Abstract
One of the reasons for the progressive yield decline observed in aerobic rice production is the rapid build-up of populations of the rice root knot nematode Meloidogyne graminicola. These nematodes induce specialized feeding cells inside root tissue, called giant cells. By injecting effectors in and sipping metabolites out of these cells, they reprogramme normal cell development and deprive the plant of its nutrients. In this research we have studied the transcriptome of giant cells in rice, after isolation of these cells by laser-capture microdissection. The expression profiles revealed a general induction of primary metabolism inside the giant cells. Although the roots were shielded from light induction, we detected a remarkable induction of genes involved in chloroplast biogenesis and tetrapyrrole synthesis. The presence of chloroplast-like structures inside these dark-grown cells was confirmed by confocal microscopy. On the other hand, genes involved in secondary metabolism and more specifically, the majority of defence-related genes were strongly suppressed in the giant cells. In addition, significant induction of transcripts involved in epigenetic processes was detected inside these cells 7 days after infection.
Key words: Giant cell, laser-capture microdissection, Meloidogyne graminicola, Oryza sativa, root knot nematode, transcriptome.
Introduction
Biotrophic plant pathogens have evolved sophisticated strategies to manipulate their host. They derive all of their nutrients from living plant tissues by making intimate contact with their host while avoiding a resistance response. Rice is one of the most important crop plants worldwide and an excellent model system for studying monocotyledonous plants. Estimates of annual yield losses due to plant-parasitic nematodes on this crop range from 10 to 25% worldwide (Bridge et al., 2005). One of the agronomically most important nematodes attacking rice is the rice root knot nematode (RKN) Meloidogyne graminicola. Attack of plant roots by sedentary plant parasitic nematodes, like the RKNs (Meloidogyne spp.) leads to the development of specialized feeding cells in the vascular tissue. The second-stage juvenile of the RKN punctures selected vascular cells with its stylet and injects pharyngeal secretions, which ultimately leads to the reorganization of these cells into typical feeding structures called giant cells, from which the nematode feeds for the remainder of its sedentary life cycle (Gheysen and Mitchum, 2011). Morphological and physiological reprogramming of the initial feeding cell leads to nuclear enlargement, proliferation of organelles, metabolic activation, cell-cycle alterations, and cell-wall changes (Gheysen and Mitchum, 2011). The hyperplasia and hypertrophy of the surrounding cells leads to the formation of a root gall, which is typically formed at the root tips in the case of the rice RKN M. graminicola. In comparison with other RKNs, M. graminicola has a very fast life cycle. In well-drained soil at 22–29 °C the life cycle of M. graminicola is completed in 19 days. Swelling of the root tips is observed as early as 1 day after infection (dai). At 3 dai, terminal hook-like galls are clearly visible (Bridge et al., 2005). After three moults the nematodes are mature, around 10–12 dai. Whereas most other RKNs deposit egg masses at the gall surface, the M. graminicola females lay their eggs inside the galls, and hatched juveniles can reinfect the same or adjacent roots.
We have recently studied transcriptional reprogramming patterns in galls induced by the RKN M. graminicola in rice using deep RNA sequencing (Kyndt et al., 2012a ). The there-reported gene-expression differences reflect a combination of changes occurring in giant cells and surrounding gall tissues. Due to the technical difficulty of isolating giant cells from the root tissue most transcriptome analyses have up till now focused on the whole-gall tissue, in Arabidopsis and tomato (e.g. Bar-Or et al., 2005; Jammes et al., 2005,). Nevertheless, though technically challenging, giant cell contents can be isolated using microaspiration (Wang et al., 2003) or laser-capture microdissection (LCM; Fosu-Nyarko et al., 2009; Barcala et al., 2010; Portillo et al., 2009, 2013). The research of Barcala et al. (2010) and Portillo et al. (2013) demonstrated the molecular distinctiveness between the giant cells and the surrounding gall tissue.
The goal of our research was to study the transcriptional changes in giant cells formed in rice roots upon RKN infection. LCM was combined with mRNA sequencing (mRNA-Seq) to study the giant cell transcriptome at two time points after infection. We have compared the data with reports from giant cells and complete galls induced by RKN in rice and other plant species. Some of the reported changes were independently validated by quantitative reverse transcriptase-PCR (qRT-PCR) and confocal microscopy. Our study highlights that key metabolic pathways, hormone homeostasis, and epigenetic processes are affected during giant cell development.
