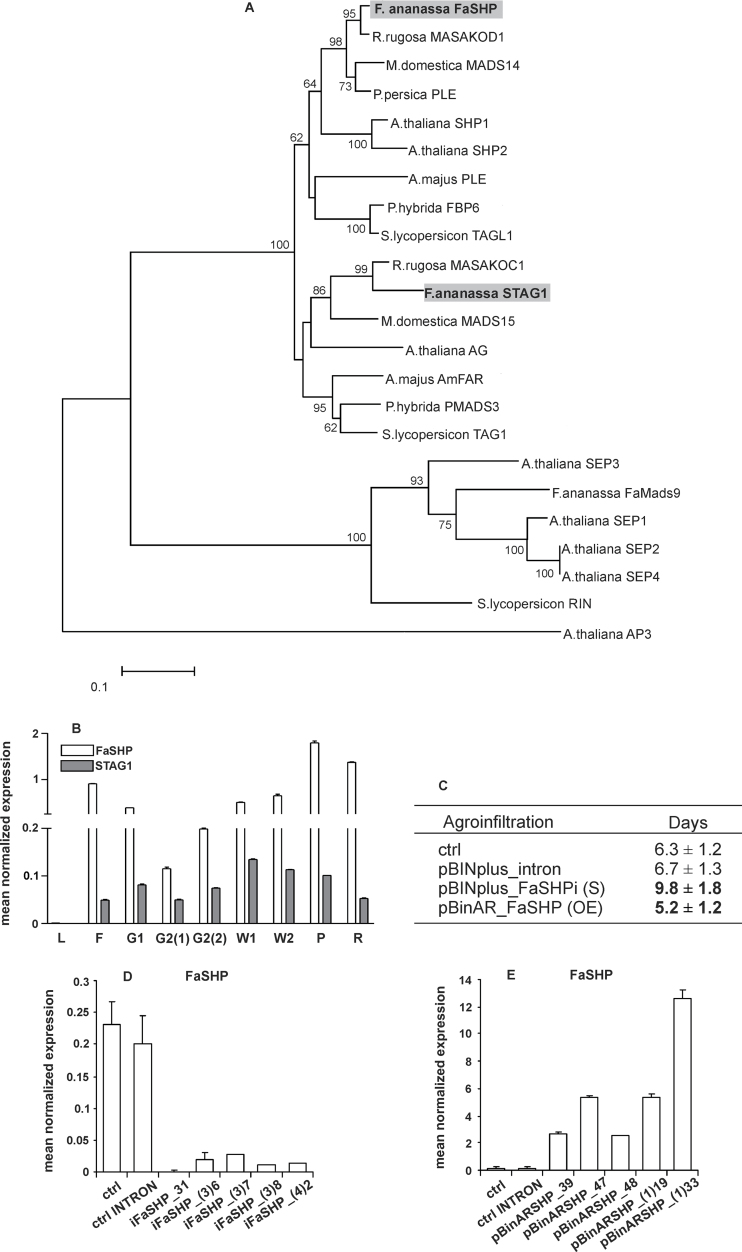
Fig. 1.
(A) Phylogenetic tree of eudicot MADS-box proteins. The two C-type strawberry sequences, FaSHP and STAG1 (highlighted by a grey background), appear grouped with the PLENA and euAGAMOUS clades, respectively. The sequence accession numbers are listed in Supplemental Table S4. (B) Relative expression profiles of the two strawberry C-type MADS-box genes, FaSHP and STAG1, in leaf (L), flower (F), and fruits at different developmental stages: small green (G1), large green [G2(1)], large green with enlarged achenes [G2(2)], white with green achenes (W1), white with brown achenes (W2), pink (P), and red (R). (C) Number of days necessary to reach the pink stage starting from the G2(2) stage in untreated fruits (ctrl), fruits agro-injected with the control construct (pBINplus_intron), FaSHP-silenced fruits (S; pBINplus_FaSHPi), and FaSHP-over-expressing fruits (OE; pBinAR_FaSHP). Values are the mean±standard deviation of five different fruits. Values in bold are significantly different by Student’s t test from the control agro-injection (P < 0.05). (D, E) Relative expression of FaSHP in pink fruits that had previously been agro-injected with either (D) the RNA-interference construct (pBINplus_FaSHPi), or (E) the over-expression construct (pBinAR_FaSHP). ctrl and ctrl INTRON values are the means of five independent fruits; the other values are from single fruits. Expression data (means of the normalized expression) were obtained by real-time PCR analyses. Bars are the standard deviations from the means.
In general, FaSHP appeared to be more highly expressed than STAG1 (Fig. 1B). In particular, the expression of STAG1 increased during fruit growth [stages G2(1)–W1] and declined during the subsequent ripening stages, whereas the expression of FaSHP showed a continuous increase during ripening and reached a maximum in pink strawberries where the transcript amount was about 20-fold greater than that of STAG1.
Transient down-regulation and over-expression of FaSHP in strawberry fruits
The expression of the FaSHP gene was transiently altered by either down-regulation or over-expression. To do so, fruits still attached to the plants were infiltrated with a suspension of A. tumefaciens cells carrying a construct for either down-regulation through RNA interference or over-expression. Two types of control were used for these experiments, one consisting of untreated fruits and the other consisting of fruits agro-infiltrated with a construct carrying an intron placed downstream of the cauliflower mosaic virus 35S promoter (see Materials and methods). The latter was meant as a control for possible effects caused by the agro-infiltration per se.
