Abstract
Pathogenic microbes manipulate eukaryotic cells during invasion and target plant proteins to achieve host susceptibility. BAX INHIBITOR-1 (BI-1) is an endoplasmic reticulum-resident cell death suppressor in plants and animals and is required for full susceptibility of barley to the barley powdery mildew fungus Blumeria graminis f.sp. hordei. LIFEGUARD (LFG) proteins resemble BI-1 proteins in terms of predicted membrane topology and cell-death-inhibiting function in metazoans, but display clear sequence-specific distinctions. This work shows that barley (Hordeum vulgare L.) and Arabidopsis thaliana genomes harbour five LFG genes, HvLFGa–HvLFGe and AtLFG1–AtLFG5, whose functions are largely uncharacterized. As observed for HvBI-1, single-cell overexpression of HvLFGa supports penetration success of B. graminis f.sp. hordei into barley epidermal cells, while transient-induced gene silencing restricts it. In penetrated barley epidermal cells, a green fluorescent protein-tagged HvLFGa protein accumulates at the site of fungal entry, around fungal haustoria and in endosomal or vacuolar membranes. The data further suggest a role of LFG proteins in plant–powdery mildew interactions in both monocot and dicot plants, because stable overexpression or knockdown of AtLFG1 or AtLFG2 also support or delay development of the powdery mildew fungus Erysiphe cruciferarum on the respective Arabidopsis mutants. Together, this work has identified new modulators of plant–powdery mildew interactions, and the data further support functional similarities between BI-1 and LFG proteins beyond cell death regulation.
Key words: Arabidopsis thaliana, haustorial complex, Hordeum vulgare, powdery mildew fungus, programmed cell death, susceptibility.
Introduction
Plant disease occurs if a virulent pathogen encounters a susceptible host plant. Since plants are considered basically resistant to most pathogenic microbes, this requires evolutionary adaptation of the pathogen to its host (Jones and Dangl, 2006). Host adaptation involves pathogen effector-mediated suppression of defence mechanisms initiated by the plant e.g. after recognition of pathogen-associated molecular patterns (PAMPs) (da Cunha et al., 2006; Koeck et al., 2011). Biotrophic pathogens often show a highly specific adaptation to their host plant. Formae speciales of the cereal powdery mildew fungus Blumeria graminis, for example, are able to colonize only one specific cereal species, while all others remain nonhosts. This may be explained by the inability of non-adapted powdery mildew fungi to suppress PAMP-triggered immunity, possibly due to an inadequate effector repertoire or timing of delivery or differences in host effector targets for susceptibility (Niks and Marcel, 2009). For successful pathogenesis, powdery mildew fungi need an efficient suppression of the plant’s preformed and inducible penetration resistance mechanisms as well as maintenance of the compatible interaction after host cell penetration, which may include suppression of programmed cell death (PCD; Hückelhoven and Panstruga, 2011). After host cell penetration, the haustorial complex is established, which consists of the fungal haustorium, an extrahaustorial modified plant plasma membrane, and the haustorial matrix in between (Koh et al., 2005; Micali et al., 2011). The haustorial complex represents the site of the most intimate contact of the fungus with its host cell and likely functions in nutrient uptake, but may also provide a platform for the delivery of fungal effectors (Micali et al., 2011). Components of defence or susceptibility to powdery mildew fungi are conserved among monocotyledonous and dicotyledonous plants. In Arabidopsis and barley, (nonhost) penetration resistance to non-adapted powdery mildew fungi involves the functionally homologous soluble N-ethylmaleimide-sensitive factor attachment (SNARE) protein PEN1/ROR2, which form complexes with VAMP721/722 and SNAP33/34 (Collins et al., 2003; Douchkov et al., 2005; Kwon et al., 2008). Conversely, one considers the plasma membrane- resident MILDEW LOCUS O (MLO) protein as a major susceptibility factor for powdery mildew fungi in monocot and dicot plants. Lack of functional MLO protein(s) results in complete race non-specific penetration resistance to the barley powdery mildew fungus B. graminis f.sp. hordei (Bgh) in barley (Büschges et al., 1997), and abolishment of host cell penetration and conidiophore production of Golovinomyces orontii or G. cichoracearum on Arabidopsis mlo2/6/12 triple mutants (Consonni et al., 2006).
The conserved cell death suppressor protein BAX INHIBITOR-1 (BI-1) is also required for full powdery mildew susceptibility in barley. Transient or stable overexpression of BI-1 support penetration of Bgh into barley epidermal cells, while transient or stable knockdown of BI-1 expression restrict it (Hückelhoven et al., 2003; Eichmann et al., 2006, 2010; Babaeizad et al., 2009). BI-1 proteins are endoplasmic reticulum-resident proteins, which are suppressors of cell death responses to biotic and abiotic stress stimuli in plants and animals (Hückelhoven, 2004; Watanabe and Lam, 2009; Henke et al., 2011; Ishikawa et al., 2011; Robinson et al., 2011). As well as having six or seven predicted transmembrane domains, BI-1 amino acid sequences harbour the ‘BI-1-like protein family motif’ (NCBI conserved domain database motif cd06181) or ‘inhibitor of apoptosis-promoting Bax1-related motif’ (InterPro IPR006214). This motif spans the transmembrane domains of the proteins, hence represents the overall domain architecture of these proteins and is of unknown function (Hückelhoven, 2004). In mammals, various transmembrane BI-1-like protein family motif-containing proteins have been identified, which do not exhibit obvious sequence similarity to BI-1 proteins. Among them are the glutamate-binding protein (GBP, also known as glutamate-binding subunit of a NMDA receptor, GRINA), responsive to centrifugal force and shear, Golgi anti-apoptotic protein (GAAP), and FAS apoptotic inhibitory molecule 2, which is also known as lifeguard (LFG) (Reimers et al., 2006, 2008; Hu et al., 2009). Comprehensive structure prediction and phylogenetic analyses showed recently that all of these proteins belong to the evolutionarily conserved LFG protein family in eukaryotes, named after the mammalian member with the best-annotated function (Hu et al., 2009). Some members of the mammalian LFG protein family (e.g. human LFG and GAAPs), possess an anti-apoptotic activity, which they have in common with BI-1 proteins (Gubser et al., 2007; Reimers et al., 2008; Hu et al., 2009). LFG proteins are conserved in plants (Reimers et al., 2006, 2008; Hu et al., 2009), but plant LFG protein functions are largely uncharacterized.
This work shows that the barley (Hordeum vulgare L.) and Arabidopsis thaliana genomes harbour at least five LFG genes (HvLFGa–HvLFGe and AtLFG1–AtLFG5). At least HvLFGa, AtLFG1, and AtLFG2 are involved in susceptibility to adapted powdery mildew fungi, further supporting functional similarity between BI-1 and LFG proteins.
Materials and methods
Plants, pathogens, and inoculation
For gene function analysis, barley (H. vulgare L.) cv. Ingrid was used. Subcellular localization studies were conducted using barley cultivars Ingrid or Golden Promise. Plants were cultivated in a growth chamber at 18 °C under a 16/8 light/dark cycle with 150 μmol m–2 s–1 light and a relative humidity of 65%. B. graminis (DC) Speer f.sp. hordei was maintained on barley cv. Golden Promise under the same conditions. Transiently transformed barley leaf segments were inoculated at a density of 150 conidia mm–2 for subsequent functional analysis and at a density of about 50 conidia mm–2 for pathogen-dependent protein localization studies. For gene expression analyses, a density of 80–100 conidia mm–2 was used.
A. thaliana ecotype Col-0 was purchased from Lehle Seeds (Round Rock, USA) and SALK lines of AtLFG1 (SALK_147263C, SALK_111590) and AtLFG2 (SALK_052507C) were ordered from the Nottingham Arabidopsis Stock Centre (Nottingham, UK). Arabidopsis plants were grown in a 2:1 soil/sand mixture. All seeds were kept at 4 °C for 2 days for stratification prior to transfer into a controlled environment growth chamber (10/14 light/dark cycle with 120 μmol m–2 s–1 light, 22 °C during the day, 20 °C during the night, 65% relative humidity). Erysiphe cruciferarum was grown on Col-0 to maintain constant aggressiveness and on susceptible phytoalexin deficient 4 (pad4) mutants for strong conidia production. Arabidopsis plants were placed under a polyamide net (0.2mm2) and inoculated at a density of 3–4 conidia mm–2 for microscopy, by brushing conidia off of pad4 plants through the net.
Assessment of phylogenetic relationships
clustal w2 multiple sequence alignment (www.ebi.ac.uk/Tools/msa/clustalw2/, last accessed 8 July 2013) followed by generation of the best-scoring maximum-likelihood tree with support values by Randomized Axelerated Maximum Likelihood (RAxML, Stamatakis et al., 2008) was the basis for phylogenetic analysis of mouse, barley and Arabidopsis BI-1 and LFG proteins. An unrooted phylogenetic tree was drawn using the software TreeView.