Materials and methods
Infection and LCM of giant cells
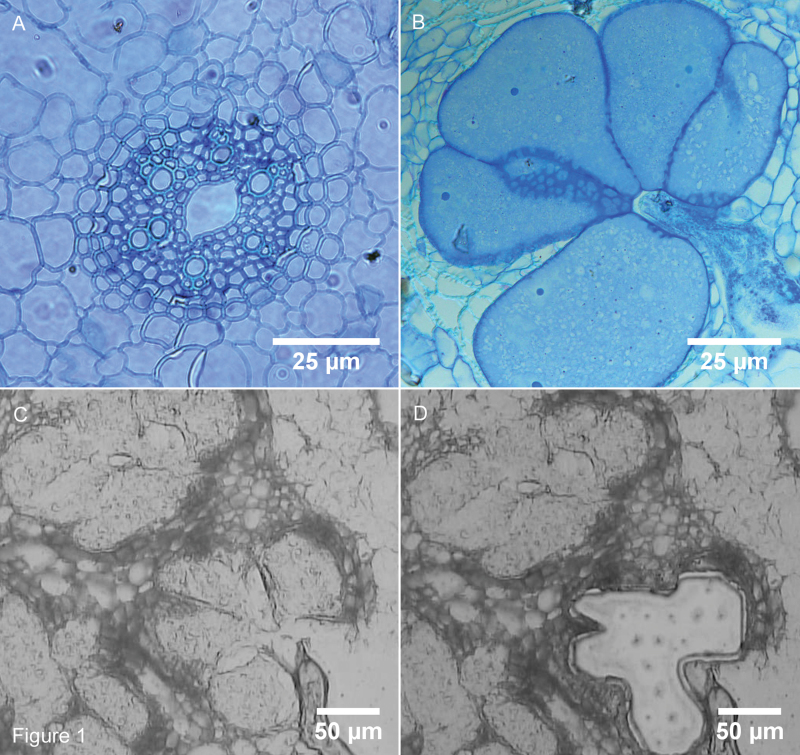
Oryza sativa cv. ‘Nipponbare’ (GSOR-100, USDA) was germinated for 6 days at 30 °C, transferred to SAP substrate (sand-absorbent polymer; Reversat et al., 1999) and further grown at 26 °C under a 16h/8h light/dark regime. Extra care was taken to prevent any light influence on the roots of the plants. Nematodes were cultured and extracted as described before (Kyndt et al., 2012b ). When 12 days old, plants were inoculated in SAP with 250 stage 2 juveniles of M. graminicola per plant. Control plants were mock-inoculated with water. One day after inoculation the plants were transferred to a hydroponic culturing system with Hoagland solution (Reversat et al., 1999) to synchronize the infection process. Infected and control roots were collected at 7 and 14 dai and fixed in Farmer’s fixative (3:1 ethanol/acetic acid). The material was dehydrated in ethanol and then cryosectioned with a cryostat at −20 °C. RKN form giant cells in the vascular tissue, but the type of cells specifically targeted is unknown and can even differ between individual nematodes in the same plant tissue (Endo, 1987). Therefore, a mixture of different types of cells from the vascular tissue of mature rice roots was used as control material in this study. Giant cells and control cells were captured using a Zeiss PALM Laser Microbeam according to the manufacturer’s instructions (Zeiss, Jena, Germany; Fig. 1). Captured cells were infiltrated in RNA extraction lysis buffer (Agilent, Waldbronn, Germany). Three independent biological replicates were taken at each time point, of which two were analysed by mRNA-Seq. The third independent biological replicate was used for qRT-PCR validation. About 150–200 giant cell sections and 300 control sections (of uninfected plants) per biological replicate were used for giant cell and control cell isolation by LCM.
Fig. 1.
Transverse sections of giant cells formed by the RKN M. graminicola in rice roots (O. sativa cv. ‘Nipponbare’). (A) Control vascular tissue; (B) 7 dai giant cells. Scale bars in A and B: 25 μm. (C) 7 dai giant cells before LCM; (D) 7 dai giant cells after LCM. Scale bars in C and D: 50 μm. This figure is available in colour at JXB online.
RNA extraction, library preparation, and Illumina GAIIx sequencing
RNA from LCM-isolated giant cells was extracted with the Absolutely RNA Nanoprep Kit (Agilent), followed by cDNA synthesis using the Ovation RNA-Seq System (NuGEN, Leek, The Netherlands). This system is based on the Ribo-SPIA® technology (NuGEN) to generate high-quality, linearly amplified cDNA from low amounts of RNA, and was specifically designed for next-generation sequencing platforms. The obtained cDNA concentrations varied between 4.8 and 6.6μg per sample. cDNA integrity was confirmed using the Agilent BioAnalyzer 2100 (Agilent) and qRT-PCR with two reference genes (Supplementary Table S1).
The full-length cDNA was fragmented by sonication with a Covaris S2 ultrasonicator (Covaris, Woburn, MA, USA). The mRNA-Seq library was constructed according to the NEB protocols E6040 (New England BioLabs, Ipswich, MA, USA). We used the multiplexing sequencing adapters provided in the Multiplexing Sample Preparation Oligo kit (Illumina, San Diego, CA, USA). Size selection of the library was performed on a 2% agarose gel (Low Range Ultra Agarose, Bio-Rad 161–3107; Bio-Rad, Nazareth Eke, Belgium). The denatured library was diluted to a final concentration of 6 pM and loaded on a paired-end read-flow cell (TruSeq v5 kit, Illumina). To minimize lane effects the samples were multiplexed. Each sample was sequenced in duplicate in two different lanes (four lanes total with eight multiplex identifier tags per lane). After cluster generation, the multiplexed library was sequenced on an Illumina Genome Analyzer IIx (36 cycles, paired-end).
Mapping reads to genome data and annotated transcripts
Reads were mapped to the O. sativa subsp. japonica reference genome (build MSU7.0) in two phases using TopHat version 1.3.1 (Trapnell et al., 2009) and Cufflinks version 1.0.3 (Trapnell et al., 2010). A detailed description of the workflow and settings used in the data analysis is given in Kyndt et al. (2012a ).
Identification of novel transcriptionally active regions
The Cufflinks program generates a GTF file including all transcripts annotated in MSU7.0 and putative novel transcripts derived from the data. All putative novel transcriptionally active regions (nTARs) marked as splice variants of known genes or located within intronic regions were disregarded and for the 18 291 remaining nTARs BLASTx searches were performed against Swiss-Prot and trEMBL and all predicted rice proteins (http://rice.plantbiology.msu.edu/). Homologues of the nTARs in rice expressed sequence tag (ESTs) were searched by tBLASTx (E<0.0001).
Calculation, normalization, and profiling of gene expression
Expression was quantified per sample and per annotated or unannotated transcript as the sum of all reads that mapped to the respective gene exons with a 16 base-pair tolerance on either side to compensate for potential errors in the gene annotation. Expression profiles were assessed using the R-package baySeq, version 1.5.1. (Hardcastle and Kelly, 2010). To compensate for artificial differences in read distributions, the original library sizes were multiplied by additional normalization factors calculated using the Trimmed Median of M-values method described in Robinson and Oshlack (2010) with standard settings as implemented in the edgeR package (version 2.0.3). A transcript was considered to be expressed if at least one sequence read was mapped to it in one of the samples. For all further analyses the expression level of each transcript for each condition was estimated as the fold change (FC) of mapped reads relative to the controls. The FC was calculated as follows: reads were normalized as described earlier and averaged over the biological replicates. Before calculating the base 2 log of the ratio of these averages, the number of reads was increased by 1 in each group (to avoid zero values).
Gene Ontology and enrichment analyses
Gene Ontology (GO) analysis and GO enrichment were performed using agriGO (Du et al., 2010). Parametric Analysis of Gene Set Enrichment (PAGE) (Kim and Volsky, 2005), based on differential gene expression levels (log2FC), was executed. Benjamini and Hochberg false discovery rate analysis (FDR) was performed using the default parameters to adjust the PAGE P values.