Since the expression of FaSHP started to increase between the large green [G2(2)] and the early white (W1) stages, we decided to agro-infiltrate fruits at the G2(2) stage (see Materials and methods). The rationale for this choice was that by interfering with FaSHP expression at such a stage we would be able to either minimize or boost the gene expression increase that normally occurs at the onset of ripening.
The agro-infiltrated fruits were left attached to the plants and harvested at the pink stage; that is, the stage when FaSHP normally shows its maximum expression. For all the agro-infiltrated fruits the days necessary to reach this stage were counted. About 30 pink fruits were analysed by real-time PCR for either down-regulation or over-expression of the FaSHP gene. The results of such analysis showed that in both cases there were various fruits with significantly altered expression of the gene.
For further analyses five independent fruits characterized by either extremely low expression of FaSHP (Fig. 1D) or by over-expression of the gene (Fig. 1E) were selected. Pictures of these fruits are available in Supplementary Fig. S1. Interestingly, the altered FaSHP expression was accompanied by differences in the number of days necessary to reach the pink stage (Fig. 1C). In both control types the fruits attained a pink colour in about 6 days, while more than 9 days were needed in the case of the down-regulated fruits, and about 5 days were enough for the fruits over-expressing FaSHP. As regards the other C-type MADS-box gene of strawberry (i.e. STAG1), its expression showed no differences between controls and agro-infiltrated fruits (Fig. 2M).
Fig. 2.
Relative expression of ripening-related genes and transcription factor-encoding genes in FaSHP-silenced (S) and -over-expressing (OE) fruits. ctrl and ctrl INTRON values are respectively from fruits either wild-type or agro-injected with the control construct pBINplus_intron. Fruits agro-injected at the G2(2) stage were left attached to the plant until attainment of pink colour. Each value represents the mean of five independent fruits. FaPG1, Polygalacturonase; FaPL, pectate lyase; FaEG1, endo-β-1,4-glucanase; FaCHS, chalcone synthase; FaPAL6, phenylalanine ammonia lyase, FaQR, quinone oxidoreductase; FaGalUR, d-galacturonate reductase. Expression data (means of the normalized expression) were obtained by real-time PCR analyses. Bars are the standard deviations from the means. Asterisks indicate values significantly different by Student’s t test from the control agro-injection (P < 0.05).
The general impact on ripening caused by the altered expression of FaSHP was then analysed in more detail by studying the expression of selected genes used as markers of various aspects of the ripening syndrome. When fleshy fruits start to ripen their tissues lose firmness and the activity of various cell-wall-modifying enzymes is required for this process (Brummel and Harpster, 2001). Three genes were used as markers of softening: two related to the degradation of pectins [a polygalacturonase gene, FaPG1 (Quesada et al., 2009) and a pectate lyase gene, FaPL (Medina-Escobar et al., 1997)] and one related to the metabolism of hemicellulose [an endo- β-1,4-glucanase gene, FaEG1 (Trainotti et al., 1999)].
Another important aspect of ripening is the change of colour that is mostly due to the breakdown of chlorophylls and the accumulation of various pigmented molecules. In the case of the ripe strawberries the red colour is due to anthocyanins, which are phenolic compounds (Perkins-Veazie, 1995). We therefore decided to use two genes as markers for the change of colour: one encoding a phenylalanine ammonia lyase (FaPAL6; Pombo et al., 2011), which is the first enzyme in the phenol biosynthetic pathway, and the other encoding a chalcone synthase (FaCHS) that has recently been shown to participate in the accumulation of the strawberry red colour (Lunkenbein et al., 2006).
During ripening, strawberries accumulate many secondary metabolites that contribute to their flavour and, in general, for the organoleptic peculiarities of the fruit (Perkins-Veazie, 1995). Furaneol is considered to be the main component of the strawberry flavour and a key enzyme for the synthesis of this compound is a quinone oxidoreductase encoded by the FaQR gene (Raab et al., 2006), which was thus selected as marker of flavour development. Ripe strawberries are also renowned for their high content of ascorbic acid (Perkins-Veazie, 1995) and a biosynthetic pathway demonstrated to be active during ripening involves the gene FaGalUR, encoding a d-galacturonate reductase (Cruz-Rus et al., 2011). Accordingly, this gene was selected as a marker for ascorbic acid accumulation.
All the genes selected as markers were ripening-specific (Supplementary Fig. S2) and the modification of FaSHP expression had an impact on their expression profile. The most striking effect on the expression of all the marker genes was observed in fruits with down-regulated FaSHP expression (Fig. 2A–2G) because their expression also appeared to be significantly reduced compared to controls, and the decrease seemed to be particularly strong in the case of the FaEG1, FaCHS, and FaQR genes (Fig. 2C, 2D, and 2F). Quite different was the result obtained with the FaSHP-over-expressing fruits where a significant increment (Fig. 2C and 2G) could be seen only for the FaEG1 and FaGalUR genes.