Gene expression analysis
Seven-day-old barley cv. Ingrid primary leaves were inoculated with Bgh conidia. At 0, 12, and 48h post inoculation (hpi), the abaxial epidermal peels and the remaining leaf tissue (mesophyll and adaxial epidermis) were collected separately, and immediately frozen in liquid nitrogen. RNA extraction and reverse-transcription (RT) PCR were performed as described in Eichmann et al. (2010). Primers 5′-TCTCGTCCCTGAGATTGC CCACAT-3′ and 5′-TTTCTCGGGACAGCAACACAATCTTCT-3′ were used to amplify a 156-bp HvUBC2 fragment (GenBank accession number AY220735; Jensen et al., 2007), primers 5′-GGCCGACATGCATTCACCAG-3′ and 5′-CATCTG ATATTGCTGGGTCTG-3′ to amplify a 506-bp HvOXLP fragment (accession number X93171, Wei et al., 1998), and primers 5′-AAGGGGGAGGTGATCCT-3′ and 5′-GGACAGGAGG AGGGGCTA-3′ to amplify a 473-bp fragment of HvLFGa.
To investigate the expression level of target genes in AtLFG SALK-lines and AtLFG overexpression mutants, two-step RT-PCR was conducted. Total RNA was extracted from frozen plant material using the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany). cDNA synthesis was primed with oligo (dT) using RevertAid Reverse Transcriptase (Fermentas, St Leon-Rot, Germany). Equal cDNA loading was confirmed by the amplification of a UBIQUITIN 5 (UBQ5, At3g62250) fragment using primers 5′-CCAAGCCGAAGAAGATCAAG-3′ and 5′-ACTCCTTCCTCAAACGCTGA-3′. AtLFG1 expression was analysed using primers 5′-GGATCCGCGATTTCAACAACAAA-3′ and 5′-CGATAAATCTATGTCTGGAA-3′, and AtLFG2 expression using primers 5′-GGATCCCACCGGCGTTGACAAA-3′ and 5′-TGGCACAGTCTTAAGAGCAA-3′.
Isolation of HvLFG and AtLFG cDNAs and cloning of expression constructs
The HvLFGa, AtLFG1, and AtLFG2 coding sequences were amplified by PCR using the primer combinations 5′-GGATCCACGCCGACGACGATGTAT′-3 and 5′-GGACAGGAGGAGGGGCTA-3′, 5′-GGATCCGCGATTTC AACAACAAA-3′ and 5′-CGATAAATCTATGTCTGGAA-3′, and 5′-GGATCCCACCGGCGTTGACAAA-3′ and 5′-TGGCACAG TCTTAAGAGCAA-3′, respectively. PCR fragments were cloned into the pGEM-T vector (Promega, Mannheim, Germany) and, after sequence confirmation, subcloned into the BamHI restriction site of the pGY-1 expression vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Schweizer et al., 1999).
For RNA interference (RNAi)-mediated transient-induced gene silencing (TIGS), a 976bp HvLFGa fragment was amplified by PCR using the primers 5′-GGATCCGAAGAACGCCGACGAC-3′ and 5′-GTCGACGAGGGGCTACGCTACG-3′, which contained a 138 bp-long intron 270bp downstream of the start codon. The blunt-ended fragment was ligated into the Gateway pENTR vector pIPKTA38 in anti-sense orientation and subcloned into the final RNAi Gateway destination vector pIPKTA30N by standard LR reaction (Douchkov et al., 2005). The final pIPKTA30N-HvLFGa construct contained the HvLFGa fragment as inverted repeats under the control of the CaMV 35S promoter, separated by the second intron of the wheat RGA2 gene.
For investigations of the subcellular localization of HvLFGa, a GFP-HvLFGa fusion construct was generated. GFP was amplified with primers 5′-GGATCCATGGTGAGCAAGGGCGAG-3′ and 5′-GGATCCTTGTACAGCTCGTCCAT-3′, and inserted in frame with the HvLFGa coding sequence into pGY-1, resulting in pGY-1-GFP-HvLFGa.
For stable overexpression in Arabidopsis, the expression cassettes of pGY-1-AtLFG1 and pGY-1-AtLFG2 were cut out and ligated into the binary vector pLH6000 (GenBank accession no. AY234328).
Transient transformation of barley epidermal cells, and gene function analysis
For gene function analysis, single epidermal cells of 7-day-old barley cv. Ingrid leaf segments were transiently transformed via ballistic delivery of expression vectors, based on a protocol originally developed for wheat (Schweizer et al., 1999). In transient overexpression experiments, 236 μg of 1-μm gold particles coated with 7 μg of the marker plasmid pGY-1-GFP (GFP under control of the CaMV 35S promoter, Schweizer et al., 1999) and 7 μg of either pGY-1- HvLFGa, pGY-1-GFP-HvLFGa, or empty pGY-1 vector were delivered per shot with a hepta adapter using a PDS-1000/He particle gun (Bio-Rad Laboratories, Munich, Germany), as described previously (Douchkov et al., 2005; Eichmann et al., 2010).
For TIGS experiments (Douchkov et al., 2005), 312 μg of 1.1 μm tungsten particles coated with 0.8 μg pGY-1-GFP together with 1 μg of either pIPKTA30N-HvLFGa or empty pIPKTA30N were delivered per shot with a single adapter using a particle inflow gun, as described previously (Hückelhoven et al., 2003).
One day after ballistic delivery of coated particles, the leaf segments were inoculated at a density of about 150 Bgh spores mm–2 (Hückelhoven et al., 2003). The penetration efficiency of Bgh into transiently transformed epidermal cells was evaluated 2 days after inoculation by dividing the number of haustoria-containing cells by the number of cells attacked by Bgh. At least five independent overexpression or TIGS experiments were conducted, whereby for each individual experiment, a minimum of 60 interaction sites were evaluated per construct. Statistical significance was analysed by means of two-sided paired Student’s t-test over absolute penetration. For data presentation, the deviation of penetration efficiency from the average control was calculated.
For transient knockdown of GFP-HvLFGa expression, tungsten particles were coated with 1 μg per shot of the RNAi construct pIPKTA30N-HvLFGa or empty vector pIPKTA30N together with 1 μg of the pGY-1-GFP-HvLFGa construct and delivered into single barley cv. Ingrid epidermal cells. As transformation marker, 0.8 μg per shot of pGY-1-RFP was co-transformed. By fluorescence microscopy, RFP-expressing cells were analysed concerning green fluorescence of GFP-HvLFGa 2 dab. In three independent experiments, 100 cells per variant were evaluated, respectively. Statistical significance was analysed by means of two-sided paired Student’s t-test.
Agrobacterium tumefaciens-mediated transformation of Arabidopsis
A. tumefaciens AGL1 was transformed using the freeze–thaw method described by Weigel and Glazebrook (2006). The floral-dip method based on the protocols of Clough and Bent (1998) and Rosso et al. (2003) was adapted to transform Arabidopsis Col-0 plants. Flower buds (5–10 cm-long) were submerged for 1min into an overnight culture of A. tumefaciens mixed 1:1 with 10% sucrose solution containing 0.02% Silwet L-77 (Lehle Seeds, Round Rock, USA).
Subcellular localization of GFP-HvLFGa
For protein localization studies, single epidermal cells of 7-day-old barley leaf segments were transiently transformed via particle bombardment. Gold particles were coated with 1 μg of pGY-1-GFP-HvLFGa plasmid together with 1 μg (or 2 μg in case of binary vectors) of a marker plasmid for the expression of red fluorescent pm-rk (Nelson et al., 2007), RFP-AtARA7 (Böhlenius et al., 2010), OsTPKb-RFP (Isayenkov et al., 2011), GmMan1-RFP (Yang et al., 2005), mCherry-AtVAM3/SYP22, or mCherry (Hoefle et al., 2011), or maize genes B-Peru and C1 for anthocyanin production (Schweizer et al., 2000). To create pGY-1-mCherry-AtVAM3/SYP22, the AtVAM3/SYP22 coding sequence was amplified from Arabidopsis Col-0 cDNA using primers 5′-TGTACAAAGGTTTCGCGAAGAAGATGAG-3′ and 5′-TGTACATGCTGTTCGAAATCAAGCTG-3′ and ligated in frame behind mCherry into pGY-1-mCherry (Hoefle et al., 2011). Imaging was done with a Leica TCS SP5 confocal laser-scanning microscope (Leica Microsystems, Mannheim, Germany) 24–72h after transformation. GFP was excited by a 488nm laser line and detected between 500 and 530nm. Red fluorescent proteins were excited by a 561nm laser line and detected between 580 and 650nm. The 561nm laser line was also used to excite anthocyanin fluorescence. Emission here was detected between 580 and 650nm.
For FM4-64 staining, barley leaf segments were vacuum infiltrated with 20 μM SynaptoRed C2 (Sigma-Aldrich, Munich, Germany) in bidistilled water, and then incubated for up to 1 hour at 25 °C in the dark. In order to visualize vacuolar membranes, leaf segments were placed on 0.5% (w/v) water agar after incubation in SynaptoRed solution and incubated for 5h at 25 °C in the dark. FM4-64 was excited with a 561nm laser line and detected between 600 and 645nm.