In addition, MapMan (Thimm et al., 2004) was used to visualize the expression of genes onto metabolic pathways and the WSR test (with Benjamini and Hochberg correction) was used to test the statistical significance of differential expression of these pathways.
Validation of mRNA-Seq by qRT-PCR
Based on potential functional importance, 13 genes were selected for validation in an independent biological sample by qRT-PCR. Locus number of these transcripts, primer sequences, and reaction efficiencies are presented in Supplementary Table S1. qRT-PCR was performed and analysed as described in Kyndt et al. (2012b ) using three technical replicates.
Confocal microscopy
Galls were collected at 7 dai and fixed in Farmer’s fixative (3:1 ethanol/acetic acid) overnight. They were dehydrated by a dilution series of 70, 90 and 100% ethanol (1h each) and stored at 4 °C for 7 days before microscopical analysis. To prepare samples for confocal microscopy, fixed galls were sliced in thin sections using a razor blade and mounted onto a slide in a drop of water. Images were acquired with a Nikon A1R confocal microscope, mounted on a Nikon Ti body and equipped with a 40× (NA=0.6) PLAN Fluor ELWD objective. Image acquisition was performed at a pinhole setting of 1 Airy unit and a pixel size of 0.6×0.6μm (zoom factor 1) or 0.2×0.2μm (zoom factor 3). To capture the spectral fingerprint (lambda stack) of chloroplast pigment autofluorescence, samples were illuminated with a 488nm argon laser and emission was detected on a spectral 32-PMT detector, set at a resolution of 2.5nm per detector within a range of 655.5–735.5nm. As a reference, lambda stacks were also acquired from fresh and fixed leaf sections using identical settings. Spectral profiles of (presumed) chloroplasts were determined by measuring the average intensity per wavelength interval (2.5nm) across the lambda stack range. Per condition, the average of the individual profiles of a minimum of four equally sized chloroplast (-like) regions was calculated.
Results
In this study, a comparative gene-expression analysis was carried out to investigate the rice response to infection with a sedentary nematode species, M. graminicola. This nematode induces the formation of specialized feeding sites called giant cells in root tissue. For an in-depth analysis of the transcriptional reprogramming induced in these cells, they were isolated by LCM at 7 and 14 dai and mRNA-Seq was carried out on the isolated RNA. Root cells from the vascular tissue of uninfected plants of the same age were used as control material. Transmitted light microscopical analysis revealed that at 7 dai the nematodes were at the juvenile 3 (J3) or J4 stage; at 14 dai most of the M. graminicola had matured and most females had laid eggs. The giant cells contained a dense cytoplasm, the shape was oval or globular, and the cell wall was thick (Fig. 1). Per biological replicate and time point 150–200 giant cell sections (Fig. 1) or 300 control sections (from uninfected plants) were used for LCM of the giant and vascular cells, respectively. After quality control, the cDNA was sequenced using the Illumina mRNA-Seq protocol.
In total 139 254 416 reads were acquired from infected and uninfected cells at the two time points. The data can be accessed through the GEO repository: GSE43577. The short reads were aligned against the whole reference genome sequence of rice cv. ‘Nipponbare’ (MSU7.0) and 79.2% of the sequenced fragments, 36-base-pair reads on each end of the fragments, could be mapped (Table 1). This mapping percentage is substantially higher than reported in our previous mRNA-Seq analysis on complete galls (Kyndt et al., 2012a ), where on average only 49.20% of the reads were mapped. The higher number of mapped reads in the current analysis can be explained by the fact that paired-end reads were used here, and because of the nature of the samples; specific isolated cells from within the root tissue have a lower chance of contaminants. The total length of mapped reads was over 10 billion bases, representing nearly 27-fold coverage of the rice genome and approximately 97-fold coverage of the annotated transcriptome. The expression of a total of 54 206 different rice transcripts was detected in the analysed tissues. Correlation between the two biological replicates sequenced from each cell type and at each time point was checked by means of Pearson correlation coefficient of the expression value of each gene after normalization. The average Pearson R between two biological replicates was 0.9959 (P<2.2e−16).
Table 1.
Overview of the obtained mRNA-Seq data from giant cells induced by nematode infection in rice and control cells from the vascular root tissue, and mapping of these sequences onto the rice genome
| Sample | Total number of sequenced fragments | Total number of paired mappings | Number of mapped fragments | Number of unmapped fragments | % Unique mapping |
|---|---|---|---|---|---|
| Uninfected vascular cells at 7 dai (1) | 17 031 983 | 32 558 679 | 13 967 317 | 3 064 666 | 82.01 |
| Uninfected vascular cells at 7 dai (2) | 16 406 474 | 30 877 769 | 12 916 523 | 3 489 951 | 78.73 |
| Giant cells at 7 dai (1) | 18 433 445 | 36 677 998 | 14 860 282 | 3 573 163 | 80.62 |
| Giant cells at 7 dai (2) | 18 253 140 | 34 825 239 | 142 94 260 | 3 958 880 | 78.31 |
| Uninfected vascular cells at 14 dai (1) | 17 135 578 | 33 563 491 | 13 585 221 | 3 550 357 | 79.28 |
| Uninfected vascular cells at 14 dai (2) | 17 600 487 | 33 773 375 | 13 622 898 | 3 977 589 | 77.40 |
| Giant cells at 14 dai (1) | 16 877 180 | 37 350 477 | 13 725 022 | 3 152 158 | 81.32 |
| Giant cells at 14 dai (2) | 17 516 129 | 37 472 359 | 13 296 867 | 4 219 262 | 75.91 |
| Total | 139 254 416 | 277 099 387 | 110 268 390 | 28 986 026 | 79.18 |
| Reads (two/fragment) | 278 508 832 | ||||
| Coverage of the rice genome | 26.86X | ||||
| Coverage of the rice transcriptome | 97.63X |
Comparative gene-expression profiling was performed by Gene Set Enrichment, pathway mapping, and statistical analysis of differential gene-expression levels between infected and uninfected cells at the different time points. In addition, a search was performed to detect nTARs not annotated in the rice genome assembly MSU7.0, and alternative splicing patterns in the isolated rice cells.