We studied also the expression of a few transcription factor-encoding genes known to be involved in the development and ripening of strawberries. These genes encode two MYB transcription factors, FaMYB10 and FaMYB1, both of them suggested to be involved in the accumulation of fruit anthocyanins (Aharoni et al., 2001; Lin-Wang et al., 2010; Salvatierra et al., 2013), and a MADS-box transcription factor, FaMADS9, belonging to the SEPALLATA group (Seymour et al., 2011). All these genes showed an increasing expression during ripening, yet the FaMYB10 gene was generally expressed at much higher levels than FaMYB1, and this difference was particularly relevant in fruits at the pink and red stages where the transcript amount of FaMYB10 was about 10-fold higher than that of FaMYB1 (Supplementary Fig. S3). Also for the transcription factor-encoding genes a significant decrease in their expression was observed in the case of FaSHP down-regulation, whereas in FaSHP-over-expressing fruits a significant parallel over-expression was observed for FaMADS9 and FaMYB10 but not for FaMYB1 (Fig. 2L, 2H, and 2I).
Treatment of fruits with either auxin or ABA
The hormonal control of ripening is still poorly understood in the case of non-climacteric fruits. However, it has been shown that auxin can negatively control the ripening of strawberries since fruits treated with the hormone showed retarded coloration and repressed expression of a number of ripening-specific genes (Given et al., 1988; Manning, 1994; Medina-Escobar et al., 1997; Trainotti et al., 1999; Aharoni and O’Connell, 2002). On the contrary, ABA has recently been shown to have a positive effect on ripening (Chen et al., 2011; Jia et al., 2011). Because of their apparently antagonistic effect on ripening, we decided to treat white fruits with either one or the other hormone to understand whether they can affect ripening by also affecting the expression of FaSHP. Treatment with auxin was effective even after 2h in preventing the increase in FaSHP expression, while ABA started to be effective in increasing the expression of FaSHP at 4h (Fig. 3A and 3B).
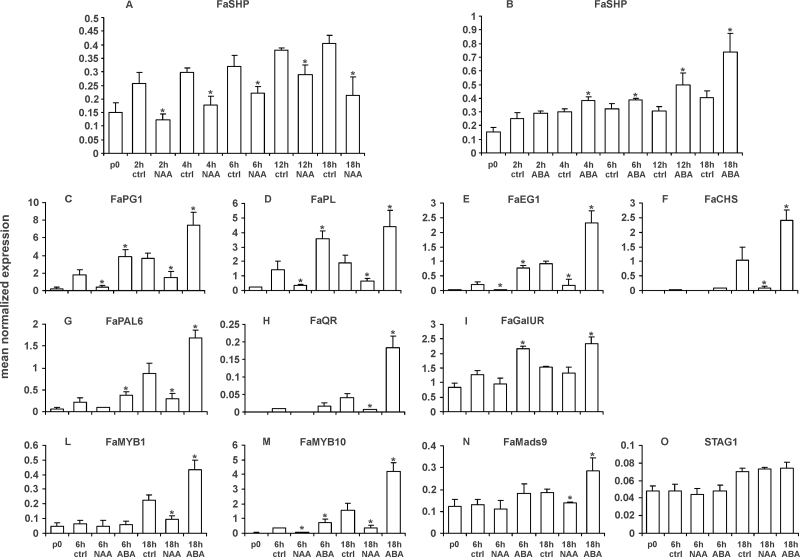
Fig. 3.
Relative expression of genes following treatment of white fruits (W2) with either NAA or ABA. (A, B) Expression of the FaSHP gene in either NAA- (A) or ABA- (B) treated fruits. Treated and control fruits were sampled at 0, 2, 4, 6, 12, and 18h. Expression of ripening-related genes (C–I) and transcription factors (L–O) in 6 and 18h fruits either untreated (ctrl) or treated with one of the two hormones (NAA and ABA). Expression data (means of the normalized expression) were obtained by real-time PCR analyses. Bars are the standard deviations from the means. Asterisks indicate values significantly different by Student’s t test from corresponding controls (P < 0.05).
Fruits treated at either 6 or 18h (see Supplementary Fig. S4) with each hormone were used to analyse the expression of the already studied marker genes. The antagonistic effect of the two hormones was evident for all of them in both the 6 and 18h samples (Fig. 3C–3I). In the case of the FaGalUR gene a positive effect of ABA was always clear, while a negative effect of auxin on its expression was better seen in the 6h sample. Interestingly, the two MYB genes and FaMADS9 also appeared to be antagonistically regulated by the two hormones, with auxin having a negative effect and ABA a positive one on their expression (Fig. 3L, 3M, and 3N). On the contrary, no effect by any of the two hormones was observed in the case of STAG1 (Fig. 3O).