Results
Identification of barley and Arabidopsis LFG proteins with predicted similarity to BI-1
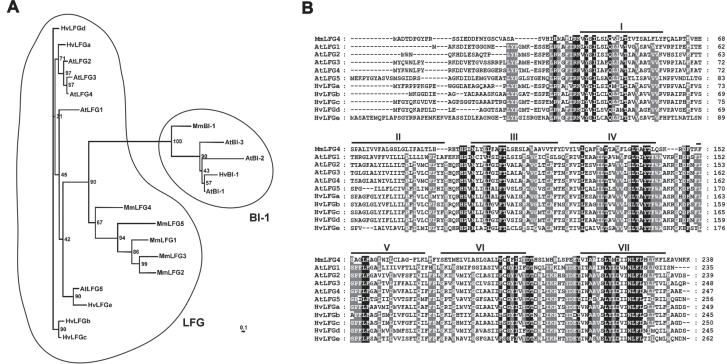
This study used the human LFG nucleotide sequence (NCBI accession NM_012306, Reimers et al., 2006) to search the TIGR plant transcript assemblies for similar sequences in barley (Childs et al., 2007). This identified the transcript assembly TA49291_4513, encoding a putative receptor-associated protein, herein named HvLFGa. Submission of this HvLFGa sequence to blast at TIGR plant transcript assemblies or NCBI revealed another four related sequences: HvLFGb (TA42670_4513), HvLFGc (TA38842_4513), HvLFGd (TA39011_4513; AK251691), and HvLFGe (TA32154_4513; AK249851). A database search for HvLFG-related Arabidopsis genes also identified five sequences: AtLFG1 (AGI code At4g14730), AtLFG2 (At3g63310), AtLFG3 (At4g02690), AtLFG4 (At1g03070), and AtLFG5 (At4g15470). Since the phylogenetic relatedness of barley and Arabidopsis orthologues is not clear (Fig. 1A), the barley LFGs were named HvLFGa to HvLFGe and the Arabidopsis LFGs AtLFG1 to AtLFG5. The deduced protein sequences were between 235 and 262 amino acids in length. InterProScan signature search (Zdobnov and Apweiler, 2001) revealed the presence of the Bax1-I (inhibitor of apoptosis-promoting Bax1) motif (formerly uncharacterized protein family motif UPF0005) (Pfam database accession PF01027) in human HsLFG and all five barley or Arabidopsis LFG protein sequences, which is also present in BI-1 proteins (Supplementary Table S1, available at JXB online). For all LFG proteins, a search against the NCBI conserved domain database (Marchler-Bauer et al., 2011) detected the BI-1-like protein family motif cd06181. However, HsLFG and barley LFG proteins do not have the short so-called BI-1 motif (PROSITE PS01243) or the C-terminal RXR and/or KKXX-like amino acid sequence, which is present in most eukaryotic BI-1 proteins. The BI-1 motif has to be distinguished from the BI-1-like protein family motif cd06181 (or InterPro IPR006214), which prokaryotic and eukaryotic BI-1 proteins have in common (Hückelhoven, 2004). Unlike in some BI-1 proteins (e.g. AtBI-1), no C-terminal coiled-coil domains are present in LFG protein sequences. For BI-1 proteins, six or seven transmembrane domains can be predicted (Hückelhoven, 2004). Similarly, seven transmembrane domains are predicted in protein sequences of the five barley and Arabidopsis LFGs and HsLFG according to sequence analysis by TOPKONS (Bernsel et al., 2009; Supplementary Table S1). No cleavable signal peptides seem to be present in HvLFG or AtLFG sequences (prediction by SignalP).
Fig. 1.
Phylogenetic analysis and sequence comparison of plant and mouse BI-1 and LFG proteins. (A) Unrooted phylogenetic tree of mouse, Arabidopsis, and barley BI-1 and LFG proteins based upon bootstrap analysis after sequence alignment using clustal w2. (B) Alignment of the five protein sequences of Arabidopsis and barley with mouse LFG4. Identical amino acids are boxed black. Amino acids conserved in all but one or two protein sequences are shaded in grey. Bars indicate the predicted positions of transmembrane domains (I–VII). At, Arabidopsis thaliana; Hv, Hordeum vulgare; Mm, Mus musculus.
According to an assessment of phylogenetic relationships by means of sequence comparison, BI-1 and LFG proteins form distinct phylogenetic clades (Fig. 1A). clustal w2 alignment scores indicate that BI-1 proteins of mouse, Arabidopsis or barley are at most 20% identical to LFG proteins of the same species (Supplementary Fig. S1). When compared to mouse MmLFG proteins, all plant LFG proteins share the highest sequence similarity with mouse MmLFG4 (31–35% identity; Supplementary Fig. S1). Sequence identity among barley LFG proteins is highest for HvLFGa-d (66–71%), whereas HvLFGe seems to be more distantly related to the other four (Supplementary Fig. S1). Arabidopsis AtLFG proteins 2, 3, and 4 are more closely related to each other (75–82% identity) than to AtLFG1 or AtLFG5 (48–53% identity). Conserved amino acids can be found throughout the transmembrane scaffold, only the N-terminal regions are highly variable (Fig. 1B). Together, structure prediction indicates similarities of LFG and BI-1 proteins, but proteins show clear sequence-specific distinctions.
Transient overexpression and knockdown experiments reveal a function of HvLFGa in susceptibility to the barley powdery mildew fungus
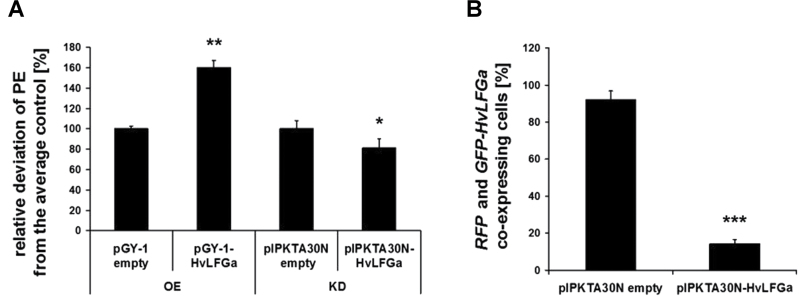
Due to the structural similarity of HvLFG and HvBI-1 proteins, this work investigated whether LFG proteins would have a function in susceptibility to powdery mildew similar to HvBI-1. Powdery mildew fungi infect the epidermis of plant leaves. Therefore, RT-PCR was used to confirm HvLFGa expression in epidermal tissue of barley leaves infected with Bgh (Supplementary Fig. S2). Expression of HvLFGa in the leaf epidermis is a prerequisite for gene function analysis using RNAi-based TIGS (Douchkov et al., 2005). The current study then tested if transient HvLFGa over- and underexpression would influence the interaction of barley epidermal cells with adapted Bgh. For overexpression experiments, single epidermal cells of the susceptible barley cv. Ingrid were transformed via particle bombardment. In five independent experiments, the transient biolistic overexpression of HvLFGa resulted in a significantly enhanced penetration efficiency (PE; i.e. the percentage of penetrated cells relative to all attacked cells) of Bgh into transformed barley epidermal cells, when compared to the empty vector control. The PE (mean ± SE) shifted from 37.6±3.0% in control cells to 60.2±7.9% in HvLFGa-overexpressing cells. The relative PE was 160.1±6.9% (Fig. 2A). Vice versa, TIGS of HvLFGa significantly reduced the penetration success of Bgh into barley epidermal cells from 37.2±3.0% to 30.2±3.3% in six independent experiments. The relative PE was 81.2±8.9% of the control (Fig. 2A). To estimate the efficiency of the HvLFGa TIGS construct, its ability to knockdown the expression of a transiently overexpressed HvLFGa fusion to the green fluorescent protein (GFP-HvLFGa) was tested. In these experiments, co-expressed red fluorescent protein (RFP) served as marker for normalizing the numbers of transformed cells. In three independent experiments, control cells showed 92.2±4.6% co-expression of GFP-HvLFGa and RFP. In contrast, co-transformation with the TIGS construct pIPKTA30N-HvLFGa reduced the co-expression rate by more than 85% to only 14.3±2.3% of cells (Fig. 2B). This confirmed functionality of the HvLFGa TIGS construct. HvLFGa thus appears to be involved in susceptibility to Bgh since the transient overexpression enhanced susceptibility to Bgh, whereas transient knockdown of HvLFGa expression limited the fungal success.
Fig. 2.
Overexpression of HvLFGa induces super-susceptibility to Bgh, while the knockdown of HvLFGa expression reduces the penetration success of the fungus. (A) HvLFGa overexpression or TIGS constructs were transiently delivered into epidermal cells of barley cv. Ingrid by particle bombardment. GFP served as transformation marker. Control cells were transformed with either pGY-1 or pIPKTA30N empty vector. Leaf segments were inoculated with spores of Bgh one day after transformation and analysed with a microscope two days later. The mean relative deviation of penetration efficiency (PE) from the average control is displayed, whereby the control was set to 100%. The effect of HvLFGa overexpression was analysed in five independent experiments, the effect of TIGS-based knockdown of HvLFGa expression in six independent experiments. (B) The efficiency of the TIGS construct to knockdown HvLFGa expression was evaluated in three independent experiments. For this, barley cv. Ingrid epidermal cells were transformed with a GFP-HvLFGa fusion construct together with the HvLFGa TIGS construct or empty pIPKTA30N vector. RFP served as marker for the identification of transformed cells. Two days after bombardment, RFP-expressing cells were analysed concerning an additional expression of GFP-HvLFGa. Data are mean and standard errors. *, **, and *** indicate significance at P < 0.05, 0.01, and 0.001, respectively, according to Student’s t-test.