Transcriptome changes in giant cells at 7 dai
At 7 dai the nematodes feeding from the giant cells in the gall tissue were at the J3/J4 stage. A total of 42 756 different transcripts was found to be expressed in the collected cells at this time point. The expression level of all rice loci was compared between 7 dai giant cells and uninfected vascular cells in roots of the same age.
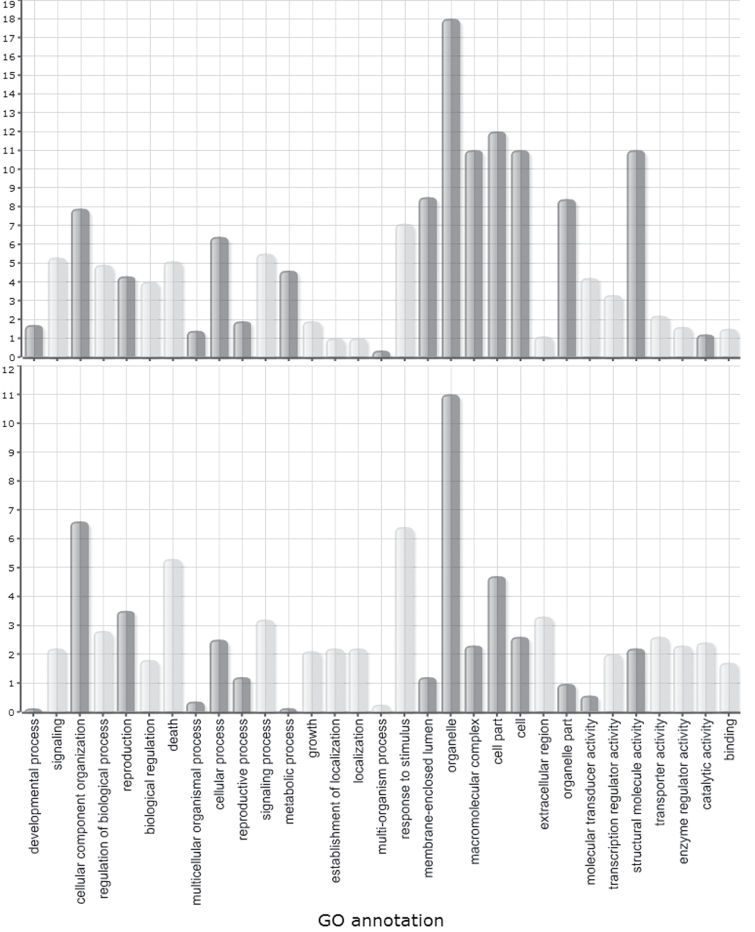
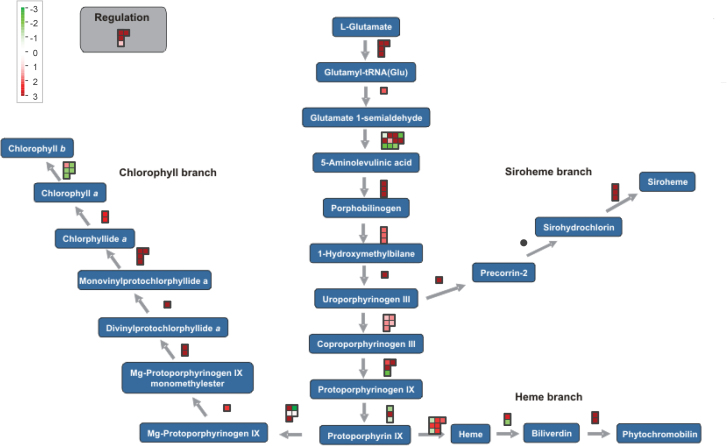
Gene Set Enrichment analysis on relative expression levels (log2FC) of all transcripts in the infected versus uninfected cells revealed that genes involved in the GO categories ‘biosynthetic process’ (mainly translation), ‘cell cycle’, ‘generation of precursor metabolites and energy’, and ‘cellular component organization’ were strongly upregulated at 7 dai, while genes involved in ‘tropism’, ‘signalling’, ‘response to stimulus’, and ‘secondary metabolic process’ were generally downregulated. Genes with a molecular function annotated as ‘structural molecule activity’ and ‘nucleic acid binding’ were generally higher expressed in 7 dai giant cells than in control cells (Fig. 2). Pathway mapping with MapMan showed a significant modification of, for instance, glycolysis, starch metabolism, trehalose metabolism, homoserine biosynthesis, tetrapyrrole synthesis, phenylpropanoid pathway, flavonoid production, cell-wall-precursor synthesis, and pectin esterases. Figure 3 shows the expression pattern of transcripts involved in tetrapyrrole biosynthesis, with a high frequency of transcripts induced in the giant cells versus control material. Higher plants contain four classes of tetrapyrroles – namely chlorophyll, haem, sirohaem, and phytochromobilin – and all of them play vital roles in various biological processes, including photosynthesis, respiration, nitrite, and sulphite reduction, as well as various cellular processes including gene expression, protein import, and the assembly of essential proteins (Tanaka and Tanaka, 2007). They are predominantly synthesized in plastids. Figure 3 shows that almost all genes involved in the biosynthesis of sirohaem, haem, phytochromobilin, and chlorophyll a, but not chlorophyll b, are expressed at higher levels in giant cells than in control cells.
Fig. 2.
Parametric Analysis of Gene Set Enrichment (PAGE) of transcriptome data from giant cells induced by RKN in rice at 7 dai (top) and 14 dai (bottom). Z scores of all secondary-level GO terms are shown. Bars in dark grey indicate GO terms that are upregulated in the infected tissue versus the corresponding control; light grey bars indicate GO terms that are downregulated in the infected tissue versus the corresponding control.
Fig. 3.
MapMan visualization of the expression profiles of genes involved in tetrapyrrole biosynthesis in 7 dai giant cells. The visualization shows the observed differential expression patterns, based on the log2FCs of mRNA levels, in giant cells versus control cells. Dots show the different paralogous genes encoding the enzyme that catalyses a certain step. Red dots indicate that the gene is upregulated in infected tissue versus the corresponding healthy control, while green indicates downregulation.