Metabolomic analysis
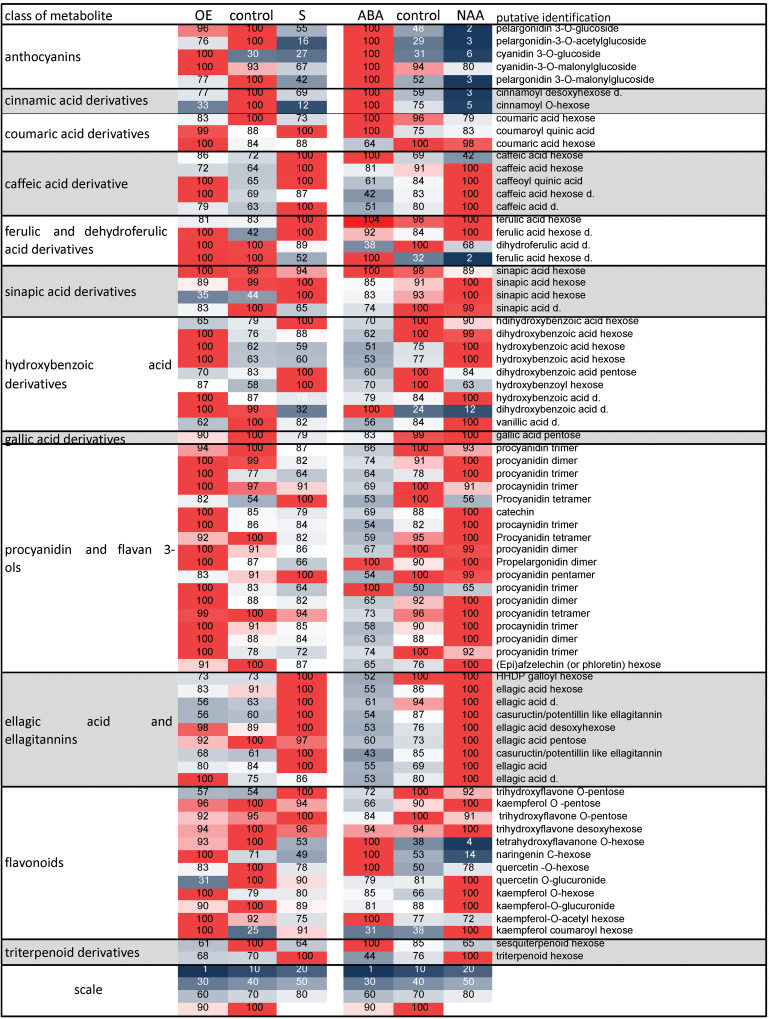
Specific shifts in secondary metabolite accumulation have been shown to occur in strawberries during development and ripening of the receptacles and, to a lesser extent, of the achenes (Fait et al., 2008). A semi-quantitative untargeted HPLC-ESI-MS analysis was carried out in strawberry fruits down- and up-regulated for FaSHP expression as well as in hormone-treated fruits, and the results are visualized as a heat map (Fig. 4). Seventy four peaks were putatively identified, including phenolic acids, anthocyanins, other flavonoids, ellagic acid derivatives, and triterpenoids. The details of metabolite putative identification are shown in Supplementary Table S1.
Fig. 4.
Heat map of the 74 putatively identified metabolites in FaSHP-over-expressing (OE) and -silenced (S) fruits compared with their control fruits (fruits agro-infiltrated with the pBINplus_intron construct), and ABA- and NAA-treated fruits compared with their controls. The average relative content of each metabolite was expressed as a percentage with respect to the samples that showed the highest content of that metabolite (=100%). Since some anthocyanins were not easily detectable in negative ionization mode, the anthocyanin data come from the positive-ionization-mode data matrix; for all other metabolites the negative-ionization-mode data matrix was used. This figure is available in colour at JXB online.
To study exactly the same material used for the analysis of gene expression, the metabolomics analyses were carried out on the same frozen and powdered fruits used for profiling the gene expression. The separation of the achenes from the receptacle was not made because the transient modification of gene expression involved only receptacles, hence the achenes were considered similar in all the examined samples.
The ripening of strawberries is marked by a colour shift from white to pink and finally to red, and this change of colour is due to anthocyanin accumulation. Both the FaSHP-down-regulated and the auxin-treated fruits showed lower levels of anthocyanins compared to the respective controls, while ABA-treated fruits accumulated higher amounts of pigments. On the contrary, the FaSHP-over-expressing fruits did not over-accumulate anthocyanins (Fig. 4). Besides anthocyanins, other metabolites either showed low levels in FaSHP-down-regulated and auxin-treated fruits or they over-accumulated after ABA treatment, including cinnamic and coumaric acid derivatives, one ferulic and one hydroxybenzoic acid derivative, and some flavonoids. Coumaric and ferulic acid derivatives have already been described as typical of red and pink receptacles, respectively (Fait et al., 2008). Some metabolites, however, showed an opposite trend of accumulation, being over-accumulated in both FaSHP-down-regulated and NAA-treated fruits and showing lower levels after ABA treatment. They included caffeic acid derivatives and ellagitannins, already described as typical of white and pink receptacles and immature receptacles and achenes, respectively (Fait et al., 2008).
Since the individual fruits showed variations in the level of the various secondary metabolites, the relationships between FaSHP expression and metabolite levels was investigated through OPLS in the various fruits, rather than by analysis of the absolute metabolite level. The OPLS analysis of control and FaSHP-down-regulated fruits resulted in a linear relationship between gene expression and the metabolite level (Supplementary Table S2). Within this analysis, the higher levels of FaSHP transcripts present in control fruits correlated positively with the accumulation of some coumaric, cinnamic, and benzoic acid derivatives and with anthocyanins (Supplementary Table S2 and Supplementary Fig. S5). The same metabolites were also strongly accumulated in ABA-treated samples and depleted in NAA-treated samples. On the other hand, the low FaSHP transcript levels present in the FaSHP-down-regulated fruits correlated with higher levels of many metabolites, including various hydroxybenzoic and hydroxycinnamic acid derivatives, ellagic acid derivatives, and flavonoids. Interestingly, with the exception of only two metabolites, the same compounds were inhibited by ABA treatment, while their level in NAA-treated fruits was variable.