Subcellular localization of a GFP-HvLFGa fusion protein
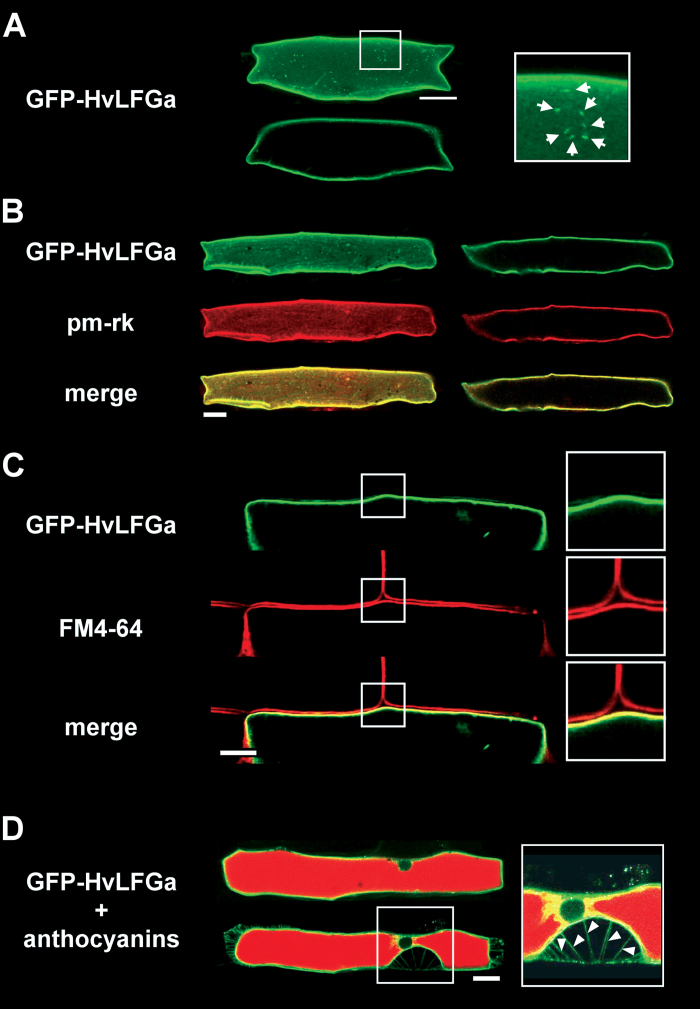
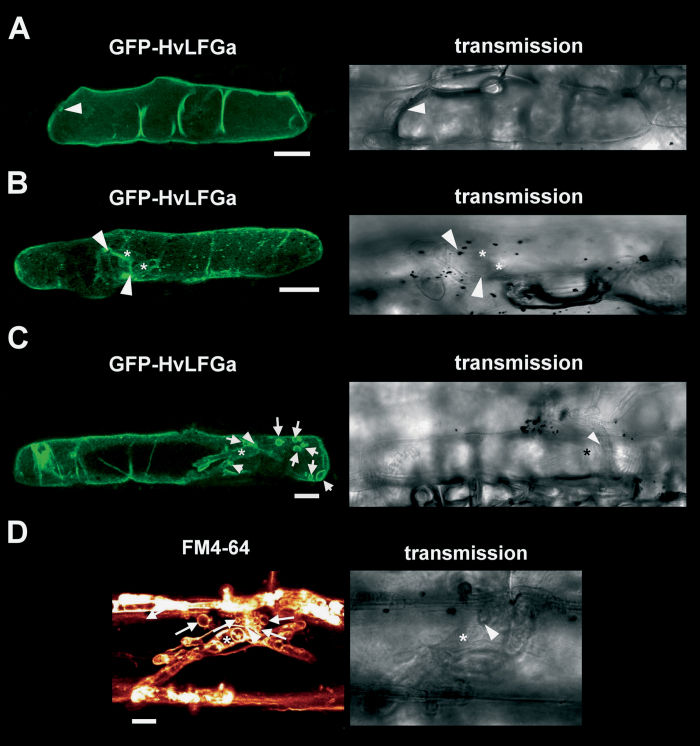
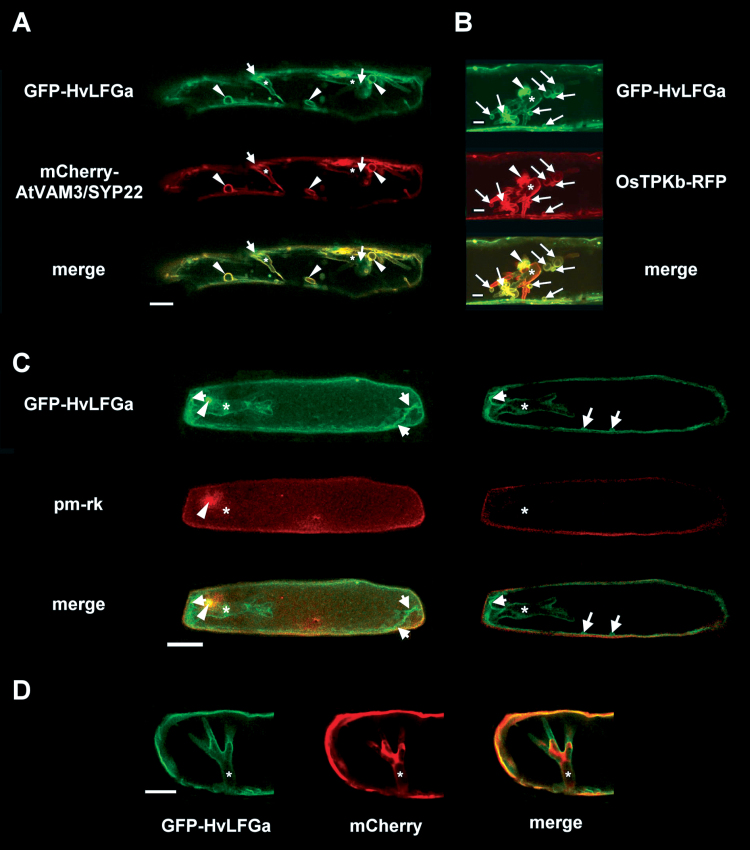
To elucidate the subcellular localization of HvLFGa, GFP was fused to the N-terminus of HvLFGa (GFP-HvLFGa) and was transiently expressed in single barley epidermal cells. To confirm functionality of the GFP-HvLFGa fusion construct, its capacity to enhance the PE of Bgh into barley epidermal cells was first tested, as shown previously for non-fused HvLFGa. In three independent experiments, transient overexpression of GFP-HvLFGa in barley cv. Ingrid epidermal cells resulted in significantly enhanced PE of Bgh, suggesting that fusion of GFP did not affect protein function (Supplementary Fig. S3A). At 48h after transformation, the GFP-HvLFGa fusion protein was visible in the periphery of transformed epidermal cells (Fig. 3A). Peripheral GFP-HvLFGa fluorescence perfectly matched plasma membrane labelling with the mCherry-tagged plasma membrane marker pm-rk (Nelson et al., 2007, Fig. 3B) or the amphiphilic styryl dye FM4-64 at 15min after staining (Fig. 3C). Additionally, initiation of plasmolysis with 20% glycerol (v/v) resulted in the occurrence of readily identifiable green-fluorescent Hechtian strands (Fig. 3D). Hence, GFP-HvLFGa clearly labelled the cell peripheral plasma membrane. At this time, there was only very faint co-localization of GFP-HvLFGa with tonoplast markers such as mCherry-AtVAM3/SYP22 (Uemura et al., 2010; Supplementary Fig. S3D), and GFP-HvLFGa did not surround transvacuolar cytoplasmic strands (Supplementary Fig. S3C). In about 70% of the green fluorescent cells, GFP-HvLFGa additionally accumulated in small punctate structures (Fig. 3A). These structures moved along with cytoplasmic streaming. Co-expression analyses indicated association of these small mobile structures with RFP-AtARA7-labelled endosomal compartments (Böhlenius et al., 2010; Supplementary Fig. S3B). GFP-HvLFGa accumulation did not co-localize with GmMAN1-RFP, a marker for Golgi organelles (Yang et al., 2005, data not shown). In order to assess GFP-HvLFGa localization during interaction with Bgh, transformed cells were inoculated one day after bombardment. GFP-HvLFGa still localized in the cell periphery and did not strongly accumulate at the site of attempted penetration when the penetration attempt of Bgh had been stopped before haustorium formation (Fig. 4A). In successfully penetrated epidermal cells, GFP-HvLFGa was found in the cell periphery and weakly around immature haustoria at 24 hpi (Fig. 4B). Two days after inoculation, GFP-HvLFGa surrounded fully developed haustoria, but additionally accumulated strongly near the haustorial neck. Furthermore, GFP-HvLFGa-derived fluorescence was observed in tubular or large round vesicle-like mobile structures of about 2–7 μm in diameter (Fig. 4C and Supplementary Video S1), which were absent in non-penetrated cells (e.g. Fig. 3). Similar structures were found in penetrated barley epidermal cells 5h after staining with the lipophilic dye FM4-64 at 48 hpi (Fig. 4D). These large vesicle-like structures also became visible upon expression of OsTPKb-RFP, a rice two-pore K+ channel, which labels vacuolar membranes (Isayenkov et al., 2011), or mCherry-tagged AtVAM3/SYP22, which also marks tonoplast and, additionally, prevacuolar compartments in plant cells (Uemura et al., 2010, Fig. 5A, B). Both markers clearly co-localized with GFP-HvLFGa in these structures. Although both GFP-HvLFGa and pm-rk labelled the plasma membrane in non-infected cells, GFP-HvLFGa but not pm-rk accumulated around the fungal haustorium or in large vesicle-like mobile structures in penetrated cells (Fig. 5C). Furthermore, the green fluorescence of GFP-HvLFGa surrounded the mCherry-labelled cytoplasm around haustorial complexes (Fig. 5D). Together, the plasma membrane-localized GFP-HvLFGa fusion protein appears to undergo remarkable relocalization to endosomes and the tonoplast in barley epidermal cells during successful establishment of biotrophy by Bgh.