At 7 dai 7709 transcripts were found to be significantly differentially expressed (FDR<0.05), with 650 transcripts downregulated and 7059 upregulated in the giant cells (Supplementary Table S2). Thirteen of them were chosen for independent validation based on their potentially interesting function, and their expression pattern in the giant cells was evaluated on an independent biological sample by qRT-PCR. The expression trend was confirmed in all but one case (Supplementary Table S3). Differences in log2FC values as obtained from mRNA-Seq and qRT-PCR are largely due to biological variation and/or to differences in the applied algorithms for estimating expression levels.
The genes with strongest downregulation in giant cells versus uninfected cells included transcripts encoding gibberellin 2-β-dioxygenase, involved in gibberellin catabolism; chalcone synthase, involved in flavonoid production; protein disulphide isomerase, that catalyses post-translational protein modifications through formation and breakage of disulphide bonds; and two NBS-LRR disease-resistance proteins. Among the strongest upregulated transcripts were those encoding 78 transporter proteins; OsBAK1, the BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1; ABIL2, known to be involved in regulation of actin and microtubule organization; 17 cell-cycle-regulating cyclins; 10 tubulins; six members of the auxin-responsive OsIAA-gene family; six other auxin-response factors; and a remorin C-terminal domain-containing protein.
Experimental validation of chloroplast and chlorophyll content in 7 dai giant cells
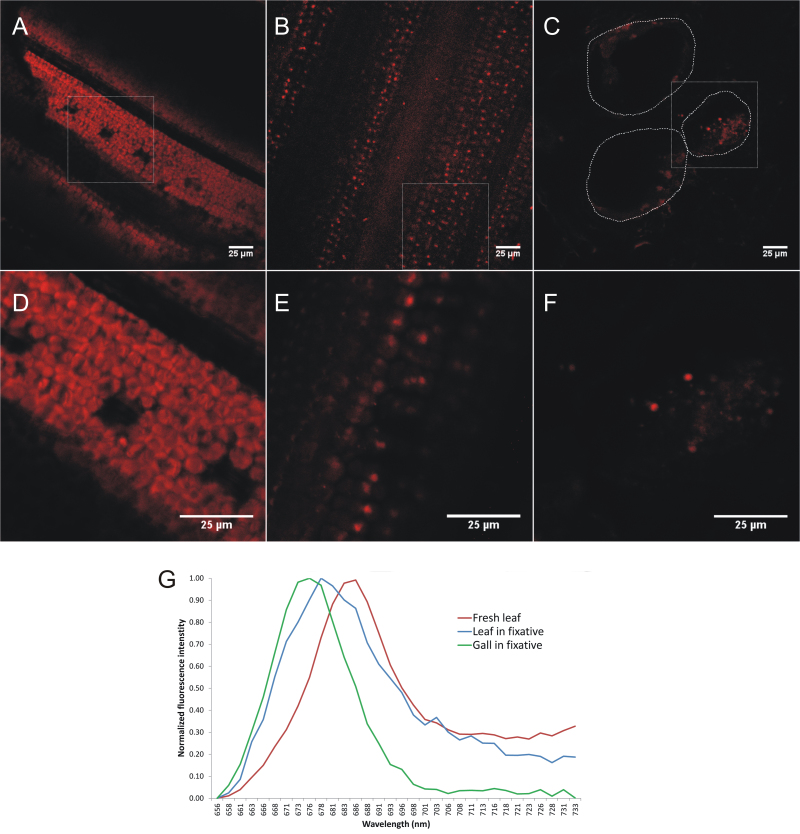
Many reports on plant–nematode interactions have analysed plants growing in Petri dishes in a day/night cycle (Sijmons et al., 1991). It is generally known that the developed nematode feeding sites contain chloroplasts under light influence (Orion and Wergin, 1982; Golinowski et al., 1996; Sijmons et al., 1991). Orion and Wergin studied this with electron microscopy and revealed the differentiation of chloroplasts from amyloplasts inside these light-exposed giant cells in tomato. However, in the current study, roots were completely shielded from light to preclude any artificial induction of chlorophyll and chloroplast formation. Interestingly, the results of the transcriptome data (Fig. 3) reveal a strong induction of photosynthesis-related genes in dark-grown giant cells, and hence we decided to confirm the presence of chlorophyll a inside the giant cells using confocal microscopy. To this end, lambda stacks of fixed giant cell sections were acquired using spectrally resolved confocal microscopy and they were compared with images from fixed and fresh leaf sections. The confocal images demonstrated that fresh rice leaves contain dense arrays of strongly autofluorescent chloroplasts, about 5μm in size and shaped as flattened disc (Fig. 4A, 4D). After fixation, the fluorescent pattern in the leaf had changed into a more punctate pattern, with chloroplast fluorescence limited to ≈1 μm-sized foci. This punctate effect is plausibly due to the fixation and (especially) the dehydration procedure (Fig. 4B, 4C). Remarkably, similar fluorescent foci were observed inside the giant cells, albeit to a lesser extent (Fig. 4C, 4F) and some autofluorescence was also seen in the neighbouring cells of the giant cells. However, the fluorescent foci and autofluorescence were never observed in healthy control roots (data not shown). To determine whether these foci in the giant cells contained chlorophyll a, we compared their spectral profile with that of chloroplasts in fixed and fresh leaf sections (Fig. 4G). Pedrós et al. (2008) reported that the chlorophyll a fluorescence emission spectrum is characterized by a major peak at 683nm, which dominates the autofluorescence spectrum of chloroplasts in the deep red region. Indeed, inside chloroplasts from fresh leaf material a fluorescent peak with a maximum intensity at 683nm was obtained upon excitation at 488nm. In chloroplasts from fixed leaf sections this peak was slightly shifted towards 678nm, possibly due to quenching effects. A very similar, although slightly shifted, peak was observed in the presumed chloroplasts from the giant cell sections, suggesting the presence of chlorophyll a in these structures. The slight shift of the peak and the deviation from the reference curves at the longer wavelengths may be due to the absence of other accessory pigments (e.g. carotenoids, xanthophylls, chlorophyll b) that contribute to the spectral profile.