The OPLS analysis of control and FaSHP-up-regulated fruits did not result in any valid OPLS model, indicating the absence of any linear relationships between gene expression in up-regulated fruits and the metabolome. However, the heat map showed that FaSHP-up-regulated fruits accumulated higher levels of procyanidins. These metabolites, induced by NAA treatment and inhibited by ABA treatment, have been described to be typical of unripe receptacles (Fait et al., 2008). Accordingly, their presence in the FaSHP-up-regulated fruits could be the consequence of a too-rapid increase in the FaSHP transcription factor that led to the appearance of the pink colour when the fruits were not yet “physiologically” pink.
Discussion
A C-type MADS-box gene (i.e. STAG1), belonging to the euAGAMOUS subgroup according to the definition by Kramer et al. (2004), was already known in strawberry (Aharoni et al., 1999; Rosin et al., 2003). In this work a second C-type gene belonging to the PLENA subgroup (a SHATTERPROOF-like one named by us FaSHP) was isolated and characterized. While STAG1 was preferentially expressed during growth of the fruit, FaSHP showed ripening-specific expression and was generally expressed at much higher levels than STAG1. Modifications of the FaSHP gene expression were therefore performed to gain information about its role in the ripening of non-climacteric strawberries. The modifications consisted of either down-regulation or over-expression obtained by infiltrating strawberries still attached to the plant with a suspension of specifically modified A. tumefaciens cells.
Contrary to what had been found in tomato (Vrebalov et al., 2009), in which the down-regulation of TAGL1 (i.e. the orthologue of FaSHP) led to increased expression of TAG1 (i.e. the orthologue of STAG1), neither the down-regulation nor the over-expression of FaSHP caused changes in the expression of STAG1. Therefore, under our experimental conditions the observed effects on ripening can be ascribed solely to the altered expression of FaSHP. Phenotypic macroscopic peculiarities were not observed but for variations in the number of days necessary to attain the pink colour starting from the moment of the agro-infiltration. This change was particularly significant in the case of the FaSHP down-regulation since about 9 days were necessary instead of the about 6 counted for the controls. Although smaller, a variation was also found in the case of the FaSHP over-expression which caused a slight acceleration of ripening with about 5 days taken to reach the pink stage. Thus, FaSHP appears to play a general positive role on the ripening of strawberries.
The significant delay in attaining the pink colour suggested a slackened expression of the genes involved in anthocyanin biosynthesis and a significantly down-regulated expression was actually observed for the FaCHS gene, which is involved in the synthesis of these coloured molecules in strawberry (Lunkenbein et al., 2006). Also the two MYB genes, reported to be involved in the regulation of the pigment synthesis in strawberry fruits (Aharoni et al., 2001; Lin-Wang et al., 2010; Salvatierra et al., 2013), were significantly down-regulated although this effect appeared particularly strong for FaMYB10.
A relevant characteristic of ripening is the softening of the fruit tissues. As evidenced in tomato (Brummell and Harpster, 2001; Carrari and Fernie, 2006) and peach (Trainotti et al., 2003), many different genes are expressed at different moments throughout this process. In the FaSHP-down-regulated fruits the three softening-related genes appeared to be significantly repressed, especially FaEG1. Therefore, it seems that the process of softening may also be positively regulated by the FaSHP gene.
Two ripening-related genes involved in the production of secondary metabolites were also analysed. One of them (FaQR) is involved in the production of furaneol which is one of the main components of the strawberry aroma (Raab et al., 2006), while the other gene (FaGalUR) is involved in the production of ascorbic acid (Cruz-Rus et al., 2011). Both genes were significantly down-regulated, thus strengthening the idea that the FaSHP gene may have a wide regulatory function during strawberry ripening.
The down-regulation of FaSHP resulted in a metabolome typical of less-ripe fruits, characterized by lower levels of metabolites, such as coumaric acid derivatives and anthocyanins, that have been previously reported as typical of ripe receptacles, and higher levels of metabolites such as caffeic acid derivatives and ellagitannins, described as typical of unripe fruits (Fait et al., 2008). Consistent with their characteristics as metabolites typical of ripe fruits, and with their relationship with FaSHP expression, coumaric acid derivatives and anthocyanins were also induced by ABA and repressed by NAA. Other metabolites, especially cinnamic acid derivatives, showed lower levels of accumulation in FaSHP-down-regulated fruits, and were induced by ABA and repressed by NAA. The latter metabolites behaved like those previously described as typical of ripe fruits, and could therefore be considered as markers of the strawberry ripe fruit. On the other hand, the metabolites previously described as typical of unripe fruit were repressed by ABA but showed different responses to NAA. Thus, a complex relationship emerged between metabolites, FaSHP, ABA, and NAA. The metabolites whose accumulation was induced by the physiological expression of FaSHP in control fruits were also induced by ABA and inhibited by NAA. On the other hand, those metabolites whose accumulation increased in FaSHP-down-regulated fruits were inhibited by ABA but not necessarily by NAA.