Fig. 3.
Localization of a GFP-HvLFGa fusion protein in barley epidermal cells after transient transformation as visualized by confocal laser-scanning microscopy. (A) The upper picture is a whole-cell projection of a transformed cell; the lower picture shows a single longitudinal section (top view) through the centre of the same cell. The framed picture in the right panel represents an enlargement of the marked area in the whole-cell projection. (B) Co-expression of GFP-HvLFGa with the red fluorescent plasma membrane marker pm-rk; left panel: whole-cell projection; right panel: single longitudinal section (top view) through the centre of the same cell; yellow pixels in the merged images indicate co-localization. (C) Longitudinal cross-section of a part of a single GFP-HvLFGa-expressing cell after staining with FM4-64 for 15 minutes; framed images on the right are enlargements of the framed areas in the left panel; yellow pixels in the merged images indicate co-localization. (D) GFP-HvLFGa-expressing cell that accumulates red fluorescent anthocyanins in the vacuole after co-expression of maize B-Peru and C1 genes; single longitudinal section of the cell before (top) and after (bottom) plasmolysis; arrowheads in the magnified detail on the right point to Hechtian strands. Bars = 25 μm.
Fig. 4.
Localization of a GFP-HvLFGa fusion protein in barley epidermal cells in interaction with B. graminis f.sp. hordei (Bgh). Right panels show transmission images of fungal structures; arrowheads indicate sites of Bgh attack; asterisks indicate fungal haustoria; arrows indicate GFP-HvLFGa-labelled bulb-like mobile structures. (A) GFP-HvLFGa accumulation (left panel) in an attacked but non-penetrated cell 1 d post inoculation (dpi). (B, C) Localization of GFP-HvLFGa in penetrated cells at 1 (B) and 2 dpi (C) with Bgh. (D) Detail of a penetrated epidermal cell of barley 48h after inoculation with Bgh and 5h after FM4-64 staining. Bars = 25 μm (A–C), 10 μm (D).
Fig. 5.
Co-localization of the GFP-HvLFGa fusion protein with cellular markers in haustoria-containing barley epidermal cells 48h after inoculation with Bgh. Confocal laser-scanning micrographs of barley epidermal cells transiently co-transformed with GFP-HvLFGa and red fluorescent cellular markers, and inoculated with spores of the barley powdery mildew fungus 1 day later; yellow pixels in the merge image indicate co-localization of green and red fluorescent proteins. (A, B) Overlays of optical sections containing fungal haustoria (asterisks): (A) co-localization of GFP-HvLFGa (green) with mCherry-AtVAM3/SYP22 (red) in a penetrated cell; (B) detail of a penetrated cell co-expressing GFP-HvLFGa (green) and OsTPKb-RFP (red). (C) Co-localization of GFP-HvLFGa (green) with pm-rk (red) in a penetrated cell; left panel shows maximum projections of 20 optical sections at 2 μm increments; right panel shows a single longitudinal section through the centre of the cell; asterisk indicates the fungal haustorium; note that green but not red fluorescence surrounds the fungal haustorium. (D) Detail of a penetrated cell co-expressing GFP-HvLFGa (green) and cytosolic mCherry (red); images represent a single longitudinal section through the haustorium (asterisk); note that green fluorescence of GFP-HvLFGa surrounds red fluorescence of mCherry. (A–C) Arrows indicate the site of fungal penetration; arrowheads mobile bulb-like or tubular structures. Bars = 25 μm (A, C, D), 10 μm (B).
Function of LFG proteins in powdery mildew susceptibility in monocots and dicots
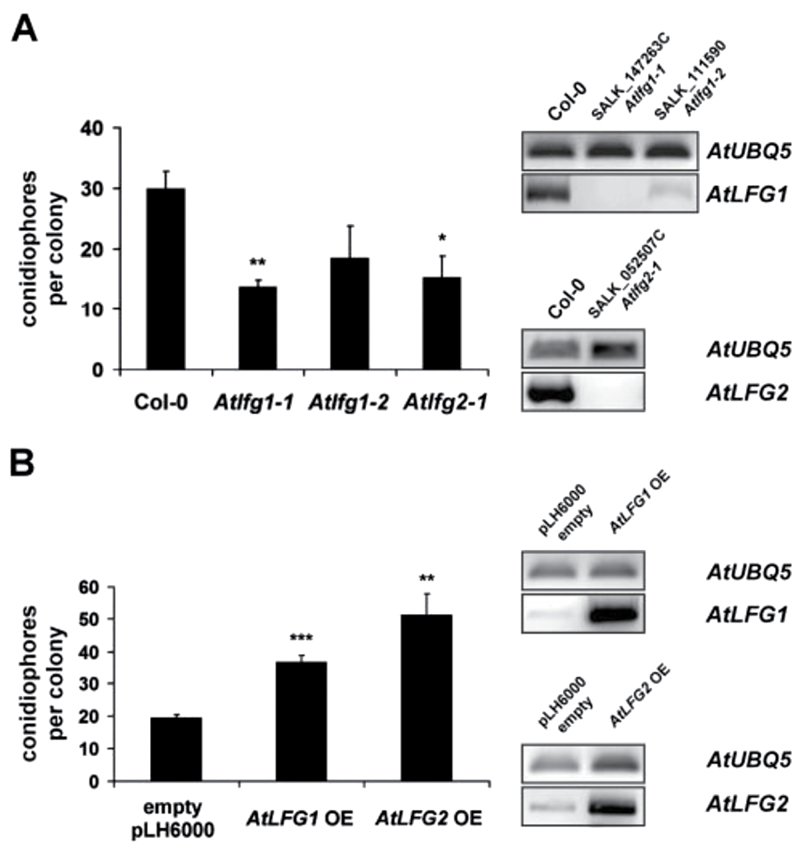
In order to examine the function of AtLFG proteins in powdery mildew susceptibility, powdery mildew disease development on Arabidopsis LFG T-DNA insertion mutants was analysed. Two T-DNA insertion mutants were available for AtLFG1 (SALK_147263C and SALK_111590) and one for AtLFG2 (SALK_052507C). The integration and positions of T-DNA insertions were confirmed by PCR (Supplementary Fig. S4A, B). Both AtLFG1 mutants carry their T-DNA insertion at different positions in the promoter region. The T-DNA position in the AtLFG2 mutant is in the 3′ UTR. Expression data retrieved from Genevestigator (https://www.genevestigator.com/gv/, Hruz et al., 2008) indicated a low level of expression of AtLFG1 and a higher expression of AtLFG2 in Arabidopsis leaves (data not shown). To confirm suppression of AtLFG expression in T-DNA insertion mutants, RT-PCR was performed. AtLFG1 expression was very low in leaves of Col-0 wild-type plants as indicated by high cycle numbers for PCR amplification. However, AtLFG1 expression was not detectable in T-DNA insertion line SALK_147263C and only weakly detectable in T-DNA insertion line SALK_111590. The full-length transcript of AtLFG2 could easily be amplified from Col-0 wild-type leaves, but was not detectable in the AtLFG2 T-DNA insertion line (Fig. 6A). Hereafter, AtLFG1 lines SALK_147263C and SALK_111590 were named Atlfg1-1 and Atlfg1-2, respectively, and the AtLFG2 T-DNA insertion line SALK_052507C Atlfg2-1.
Fig. 6.
Impact of AtLFG1 or AtLFG2 over- or underexpression on the outcome of the Arabidopsis–powdery mildew interaction. (A) Development of E. cruciferarum on 5-week-old Col-0 and Atlfg T-DNA insertion mutants at 5 dpi. Conidiophores per colony were counted on five individual plants per mutant, respectively; 50 colonies per line were evaluated. Expression of the full-length target genes was analysed by RT-PCR. Amplification of a UBIQUITIN 5fragment indicated similar quantity of template cDNA. The upper panel shows AtLFG1 expression in wild-type Col-0, Atlfg1-1, and Atlfg1-2 plants; the lower panel shows AtLFG2 expression in wild-type Col-0 and in Atlfg2-1. (B) Development of E. cruciferarum on 5-week-old AtLFG overexpression mutants and empty pLH6000 vector control plants at 5 dpi. Conidiophores per colony were counted on one leaf of five individual plants per mutant, respectively. AtLFG1 or 2 expression in plants transformed with the empty vector pLH6000 and AtLFG1 or 2 overexpression mutants was examined by RT-PCR. Amplification of a UBQ5 fragment indicated similar quantity of template cDNAs. The upper panel shows AtLFG1 expression in empty pLH6000 vector and Atlfg1 overexpression plants; the lower panel AtLFG2 expression in empty pLH6000 vector and in AtLFG2 overexpression plants. Values are mean and standard errors; *, ** and *** indicate significance at P < 0.05, 0.01 and 0.001, respectively, according to Student’s t-test, calculated over mean conidiophores per colony of the single plants. The experiments were repeated twice with similar results.