Fig. 4.
Confocal microscopy and spectral profiling of chloroplast (-like) autofluorescence. (A–F) Confocal images were acquired from sections of fresh leaves (A, D), fixed leaves (B, E), and fixed giant cells (C, F) at two different zoom factors – 1× (A, B, C) and 3× (D, E, F) – using a spectral detector set to capture chlorophyll a autofluorescence (655.5–735.5nm). White squares in A, B, and C, show the region that was magnified in D, E, and F, respectively. The dotted lines in C delineate the boundaries of the giant cells. Note the difference in shape between chloroplasts of fresh and fixed leaf sections and the presence of chloroplast-like structures in the giant cell section. (G) Average spectral profiles, measured as average intensity per wavelength interval across the lambda stack range for at least four (presumed) chloroplast regions.
Transcriptome changes in giant cells at 14 dai
The expression level of all rice loci was compared between 14 dai giant cells and uninfected root vascular cells at the same time point. In total, 41 179 different transcripts were found to be expressed in the collected rice cells. While still ongoing at 7 dai, the expansion of giant cells and cytoskeleton rearrangements end at 10–14 dai in the case of Meloidogyne incognita infection on Arabidopsis (de Almeida-Engler et al., 2004).
Gene set enrichment analysis of relative expression levels (log2FC) of all transcripts in the infected versus uninfected cells revealed that genes involved in ‘photosynthesis’, ‘DNA metabolic process’, ‘cellular component organization’, ‘generation of precursor metabolites and energy’, and ‘cell cycle’ were strongly upregulated at 14 dai, while genes involved in ‘secondary metabolic process’, ‘response to stimulus’, and ‘cell death’ were generally downregulated. Transcripts coding for proteins with ‘hydrolase’ and ‘nuclease’ activity were generally upregulated (Fig. 2). When comparing gene set enrichment of 7 and 14 dai giant cells (Fig. 2), similar trends are observed, although genes in the GO categories ‘metabolic processes’, ‘structural molecule activity’, ‘organelle part’, ‘membrane-enclosed lumen’, and ‘macromolecular complex’ show less strong induction at 14 dai than at 7 dai. Pathway mapping with MapMan showed, similar to the 7 dai data, a significant modification of starch and sucrose metabolism, trehalose metabolism, tetrapyrrole synthesis, and the phenylpropanoid pathway. Additionally, significant modifications were detected in light reactions, flavonoid production, and cell-wall-related pathways in 14 dai giant cells.
2884 transcripts were significantly differentially expressed at FDR<0.05. 238 of them were downregulated, while 2646 were upregulated (Supplementary Table S4). Among the most downregulated genes, transcripts were found coding for nicotianamine synthase, involved in phytosiderophore biosynthesis; a glucan endo-1,3-β-glucosidase precursor; α-DOX2, involved in the synthesis of oxylipins; transcription factor WRKY71; four thionin-like peptides; flavonol synthase, and phenylalanine ammonia lyase, both involved in the phenylpropanoid pathway. The strongest upregulated transcripts included starch synthase; Cullin-1; AP2-like ethylene-responsive transcription factor AINTEGUMENTA, which regulates growth and cell numbers during organogenesis; roothairless-1; and genes involved in cell-cycle control, such as those encoding cyclin-T1-1, cyclin-dependent kinase A-1, and cyclin-dependent kinase C-2.
Comparison between 7 and 14 dai giant cells and 7 dai galls
A total of 942 genes was found to be differentially expressed (FDR<0.05) between control and giant cells at both 7 and 14 dai. Correlation between the log2FC-values (GC versus respective control) of those 942 genes was Pearson R=0.85 (P<2.2e−16). This demonstrates that differentially expressed genes at 7 and 14 dai show a similar transcriptional profile.
In the following paragraphs the transcriptional profiles of some specific pathways are explored in the giant cells (7 and 14 dai), in comparison with previously generated transcriptome data from 7 dai whole-gall material of rice (Kyndt et al., 2012a ).
Transcripts involved in the metabolism of the phytohormones salicylic acid, jasmonic acid, ethylene, abscisic acid, and gibberellic acid
As already described by Barcala et al. (2010) and Portillo et al. (2013) for tomato and Arabidopsis, expression patterns in rice giant cells were strongly different from the whole-gall transcriptional profile (Kyndt et al., 2012a ). TableS5 shows the log2FC of genes involved in biosynthesis of different plant hormones in 7 and 14 dai giant cells and 7 dai whole-gall material (data taken from Kyndt et al., 2012a ), each in comparison with their corresponding uninfected control tissue. Results show that transcripts involved in salicylic acid (SA) biosynthesis through the phenylpropanoid pathway are suppressed in giant cells at both 7 and 14 dai, with for instance many paralogues encoding phenylalanine ammonia lyase much less expressed in giant cells in comparison with uninfected vascular cells (Supplementary Table S5). Some of these genes were found to be rather induced in 7 dai whole galls. Many transcripts involved in jasmonate biosynthesis, e.g. encoding lipoxygenases, allene oxide synthase, and 12-oxophytodienoate reductases, are suppressed in 7 and 14 dai giant cells versus uninfected vascular root cells. A lot of these transcripts are rather induced in 7 dai whole galls (Supplementary Table S3). The fact that gibberellic acid (GA) biosynthesis and signalling through DELLA proteins is generally induced in gall tissue was reported by Kyndt et al. (2012a ), and the data shown in the current study reveal that many of these enzymes were also expressed to a higher level inside giant cells at both time points. The fact that genes involved in GA degradation are also induced is potentially due to a feedback effect to control internal GA homeostasis.
A lot of transcripts involved in the production of abscisic acid are induced in 7 dai galls, while many abscisic acid-biosynthesis genes are repressed in 7 dai giant cells. In 14 dai giant cells both induced and repressed expression patterns are observed. Nevertheless, the transcript encoding 8-hydroxylase, involved in abscisic acid degradation, is very strongly induced in the gall and GC tissue both at 7 and 14 dai. In the ET biosynthesis pathway either strong induction or strong suppression was observed for different 1-aminocyclopropane-1-carboxylic acid (ACC)-synthase and ACC-oxidase paralogues, but no clear trend was seen, although slightly more transcripts encoding ACC-oxidase were found in 7 dai galls.