The results of the FaSHP over-expression experiments did not appear as straightforward as in the case of the gene down-regulation; nonetheless, they were equally interesting. Only in the case of FaEG1 and FaGalUR did the over- expression of FaSHP induce a significant increase in their expression, while all the other genes appeared unaffected. Since the acceleration of ripening was by 1 day only, it is probable that the ripening genes did not have enough time for a significant increase. Moreover, this finding suggests that the control of ripening by FaSHP is not wholly direct and might be mediated by other transcription factors.
The effect of the altered FaSHP expression on that of the two genes (FaCHS and FaMYB10) more directly involved in anthocyanin formation supports this idea. FaMYB10 encodes a transcription factor that, when over-expressed in strawberry, caused an over-accumulation of anthocyanins (Lin-Wang et al., 2010), and seems therefore to regulate more directly the downstream gene(s) coding for the enzyme(s) involved in the actual synthesis of the coloured molecules. In our case the FaMYB10 gene was either down-regulated or over-expressed in a significant manner in parallel with the variations in FaSHP expression. On the contrary, the downstream gene FaCHS (Lunkenbein et al., 2006) showed significantly altered expression only in the FaSHP-down-regulated strawberries that took 3 days more than controls to reach the pink stage. The results of the metabolomic analyses support this idea since significant variations in anthocyanin contents were only found in the FaSHP-down-regulated fruits.
Recently a negative role in anthocyanin formation has been shown for the MYB1 strawberry gene. In fact, the gene down-regulation in white Chilean strawberries (Fragaria chiloensis ssp. chiloensis) resulted in a coloration of the fruits (Salvatierra et al., 2013). Our data are in agreement with this idea since during normal ripening the FaMYB1 gene is expressed at much lower levels than FaMYB10 and, contrary to MYB10, in FaSHP-over-expressing fruits the MYB1 transcript amount did not show any variation compared to controls.
Recently it was found in tomato that the RIN MADS-box transcription factor could bind to the promoter of genes coding for cell wall hydrolases, besides to the promoter of the two 1-aminocyclopropane-1-carboxylate synthase genes ACS2 and ACS4 (Fujisawa et al., 2013). An informatic analysis carried out on the sequence of the FaEG1 promoter (Spolaore et al., 2003) evidenced the presence of CArG-box elements, the latter regarded as binding sites for MADS-box transcription factors. Moreover, using databases of known position-specific scoring matrices (detailed in Materials and methods) CArG-box elements could be evidenced also in the promoter sequence of the FaGalUR gene (not shown). Accordingly, the regulatory activity performed by FaSHP on the FaEG1 and FaGalUR genes might have been direct. Of course, we are aware that the presence of putative CArG-box elements does not demonstrate that the FaSHP protein actually binds to them.
Also in the case of the FaMADS9 gene (of the SEPALLATA group) its expression changed significantly in parallel with either the down-regulation or the over-expression of FaSHP. This result suggests that the FaMADS9 gene might be situated downstream of FaSHP along the regulatory network controlling ripening in strawberry.
It has long been known that for ripening to start the auxin content of the strawberry fleshy receptacle must decrease (Given et al., 1988). Ripening-related processes that are sensitive to auxin comprise softening, change of colour, and the formation of aroma (Aharoni and O’Connell, 2002). Concomitant with the start of ripening, ABA is synthesized and contributes to the control of the process (Chen et al., 2011; Jia et al., 2011). Therefore, the two hormones are not significantly overlapping in their control of ripening, which is negative for auxin and positive for ABA.
The study of the FaSHP expression kinetics following treatments with either auxin or ABA has evidenced that both hormones can impact the expression of this important gene, although in a manner that reflects their role in ripening: the gene is down-regulated by auxin and up-regulated by ABA. A fragment of the FaSHP promoter (1612bp upstream of the ATG codon) was obtained by RT-PCR and an informatic survey was carried out on its sequence. Interestingly, the promoter was found to harbour five putative domains for response to auxin and one putative domain for response to ABA (Supplementary Fig. S6), and this might explain why FaSHP can be regulated by both hormones.
The auxin effect on the expression of FaSHP was already clear after 2h of treatment, while for ABA a clear effect was visible in the 4 h-treated sample. The ripening-related genes responded in the expected manner to the hormones but for FaGalUR which appeared positively regulated by ABA and only weakly repressed by auxin under our experimental conditions. Also three of the transcription factor-encoding genes (i.e. FaMYB1, FaMYB10, and FaMADS9) responded in the expected manner to the hormones although not in the same measure. On the contrary, the other C-type MADS-box gene (STAG1) appeared to be insensitive to both hormones, thus suggesting once more its lack of involvement in the ripening of strawberries.