We inoculated Atlfg1-1, Atlfg1-2, and Atlfg2-1 mutants with E. cruciferarum, a powdery mildew fungus, which is adapted to Arabidopsis, and analysed the infection by macroscopy as well as microscopy. Three independent experiments did not observe strong differences in powdery mildew symptoms on Atlfg1 and Atlfg2 mutants compared to Col-0 at 9 d post inoculation (dpi, data not shown). However, on Atlfg T-DNA insertion mutants, fungal development was reduced at 5 dpi, as assessed by counting the number of conidiophores per colony. E. cruciferarum developed 30±3 conidiophores per colony on Col-0 wild-type leaves, but only 14±1 conidiophores per colony on Atlfg1-1, 18±5 on Atlfg1-2, and 15±4 conidiophores per colony on Atlfg2-1 (Fig. 6A). This effect was statistically significant for the null mutants Atlfg1-1 and Atlfg2-1 but not for Atlfg1-2, which has residual expression of AtLFG1. Similar results were obtained in two further biological replications. Hence, loss of AtLFG1 or AtLFG2 delayed fungal development on susceptible Arabidopsis. As assessed by microscopy, impaired development of E. cruciferarum on Atlfg mutants did not seem to be associated with accelerated cell death responses (data not shown). However, all Atlfg mutants showed a reproducible tendency to more cell death upon Alternaria alternata f.sp. lycopersici (AAL) toxin treatment (Supplementary Fig. S5), suggesting some potential of AtLFG proteins to inhibit cell death.
By means of A. tumefaciens-mediated transformation, stable transgenic Arabidopsis plants were created, that express AtLFG1 or AtLFG2 coding sequences under the control of the CaMV 35S promoter. Control plants were transformed with the empty vector pLH6000. Presence of the integrated T-DNA was confirmed by PCR. Transgenic plants showed elevated levels of AtLFG1 or AtLFG2 transcripts, respectively, when compared to empty vector control plants as analysed by RT-PCR (Fig. 6B). On AtLFG1 as well as on AtLFG2 overexpression mutants, E. cruciferarum showed an accelerated development when compared to the empty vector control plants. The number of conidiophores per colony increased from 19±1 on empty vector control plants to 37±2 on AtLFG1 and to 51±7 on AtLFG2 overexpressing plants (Fig. 6B). Another two biological replications gave similar results. Together, data indicate a function of AtLFG1 and AtLFG2 in supporting development of E. cruciferarum in a compatible interaction with Arabidopsis.
Discussion
Regulation of cell death is crucial for the development and functional maintenance of multicellular organisms and plays an important role in their response to stress (Williams and Dickman, 2008; Coll et al., 2011; Fuchs and Steller, 2011). In mammals, different kinds of signalling pathways end up in apoptosis type PCD. BI-1 as well as LFG proteins can inhibit these types of PCD (Hu et al., 2009; Henke et al., 2011; Robinson et al., 2011). Here, this work shows that, besides their functions in inhibition of cell death, BI-1 and LFG proteins share similar functions in plant susceptibility to powdery mildew fungi.
In addition to the obvious functional relationship in cell death suppression in metazoans, there are striking similarities in the transmembrane scaffold of BI-1 and LFG proteins. Most computational analyses predicted six or seven transmembrane domains for BI-1 or LFG proteins (e.g. Reimers et al., 2006, 2008; Gubser et al., 2007; Nielsen et al., 2011; Supplementary Table S1). Only recently, Carrara et al. (2012) suggested a six transmembrane topology for both, BI-1 and GAAPs, with their C-terminal portions extending into the cytosol or forming a membrane reentrant loop.
Mammalian LFG proteins split up into five subfamilies (Fig. 1A). Phylogenetic analyses by Hu et al. (2009) indicate that all mammalian members of the LFG family likely arose from an LFG4-like ancestor, which, according to analysis of available genomic data, may also be the progenitor of plant LFGs. In accordance with that, all barley and Arabidopsis LFG protein sequences share the highest similarity with mouse MmLFG4 (Fig. 1). Since phylogenetic relatedness among barley and Arabidopsis LFG proteins is not clear, barley proteins were designated HvLFGa–HvLFGe and Arabidopsis proteins AtLFG1–AtLFG5.
Except in yeast cells, which apparently do not have BI-1 (only the GAAP/TMBIM4/LFG4-related protein YNL305C, Henke et al., 2011), both BI-1 and LFG proteins are supposedly generally present in eukaryotic cells, whereas other important metazoan cell death regulators are missing in plants. The high degree of structural conservation and sequence similarity within the LFG protein family may indicate an evolutionarily important function. At least human GAAP/TMBIM4/LFG4 seems to be necessary for cell survival in HeLa cells (Gubser et al., 2007). However, the expression of single LFG genes does not seem to be mandatory for whole organisms, since for example LFG knockout mice or Drosophila melanogaster larvae do not show obvious impairment in vitality or fertility (Hurtado de Mendoza et al., 2011; Nielsen et al., 2011; Rojas-Rivera et al., 2012). Similarly, knockout or knockdown of single AtLFG family members did not obviously affect viability or development of Arabidopsis plants in general (data not shown), possibly due to redundant functions of AtLFG proteins. Functional redundancy may also be the reason for only weakly enhanced cell-death-related phenotypes in Atlfg mutants after treatment with the cell-death-inducing AAL toxin, which is a ceramide synthase inhibitor interfering with sphingolipid biosynthesis (Möbius and Hertweck, 2009). Similarly, Atbi-1 knockout mutants showed accelerated cell death reactions upon treatment with the fungal toxin fumonisin B1, which acts similar to the AAL toxin (Watanabe and Lam, 2006; Ishikawa et al., 2011).
Besides direct interference with the formation of cell-death-inducing protein complexes or cell-death-related signal transduction, the ability to regulate intracellular Ca2+ fluxes may account for the cell-death-inhibiting capacities of LFG proteins (Somia et al., 1999; Fernández et al., 2007; Gubser et al., 2007; de Mattia et al., 2009; Hu et al., 2009; Yamaji et al., 2010; Hurtado de Mendoza et al., 2011; Nielsen et al., 2011; Shukla et al., 2011; Rojas-Rivera et al., 2012). Similarly, BI-1 may inhibit cell death by modulating Ca2+ concentrations in the endoplasmic reticulum and cytosol possibly by regulating ion channel function or by forming an ion channel itself (Robinson et al., 2011). GFP-tagged BI-1 proteins largely localize to the endoplasmic reticulum and the nuclear envelope in mammalian and plant cells (Hückelhoven, 2004; Robinson et al., 2011). LFG proteins can also be endoplasmic reticulum-resident (Gubser et al., 2007; Yamaji et al., 2010; Rojas-Rivera et al., 2012), but often accumulate in other membrane structures such as the plasma membrane, Golgi organelles, or edosomal/lysosomal compartments (Somia et al., 1999; Fernández et al., 2007; Yamaji et al., 2010; Nielsen et al., 2011; Shukla et al., 2011). In line with that, a GFP-HvLFGa protein accumulated in the plasma membrane of non-inoculated barley epidermal cells, with a small portion being associated with endosomal compartments (Fig. 3A and Supplementary Fig. S3B).
Importantly, HvBI-1 is a susceptibility factor in barley interaction with Bgh (Eichmann et al., 2010), and plant LFG proteins seem to share this function with BI-1. Overexpression of either HvBI-1 or HvLFGa promotes the penetration success of the fungus, while their knockdown reduces it (Hückelhoven et al., 2003; Eichmann et al., 2004, 2006, 2010; Fig. 2B). The apparent discrepancy between the strong ability of the HvLFGa TIGS construct to silence GFP-HvLFGa expression and its comparatively weaker ability to reduce the susceptibility to Bgh (Fig. 2B, C) supports that susceptibility to Bgh is genetically redundant and may involve other barley LFG genes.
Despite obvious differences in their subcellular localization, HvBI-1 and HvLGFa proteins are locally detectable at Bgh penetration sites, and additionally accumulate around haustorial complexes in penetrated cells of barley (Eichmann et al., 2006; Figs. 4 and 5). As for MLO, the exact cellular function of susceptibility factors and the way that powdery mildew fungi potentially use them for their own advantage is often not clear (Hückelhoven and Panstruga, 2011). Given that the proteins bear the potential to regulate Ca2+ fluxes, it might be conceivable that both BI-1 and LFG support the penetration of Bgh into epidermal cells by locally modulating (Ca2+-dependent) basal pathogen-responses at the cell wall, such as ROS production, cytoskeleton rearrangement, and polarized secretion processes (Eichmann et al., 2006, 2010; Hückelhoven, 2007). This may further be supported by the fact that both BI-1 and LFG proteins have been associated with the formation or function of lipid rafts (Fernández et al., 2007; Nagano et al., 2009), sphingolipid-containing plasma membrane microdomains, which have been discussed as putative entry portals for pathogens or pathogen effectors into eukaryotic cells (Bhat and Panstruga, 2005; Kale, 2012).