Transcriptional changes in genes involved in epigenetic modifications
Different epigenetic processes, which are not necessarily independent of one another, have been described to affect gene expression (Berger, 2007). Many transcripts involved in (post)-transcriptional gene silencing, like those encoding Dicer and ARGONAUTE proteins, are induced in the 7 dai giant cells and some also in whole 7 dai gall material (Table 2), but this trend does not persist in the 14 dai giant cell. Histone-modifying enzymes are also dramatically upregulated in the 7 dai giant cells, whereas they show varying expression profiles in the 14 dai giant cells (Table 2). In complete gall tissue the transcriptional profile is diluted by the neighbouring cells, but one of these genes (LOC_Os01g59620) is significantly more expressed in comparison with the uninfected root tips.
Table 2.
Transcriptional changes in genes involved in epigenetic mechanisms as obtained by mRNA-Seq on 7 and 14 dai giant cells (versus control vascular root cells) and 7 dai galls (versus control root tips; data extracted from Kyndt et al., 2012a )
| Log2FC gall 7 dai vs control root tips | Log2FC giant cell 7 dai vs control root cells | Log2FC giant cell 14 dai vs control root cells | Annotation | |
|---|---|---|---|---|
| Post-transcriptional gene silencing | ||||
| LOC_Os01g16870 | 0.02 | 1.98 | 0.73 | Argonaute |
| LOC_Os01g16860 | 1.58 | 1.37 | NE | AGO4-2 |
| LOC_Os10g34430 | 5.35 | 7.03 | 0.01 | Dicer |
| LOC_Os04g43050 | 2.45 | 1.90 | 0.76 | Dicer |
| LOC_Os04g39160 | 2.78 | 4.59 | 2.04 | RNA-dependent RNA polymerase |
| LOC_Os01g34350 | 2.19 | 0.54 | −6.89 | RNA-dependent RNA polymerase |
| LOC_Os02g58490 | −0.26 | 4.70 | 3.79 | PINHEAD |
| LOC_Os04g47870 | 1.91 | 2.45 | 0.35 | PINHEAD |
| Histone modification | ||||
| LOC_Os01g56540 | 1.00 | 3.99 | NE | Histone-lysine N-methyltransferase SUVR3 |
| LOC_Os01g59620 | 2.16 | 4.25 | 4.25 | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH1 |
| LOC_Os07g25450 | −0.09 | 5.87 | −0.15 | Histone-lysine N-methyltransferase |
| LOC_Os08g10470 | 0.03 | 7.56 | 4.91 | Histone-lysine N-methyltransferase ASHR2 |
| LOC_Os12g32374 | −0.88 | 4.89 | 2.20 | Histone deacetylase 6 |
| LOC_Os05g36920 | 0.88 | 5.53 | −3.11 | Histone deacetylase |
| LOC_Os10g28040 | −0.45 | 5.58 | −2.98 | Histone acetyltransferase GCN5 |
| LOC_Os06g38470 | 0.97 | 3.19 | 1.10 | Histone deacetylase HDAC1 |
NE, not expressed.
Novel transcripts and alternatively spliced transcripts
A total of 18 291 nTARs were detected in the isolated cells (Supplementary Table S6). A BLAST search was done against all ESTs from O. sativa, and all proteins predicted from the O. sativa genome project. 7383 nTARs gave a significant hit (E<0.0001) against at least one EST from O. sativa. tBLASTx against all proteins of O. sativa resulted in hits for 3587 of the nTARs, indicating potential paralogy to a known rice transcript (Supplementary Table S6).
To predict a potential function of these nTARs, a SwissProt/Trembl search was done, and although this was successful for 4719 transcripts many of them were annotated as ‘Putative uncharacterized protein, O. sativa’. Nevertheless, among the nTARS the following rice orthologues were detected: a Bowman–Birk-type wound-induced proteinase inhibitor WIP1 (Zea mays), a receptor-like protein 12 (Arabidopsis thaliana), disease-resistance protein RGA2 (Solanum bulbocastanum), regulatory protein NPR3 (A. thaliana), and gibberellin 20 oxidase 1 (A. thaliana) (Supplementary Table S4). Among the nTARs, 12 185 nTARs showed a significant differential expression pattern in the giant cells versus the uninfected vascular cells, with 2214 nTARs significantly downregulated in the giant cells, and 9971 significantly upregulated (FDR<0.05). Among the downregulated nTARs were for example transcripts showing homology to isoflavone reductase and carotenoid 9,10(9′,10′)-cleavage dioxygenase. Potential novel orthologues of callose synthase 3 and polygalacturonase are both significantly induced in the giant cells (Supplementary Table S6). In addition to the nTARs, 16 063 alternatively spliced transcripts were detected (Supplementary Table S6), among which 8374 have significant differential expression patterns (2465 down- and 5909 upregulated).
Discussion
During pathogen attack, a plant host modulates its gene expression to ward off the invader, while the pathogen produces effector proteins to manipulate the molecular machinery of the plant aiming at increased susceptibility. Characterizing gene expression in the cells that are specifically targeted by the pathogens provides insights into this complex molecular arms race. To the best of our knowledge, this report represents the first successful combination of LCM with mRNA-Seq for expression profiling in the field of plant–nematode interactions. Recently, we have reported an mRNA-seq-based transcriptome analysis of galls induced by M. graminicola in rice (Kyndt et al., 2012a ). Within this gall, developing giant cells undergo repeated nuclear divisions without cytokinesis to form large multinucleate cells. As giant cells comprise only a small fraction of the gall material the analysis of gene expression at the single-cell-type level was required for a further in-depth study on these highly specialized nematode feeding cells. In the current study giant cells at 7 and 14 dai and the corresponding control cells were isolated by LCM, and their transcriptomes were compared using mRNA-Seq.