In the climacteric tomato and peach fruits the initial growth stage is characterized by high auxin content while during ripening ethylene plays a fundamental role (Gillaspy et al.1993; Miller et al., 1987). Recently, ABA has also been shown to participate in the control of ripening together with ethylene (Sun et al., 2010). During the passage from auxin to ethylene control the tomato gene TAGL1 increases its expression and has a role in ripening (Itkin et al., 2009; Vrebalov et al., 2009). In non-climacteric strawberries there is an early growth stage under the control of auxin followed by ripening that seems to be under ABA control (Perkins-Veazie 1995; Chen et al., 2011; Jia et al., 2011). In this fruit, during the passage from auxin to ABA control, the FaSHP gene (orthologous of TAGL1) increases its expression and begins to exert its important regulatory role on the ripening process. Thus the characteristic changes of the secondary metabolome that accompany ripening (i.e. accumulation of metabolites typical of ripe fruit and depletion of those typical of unripe fruit) appear to be under FaSHP, ABA, and NAA control, where ABA and FaSHP act as inducers of ripe-typical metabolite accumulation and NAA as repressor of ripe-typical metabolites. On the other hand, the unripe-typical metabolome depends on the low level of expression of FaSHP and the low level of ABA, while only some metabolites typical of unripe fruit seem to be under NAA control.
TAGL1 and FaSHP belong to the PLENA subclade of the C-type MADS-box genes, and both of them appear to increase their expression and to positively impact fruit ripening when the initial control on fruit growth exerted by auxin is handed over to either ethylene or ABA. However, TAGL1 is active in tomato which is a botanical climacteric fruit while FaSHP is active in strawberry which is a non-climacteric receptacle-derived fruit. This suggests that the MADS-box genes of the PLENA subgroup may impact the ripening of fleshy structures with fruit function independently of their anatomical origin.
Supplementary material
Supplementary material is available at JXB online.
Supplementary File 1. Supplementary materials and methods.
Supplementary Fig. S1. Pictures of FaSHP-down-regulated, FaSHP-up-regulated, and control fruits.
Supplementary Fig. S2. Relative expression profiles of ripening-related genes.
Supplementary Fig. S3. Relative expression profiles of ripening -related transcription factor-encoding genes.
Supplementary Fig. S4. Pictures of NAA- and ABA-treated fruits.
Supplementary Fig. S5. OPLS analysis of FaSHP-down-regulated and control fruits.
Supplementary Fig. S6. Bioinformatic analysis of the FaSHP gene promoter.
Supplementary Table S1. Metabolite putative identification.
Supplementary Table S2. Pq(corr) loadings of OPLS analysis and comparison with metabolite levels in agro-injected and hormone-treated fruits.
Supplementary Table S3. Sequences of the oligonucleotides used in the real-time PCR experiments.
Supplementary Table S4. GenBank accession numbers of the sequences used for the construction of the phylogenetic tree.
Acknowledgements
We thank Mr D. Denicolò (Cooperativa Sant’Orsola, Pergine, Trento, Italy) for the generous gift of the strawberry plants and fruits used in this work. We also thank Drs A. Pavanello and A. Lovisetto for help with the screening of the strawberry cDNA library. Finally, we want to thank CSIRO for providing the pKANNIBAL vector. This work was supported by a grant from the University of Padua and, partly, from the Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR), Italy, to G.C.
Glossary
Abbreviations:
- ABA
abscisic acid
- EST
expressed sequence tag
- FW
forward
- HPLC-ESI-MS
high-performance liquid chromatography-electrospray ionization-mass spectrometry
- NAA
1-naphthalene acetic acid
- OPLS
orthogonal projections to latent structures
- RT
reverse transcription
- RV
reverse.
References
- Aharoni A, O’Connell AP. 2002. Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. Journal of Experimental Botany 53, 2073–2087. [DOI] [PubMed] [Google Scholar]
- Aharoni A, van Tunen AJ, Rosin FM, Hannapel DJ. 1999. Isolation of an AGAMOUS cDNA (STAG1) from strawberry (Fragaria×ananassa cv. Elsanta) (Accession No. AF168468) (PGR99-153). Plant Physiology 121, 686 [Google Scholar]
- Aharoni A, Vos CHRD, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O’Connell AP. 2001. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal 28, 319–332. [DOI] [PubMed] [Google Scholar]
- Alexander L, Grierson D. 2002. Ethylene biosynthesis and action in tomato: a model for climateric fruit ripening. Journal of Experimental Botany 53, 2037–2055. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH. 2001. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plant. Plant Molecular Biology 47, 311–339. [PubMed] [Google Scholar]
- Carrari F, Fernie AR. 2006. Metabolic regulation underlying tomato fruit development. Journal of Experimental Botany 57, 1883–1897. [DOI] [PubMed] [Google Scholar]
- Chen JY, Liu DJ, Jiang YM, Zhao ML, Shan W, Kuang JF, Lu WJ. 2011. Molecular characterization of a strawberry FaASR gene in relation to fruit ripening. PLoS One 6, e24649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Rus E, Amaya I, Sánchez-Sevilla JF, Botella MA, Valpuesta V. 2011. Regulation of L–ascorbic acid content in strawberry fruits. Journal of Experimental Botany 62, 4191–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H. 2010. The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. Journal of Experimental Botany 61, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. 2008. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiology 148, 730–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y. 2013. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. The Plant Cell 25, 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. 1993. Fruits: a developmental perspective. The Plant Cell 5, 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. 