Similarly to HvLFGa, AtLFG1 and AtLFG2 proteins may support powdery mildew development (Fig. 6). Although thus far only one null mutant could be identified each for AtLFG1 and AtLFG2, opposite powdery mildew phenotypes of mutants and overexpressors support a function of these AtLFG proteins in the interaction with powdery mildew fungi. This may suggest a common function of plant LFG proteins in powdery mildew susceptibility. Inspections of interaction sites on Atlfg mutants did not indicate obvious differences in penetration rates or frequencies of defence reactions such as local or whole-cell callose depositions (data not shown). However, impaired development of E. cruciferarum on Atlfg mutants might be explained by a reduced supply of nutrients to the fungus. Powdery mildew fungi establish haustorial complexes inside their host cells likely in order to take up nutrients, and are thought to reprogramme host cell metabolism towards fungal nutrition (O’Connell and Pantruga, 2006; Eichmann and Hückelhoven, 2008; Micali et al., 2011). Green fluorescence of GFP-HvLFGa surrounded the mCherry-labelled host cytoplasm around the haustorial complex (Fig. 5D). This may indicate that the GFP-HvLFGa-labelled membrane likely represents the tonoplast invaginated by the fungal haustorium. Exo- and endocytic processes appear to participate in the formation and functional maintenance of fungal feeding structures, since they involve enlargement and modifications of the plant plasma membrane (Mendgen et al., 1995; Koh et al., 2005; O’Connell and Pantruga, 2006; Leborgne-Castel et al., 2010; Micali et al., 2011). The current study observed a dramatic relocalization of GFP-HvLFGa (but not the plasma membrane marker pm-rk) into mobile tubular and big round vesicle-like structures in barley cells containing mature fungal haustoria (Figs. 4C and 5A–C). Similar structures became visible after expression of vacuolar membrane markers (Fig. 5A, B) or after staining with the endocytosis tracer FM4-64 near powdery mildew haustoria (Fig. 4D). FM4-64-labelled tubular and round vesicle-like structures also occurred in rice epidermal cells containing biotrophic invasive hyphae of Magnaporthe oryzae and might represent endocytic compartments derived from the plant plasma membrane. In addition, cells containing biotrophic invasive hyphae showed elevated vacuolar dynamics (Kankanala et al., 2007). In Arabidopsis, bulb-like vacuolar structures occurred predominantly in young, metabolically active tissues (Saito et al., 2002; Uemura et al., 2002). Endosomes/multivesicular bodies also accumulate in barley epidermal cells penetrated by Bgh as observed by transmission electron microscopy (An et al., 2006). It is not yet clear whether GFP-HvLFGa only labels or even supports endocytosis or vacuolar remodelling in penetrated epidermal cells. Vacuolar reorganization associated with enhanced endocytosis might promote nutrient uptake through fungal haustoria.
Possible roles in suppression of cell death or cell-wall-associated defence as well as in nutrient supply render BI-1 and LFG proteins possible targets for fungal effectors. It is assumed that the other members of the barley or Arabidopsis LFG family have similar functions as the ones tested here. According to publicly available gene expression data in Genevestigator (Hruz et al., 2008) and PlexDB (Dash et al., 2012), all barley and Arabidopsis LFG genes seem to be expressed in leaves. Given the high sequence similarity of AtLFG genes, functional redundancy possibly prevent stronger cell death and pathogenicity-related phenotypes in single Atlfg mutants, which would become more obvious in multiple knockout or knockdown mutants. In animal cells, GBP/GRINA/TMBIM3/LFG1 and BI-1 have synergistic anti-apoptotic activities and might even physically interact (Rojas-Rivera et al., 2012). According to 6503 publicly available gene expression samples analysed in Genevestigator 4, AtBI-1 and AtLFG2 are co-expressed in Arabidopsis with a Pearson′s correlation coefficient of 0.67 (position number 5 among the AtLFG2 co-expressed genes, data not shown). Further experiments are needed to show whether BI-1 and LFG proteins jointly regulate a converging signalling pathway to enhanced susceptibility or regulate similar processes independently.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Comparison of plant and mouse BI-1 and LFG proteins.
Supplementary Fig. S2. Expression of HvLFGa in barley epidermal cells.
Supplementary Fig. S3. Subcellular localization of a functional GFP-HvLFGa fusion protein in unchallenged barley epidermal cells.
Supplementary Fig. S4. T-DNA position in AtLFG1 and 2 T-DNA insertion mutants.
Supplementary Fig. S5. Atlfg mutants have a tendency to more sensitivity to cell-death-inducing AAL toxin.
Supplementary Table S1. Amino acid positions of protein domains in BI-1 and BI-1-related LFG proteins of human, barley, and Arabidopsis thaliana.
Supplementary Video S1. GFP-HvLFGa-labelled bulb-like mobile structures near a haustorium of the barley powdery mildew fungus in a barley epidermal cell.
Acknowledgements
The authors are grateful to Angela Alkofer, Marina Daake, Simone Rankl, Simon Selleneit, and Katharina Beckenbauer for technical assistance, to David Robinson (University of Heidelberg, Germany) for providing the GmMAN1-RFP construct, to Frans JM Maathuis (University of York, UK) for providing the OsTPKb-RFP construct, to Hans Thordal-Christensen (University of Copenhagen, Denmark) for providing the RFP-AtARA7 construct, and to Gregor Langen (University of Giessen, Germany) for providing pGY-1-mCherry. The authors also thank the Salk Institute Genomic Analysis Laboratory and the Nottingham Arabidopsis Stock Centre for generating and providing the sequence-indexed Arabidopsis T-DNA insertion mutants. This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft) (grant number EI835/1-1) to R.E. and R.H.
Glossary
Abbreviations:
- AAL
Alternaria alternata f.sp. lycopersici
- At
Arabidopsis thaliana
- BI-1
BAX INHIBITOR-1
- CaMV
cauliflower mosaic virus
- dpi
d after inoculation
- GAAP
Golgi anti-apoptotic protein
- hpi
h post inoculation
- Hv
Hordeum vulgare
- LFG
LIFEGUARD
- MLO
MILDEW LOCUS O
- OXLP
OXALATE OXIDASE-LIKE PROTEIN
- PAMP
pathogen-associated molecular pattern
- PCD
programmed cell death
- RFP
red fluorescent protein
- RNAi
RNA interference
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment
- T-DNA
transferred DNA
- TIGS
transient-induced gene silencing
- TMBIM
transmembrane Bax Inhibitor-motif
- UBC2
UBIQUITIN CONJUGATING ENZYME 2
- UBQ5
UBIQUITIN 5.
References
- An Q, Ehlers K, Kogel KH, van Bel AJE, Hückelhoven R. 2006. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytologist 172, 563–576. [DOI] [PubMed] [Google Scholar]
- Babaeizad V, Imani J, Kogel KH, Eichmann R, Hückelhoven R. 2009. Over-expression of the cell death regulator BAX inhibitor-1 in barley confers reduced or enhanced susceptibility to distinct fungal pathogens. Theoretical and Applied Genetics 118, 455–463. [DOI] [PubMed] [Google Scholar]
- Bernsel A, Viklund H, Hennerdal A, Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Research 37, W465–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RA, Panstruga R. 2005. Lipid rafts in plants. Planta 223, 5–19. [DOI] [PubMed] [Google Scholar]
- Böhlenius H, Mørch SM, Godfrey D, Nielsen ME, Thordal-Christensen H. 2010. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. The Plant Cell 22, 3831–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, et al. 1997. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Carrara G, Saraiva N, Gubser C, Johnson BF, Smith GL. 2012. Six-transmembrane topology for Golgi anti-apoptotic protein (GAAP) and Bax inhibitor 1 (BI-1) provides model for the transmembrane Bax inhibitor-containing motif (TMBIM) family. Journal of Biological Chemistry 287, 15896–15905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP. 2007. The TIGR Plant Transcript Assemblies database. Nucleic Acids Research 35, D846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. 2011. Programmed cell death in the plant immune system. Cell Death and Differentiation 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, et al. 2003. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. [DOI] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, et al. 2006. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nature Genetics 38, 716–720. [DOI] [PubMed] [Google Scholar]
- da Cunha L, McFall AJ, Mackey D. 2006. Innate immunity in plants: a continuum of layered defenses. Microbes and Infection 8, 1372–1381. [DOI] [PubMed] [Google Scholar]
- de Mattia F, Gubser C, van Dommelen MM, et al. 2009. Human Golgi antiapoptotic protein modulates intracellular calcium fluxes. Molecular Biology of the Cell 20, 3638–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. 2012. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Research 40, D1194–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P. 2005. A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Molecular Plant–Microbe Interactions 18, 755–761. [DOI] [PubMed] [Google Scholar]
- Eichmann R, Bischof M, Weis C, Shaw J, Lacomme C, Schweizer P, Duchkov D, Hensel G, Kumlehn J, Hückelhoven R. 2010. BAX INHIBITOR-1 is required for full susceptibility of barley to powdery mildew. Molecular Plant–Microbe Interactions 23, 1217–1227. [DOI] [PubMed] [Google Scholar]
- Eichmann R, Dechert C, Kogel KH, Hückelhoven R. 2006. Transient over-expression of barley BAX inhibitor-1 weakens oxidative defence and MLA12-mediated resistance to Blumeria graminis f.sp. hordei . Molecular Plant Pathology 7, 543–552. [DOI] [PubMed] [Google Scholar]
- Eichmann R, Hückelhoven R. 2008. Accommodation of powdery mildew fungi in intact plant cells. Journal of Plant Physiology 165, 5–18. [DOI] [PubMed] [Google Scholar]
- Eichmann R, Schultheiss H, Kogel KH, Hückelhoven R. 2004. The barley apoptosis suppressor homologue BAX inhibitor-1 compromises nonhost penetration resistance of barley to the inappropriate pathogen Blumeria graminis f. sp. tritici . Molecular Plant–Microbe Interactions 17, 484–490. [DOI] [PubMed] [Google Scholar]
- Fernández M, Segura MF, Solé C, Colino A, Comella JX, Ceña V. 2007. Lifeguard/neuronal membrane protein 35 regulates Fas ligand-mediated apoptosis in neurons via microdomain recruitment. Journal of Neurochemistry 103, 190–203. [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. 2011. Programmed cell death in animal development and disease. Cell 147, 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser C, Bergamaschi D, Hollinshead M, Lu X, van Kuppeveld FJ, Smith GL. 2007. A new inhibitor of apoptosis from vaccinia virus and eukaryotes. PLoS Pathogens 3, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke N, Lisak DA, Schneider L, Habicht J, Pergande M, Methner A. 2011. The ancient cell death suppressor BAX inhibitor-1. Cell Calcium 50, 251–260. [DOI] [PubMed] [Google Scholar]
- Hoefle C, Huesmann C, Schultheiss H, Börnke F, Hensel G, Kumlehn J, Hückelhoven R. 2011. A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. The Plant Cell 23, 2422–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Smith TF, Goldberger G. 2009. LFG: a candidate apoptosis regulatory gene family. Apoptosis 14, 1255–1265. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. 2004. BAX inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 9, 299–307. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. 2007. Cell wall-associated mechanisms of disease resistance and susceptibility. Annual Reviews of Phytopathology 45, 101–127. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Dechert C, Kogel KH. 2003. Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis . Proceedings of the National Academy of Sciences, USA 100, 5555–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R, Panstruga R. 2011. Cell biology of the plant–powdery mildew interaction. Current Opinion in Plant Biology 14, 738–746. [DOI] [PubMed] [Google Scholar]
- Hurtado de Mendoza T, Perez-Garcia CG, Kroll TT, Hoong NH, O’Leary DD, Varma IM. 2011. Antiapoptotic protein Lifeguard is required for survival and maintenance of Purkinje and granular cells. Proceedings of the National Academy of Sciences, USA 108, 17189–17194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isayenkov S, Isner JC, Maathuis FJ. 2011. Rice two-pore K+ channels are expressed in different types of vacuoles. The Plant Cell 23, 756–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M, Lam E. 2011. Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death and Differentiation 18, 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Hansen M, Joehnk N, Lyngkjaer MF, Collinge DB. 2007. The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis . Plant Molecular Biology 65, 137–150. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kale SD. 2012. Oomycete and fungal effector entry, a microbial Trojan horse. New Phytologist 193, 874–881. [DOI] [PubMed] [Google Scholar]
- Kankanala P, Czymmek K, Valent B. 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. The Plant Cell 19, 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck M, Hardham AR, Dodds PN. 2011. The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cellular Microbiology 13, 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S. 2005. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. The Plant Journal 44, 516–529. [DOI] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, et al. 2008. Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N, Adam T, Bouhidel K. 2010. Endocytosis in plant-microbe interactions. Protoplasma 247, 177–193. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research 39, D225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendgen K, Bachem U, Stark-Urnau M, Xu H. 1995. Secretion and endocytosis at the interface of plants and fungi. Canadian Journal of Botany 73, 640–648. [Google Scholar]
- Micali CO, Neumann U, Grunewald D, Panstruga R, O’Connell R. 2011. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cellular Microbiology 13, 210–226. [DOI] [PubMed] [Google Scholar]
- Möbius N, Hertweck C. 2009. Fungal phytotoxins as mediators of virulence. Current Opinion in Plant Biology 12, 390–398. [DOI] [PubMed] [Google Scholar]
- Nagano M, Ihara-Ohori Y, Imai H, Inada N, Fujimoto M, Tsutsumi N, Uchimiya H, Kawai-Yamada M. 2009. Functional association of cell death suppressor, Arabidopsis Bax inhibitor-1, with fatty acid 2-hydroxylation through cytochrome b5 . The Plant Journal 58, 122–134. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Chambers MA, Romm E, Lee LY, Berndt JA, Hudson LD. 2011. Mouse transmembrane BAX inhibitor motif 3 (Tmbim3) encodes a 38kDa transmembrane protein expressed in the central nervous system. Molecular and Cellular Biochemistry 357, 73–81. [DOI] [PubMed] [Google Scholar]
- Niks RE, Marcel TC. 2009. Nonhost and basal resistance: how to explain specificity? New Phytologist 182, 817–828. [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Panstruga R. 2006. Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytologist 171, 699–718. [DOI] [PubMed] [Google Scholar]
- Reimers K, Choi CY, Bucan V, Vogt PM. 2008. The Bax Inhibitor-1 (BI-1) family in apoptosis and tumorigenesis. Current Molecular Medicine 8, 148–156. [DOI] [PubMed] [Google Scholar]
- Reimers K, Choi CY, Mau-Thek E, Vogt PM. 2006. Sequence analysis shows that Lifeguard belongs to a new evolutionarily conserved cytoprotective family. International Journal of Molecular Medicine 18, 729–734. [PubMed] [Google Scholar]
- Robinson KS, Clements A, Williams AC, Berger CN, Frankel G. 2011. Bax inhibitor 1 in apoptosis and disease. Oncogene 30, 2391–2400. [DOI] [PubMed] [Google Scholar]
- Rojas-Rivera D, Armisén R, Colombo A, et al. 2012. TMBIM3/GRINA is a novel unfolded protein response (UPR) target gene that controls apoptosis through the modulation of ER calcium homeostasis. Cell Death and Differentiation 19, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Saito C, Ueda T, Abe H, Wada Y, Kuroiwa T, Hisada A, Furuya M, Nakano A. 2002. A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis . The Plant Journal 29, 245–255. [DOI] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Abderhalden O, Dudler R. 1999. A transient assay system for the functional assessment of defense-related genes in wheat. Molecular Plant–Microbe Interactions 12, 647–654. [DOI] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Schulze-Lefert P, Dudler R. 2000. Double-stranded RNA interferes with gene function at the single-cell level in cereals. The Plant Journal 24, 895–903. [DOI] [PubMed] [Google Scholar]
- Shukla S, Fujita K, Xiao Q, Liao Z, Garfield S, Srinivasula SM. 2011. A shear stress responsive gene product PP1201 protects against Fas-mediated apoptosis by reducing Fas expression on the cell surface. Apoptosis 16, 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somia NV, Schmitt MJ, Vetter DE, Van, Antwerp D, Heinemann SF, Verma IM. 1999. LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proceedings of the National Academy of Sciences, USA 96, 12667–12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57, 758–771. [DOI] [PubMed] [Google Scholar]
- Uemura T, Morita MT, Ebine K, Okatani Y, Yano D, Saito C, Ueda T, Nakano A. 2010. Vacuolar/pre-vacuolar compartment Qa-SNAREs VAM3/SYP22 and PEP12/SYP21 have interchangeable functions in Arabidopsis . The Plant Journal 64, 864–873. [DOI] [PubMed] [Google Scholar]
- Uemura T, Yoshimura SH, Takeyasu K, Sato MH. 2002. Vacuolar membrane dynamics revealed by GFP-AtVam3 fusion protein. Genes to Cells 7, 743–753. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. 2006. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. The Plant Journal 45, 884–894. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. 2009. Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. International Journal of Molecular Sciences 10, 3149–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YD, Zhang Z, Andersen CH, Schmelzer E, Gregersen PL, Collinge DB, Smedegaard-Petersen V, Thordal-Christensen H. 1998. An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Molecular Biology 36, 101–112. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. 2006. Transformation of agrobacterium using the freeze-thaw method. CSH Protocols 2006, 10.1101/pdb.prot4666. [DOI] [PubMed] [Google Scholar]
- Williams B, Dickman M. 2008. Plant programmed cell death: can’t live with it; can’t live without it. Molecular Plant Pathology 9, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji T, Nishikawa K, Hanada K. 2010. Transmembrane BAX inhibitor motif containing (TMBIM) family proteins perturbs a trans-Golgi network enzyme, Gb3 synthase, and reduces Gb3 biosynthesis. Journal of Biological Chemistry 285, 35505–35518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG. 2005. Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. The Plant Cell 17, 1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. 2001. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.