2004. Genetic regulation of fruit development and ripening. The Plant Cell 16, S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given NK, Venis NA, Grierson D. 1988. Hormonal regulation of ripening in the strawberry, a non-climateric fruit. Planta 174, 402–404. [DOI] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W. 2006. RNAi-induced silencing of gene expression in strawberry fruit (Fragaria×ananassa) by agroinfiltration: a rapid assay for gene function analysis. The Plant Journal 48, 818–826. [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. 2009. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. The Plant Journal 60, 1081–1095. [DOI] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, et al. 2010. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiology 153, 1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. 2011. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiology 157, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. 2011. Genetics and control of tomato fruit ripening and quality attributes. Annual Review of Genetics 45, 41–59. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA, Di, Stilio VS. 2004. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. 2010. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae . BMC Plant Biology 10, 50–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XJ, Prat S, Willmitzer L, Frommer WB. 1990. Cis regulatory elements directing tuber-specific and sucrose-inducible expression of a chimeric class I patatin promoter/GUS-gene fusion. Molecular Genetics and Genomics 223, 401–406. [DOI] [PubMed] [Google Scholar]
- Lovisetto A, Guzzo F, Tadiello A, Toffali K, Favretto A, Casadoro G. 2012. Molecular analyses of MADS-box genes trace back to Gymnosperms the invention of fleshy fruits. Molecular Biology and Evolution 29, 409–419. [DOI] [PubMed] [Google Scholar]
- Lunkenbein S, Coiner HA, Ric de Vos CH, Schaart JG, Boone MJ, Krens FA, Schwab W, Salentijn EMJ. 2006. Molecular characterization of a stable antisense chalcone synthase phenotype in strawberry (Fragaria×ananassa). Journal of Agricultural and Food Chemistry 54, 2145–2153. [DOI] [PubMed] [Google Scholar]
- Manning K. 1994. Changes in gene expression during strawberry fruit ripening and their regulation by auxin. Planta 194, 62–68. [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. 2006. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics 38, 948–952. [DOI] [PubMed] [Google Scholar]
- Medina-Escobar N, Cárdenas J, Moyano E, Caballero JL, Muñoz-Blanco J. 1997. Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Molecular Biology 34, 867–877. [DOI] [PubMed] [Google Scholar]
- Miller AN, Walsh CS, Cohen JD. 1987. Measurement of indole-3acetic acid in peach fruits (Prunus persica L. Batsch cv. Redhaven) during development. Plant Physiology 84, 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins-Veazie P. 1995. Growth and ripening of strawberry fruit. Horticultural Reviews 17, 267–297. [Google Scholar]
- Pombo MA, Martìnez GA, Civello PM. 2011. Cloning of FaPAL6 gene from strawberry fruit and characterization of its expression and enzymatic activity in two cultivars with different anthocyanin accumulation. Plant Science 181, 111–118. [DOI] [PubMed] [Google Scholar]
- Quesada MA, Blanco-Portales R, Posé S, García-Gago JA, Jiménez-Bermúdez S, Muñoz-Serrano A, Caballero JL, Pliego-Alfaro F, Mercado JA, Muñoz-Blanco J. 2009. Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in strawberry fruit softening. Plant Physiology 150, 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab T, López-Ráez JA, Klein D, Caballero JL, Moyano E, Schwab W, Muñoz-Blanco J. 2006. FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. The Plant Cell 18, 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin FM, Aharoni A, Salentijn EMJ, Schaart JG, Boone MJ, Hannapel DJ. 2003. Expression patterns of a putative homolog of AGAMOUS, STAG1, from strawberry. Plant Science 165, 959–968. [Google Scholar]
- Salvatierra A, Pimentel P, Moya-León MA, Herrera R. 2013. Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90, 25–36. [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K. 2011. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria×ananassa Duch.) fruit, a non-climacteric tissue. Journal of Experimental Botany 62, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Chapman NH, Chew BL, Rose JK. 2012. Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnology Journal 11, 269–278. [DOI] [PubMed] [Google Scholar]
- Spolaore S, Trainotti L, Pavanello A, Casadoro G. 2003. Isolation and promoter analysis of two genes encoding different endo-beta-1,4-glucanases in the non-climacteric strawberry. Journal of Experimental Botany 54, 271–277. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang M, Ren J, Qi J, Zhang G, Leng P. 2010. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biology 10, 257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadiello A, Pavanello A, Zanin D, Caporali E, Colombo L, Rotino GL, Trainotti L, Casadoro G. 2009. A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. Journal of Experimental Botany 60, 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti L, Spolaore S, Pavanello A, Baldan B, Casadoro G. 1999. A novel E-type endo-beta-1,4-glucanase with a putative cellulose-binding domain is highly expressed in ripening strawberry fruits. Plant Molecular Biology 40, 323–332. [DOI] [PubMed] [Google Scholar]
- Trainotti L, Zanin D, Casadoro G. 2003. A cell wall-oriented genomic approach reveals a new and unexpected complexity of the softening in peaches. Journal of Experimental Botany 54, 1821–1832. [DOI] [PubMed] [Google Scholar]
- Tranbarger TJ, Dussert S, Joët T, Argout X, Summo M, Champion A, Cros D, Omore A, Nouy B, Morcillo F. 2011. Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiology 156, 564–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. 1995. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Research 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan I, Arroyo AJM, et al. 2009. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1 . The Plant Cell 21, 3041–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Mediano D, Drake R, Schuch W, Giovannoni J. 2002. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal 7, 581–590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.