In total 278 billion paired-end mRNA-Seq reads were generated, leading to estimated expression levels of more than 50 000 annotated and novel rice loci, with a coverage of approximately 100-fold the annotated transcriptome. Previous studies on complete gall tissue and microdissected giant cells from Arabidopsis, tomato, and soybean (Bar-Or et al., 2005; Jammes et al., 2005; Barcala et al., 2010; Ibrahim et al., 2011) applied microarray analysis, and hence only transcripts represented on the arrays could be detected. The benefit of mRNA-Seq lies in the fact that next to annotated transcripts this technique allows the detection of novel splice junctions, novel transcripts, paralogues, and rare transcripts (Sultan et al., 2008; Wilhelm et al., 2008; Zhang et al., 2010). Indeed, our analysis uncovered 18 291 putative nTARs and 16 063 alternatively spliced transcripts expressed in the analysed LCM-isolated rice cell types. Similar high levels of alternative splicing in rice have been reported by Lu et al. (2010) who estimated that ≈48% of rice genes show alternative splicing patterns. Among the nTARs, 12 185 are differentially expressed, with 2214 nTARs significantly downregulated in the giant cells and 9971 significantly upregulated (FDR<0.05). These represent transcripts that might only be expressed at very low levels and/or under specific circumstances, making them undetected in previous studies.
In the following paragraphs transcriptional changes observed in rice giant cells are being compared to expression results from Arabidopsis and tomato giant cells (Barcala et al., 2010; Portillo et al., 2013), complete galls from tomato, Arabidopsis, and rice (Bar-Or et al., 2005; Barcala et al., 2010; Kyndt et al., 2012a ; Portillo et al., 2013), and above-ground tissues of nematode-infected rice plants (Kyndt et al., 2012b ). Our results were also compared with data obtained from plants infected with cyst nematodes, another type of sedentary plant-parasitic nematode that forms feeding sites called syncytia (Ithal et al., 2007a , 2007b ; Klink et al., 2007; Szakasits et al., 2009).
Metabolic changes in giant cells induced by RKN infection in rice
Feeding sites of sedentary endoparasitic nematodes, like Meloidogyne spp. and cyst nematodes, are the only source of nutrients for these root parasites throughout their life (Jammes et al., 2005; Szakasits et al., 2009). This high demand for resources is reflected in the upregulation of genes involved in the primary metabolism of the plant cell, with a prominent induction of starch production, something already observed by Barcala et al. (2010) in Arabidopsis giant cells. Also, syncytia formed by cyst nematodes store carbohydrates by starch accumulation in the plastids (Hofmann et al., 2008), probably as a carbohydrate buffer and long-term storage to compensate for changing solute uptake by the nematode. In contrast with the induction of the primary metabolism, the secondary metabolism of the giant cells is strongly impaired, with for instance many downregulated transcripts involved in phenylpropanoid production.
The flavonoid side chain of the phenylpropanoid pathway
The first step in the phenylpropanoid pathway is catalysed by phenylalanine ammonia lyase. Strikingly, all phenylalanine ammonia lyase homologues were suppressed in giant cells at both investigated time points. However, when looking at whole-gall tissue many of them were repressed at 3 dai (Kyndt et al., 2012a ), but induced at 7 dai (Supplementary Table S5). After non-oxidative deamination of l-phenylalanine to trans-cinnamic acid by phenylalanine ammonia lyase, the phenylpropanoid pathway branches into different side chains responsible for the biosynthesis of different metabolites, like lignin precursors, flavonoids, hydroxycinnamic acid esters, and salicylic acid (Boudet, 2000).
Active suppression of the flavonoid pathway is an important feature of pathogenicity in many other plant–pathogen interactions (Oh and Collmer, 2005). In line with this view, we found with both mRNA-Seq and qRT-PCR that chalcone synthase, the key enzyme in the flavonoid side branch of the phenylpropanoid pathway, is significantly suppressed in rice giant cells (Supplementary Table S3). Nevertheless, Hutangura et al. (1999) showed that chalcone synthase is induced around the invading nematode at 24h after Meloidogyne javanica infection in white clover (Trifolium repens) and that flavonoids were detectable in and around the feeding site within 48h of the start of the infection process. Similarly, in other plant–nematode interactions, genes involved in flavonoid biosynthesis have been reported to be induced, e.g. upon migratory nematode infection in rice (Kyndt et al., 2012a ) and cyst nematode infection in soybean and Arabidopsis (Ithal et al., 2007a ; Jones et al., 2007). Data from mutant lines impaired in flavonoid biosynthesis showed that they were either equally or more susceptible to Heterodera schachtii (Jones et al., 2007), supporting the view that flavonoids are produced by the plant as part of the defence response against nematodes. These metabolites have indeed been shown to have a direct negative effect on many nematode species (Wuyts et al., 2006). Hence, local suppression of flavonoid biosynthesis in the giant cells might be an important strategy for the RKN to overcome host defence responses.
Trehalose
Prior to the current study, an activation of the trehalose metabolism was observed in whole galls induced by RKN in rice (Kyndt et al., 2012a ) and systemic tissue of cyst nematode-infected plants (Hofmann et al., 2010). Remarkably, in studies focusing on isolated nematode feeding sites several genes encoding trehalose-6-phosphate synthase, an enzyme needed to form trehalose 6-phosphate, were rather downregulated (giant cells: this study; syncytia: Szakasits et al., 2009). Trehalose 6-phosphate has multiple functions in plants, not only in carbohydrate storage and metabolism but also as a stress protectant and as a metabolic signalling molecule involved in many plant–pathogen interactions (Fernandez et al., 2010) and cell-wall modification (Bae et al., 2005). Furthermore, other transcripts that are known to be responsive to abiotic and biotic stimuli, like (receptor-like) protein kinases and stress- and disease-resistance-related proteins, are generally strongly repressed in giant cells when compared to corresponding uninfected vascular root cells.
Dark-grown giant cells contain chloroplast-like organelles
Plant roots mainly develop non-photosynthetic plastids, such as starch-containing amyloplasts, but roots of several plant species have the potential to turn green when exposed to light (Flores et al., 1993). Upon light exposure, amyloplasts inside RKN-induced galls in tomato roots were reported to differentiate into chloroplasts (Orion and Wergin, 1982), and similar phenomena are generally seen in syncytia formed in nematode-infected plants under light influence (Sijmons et al., 1991; Golinowski et al., 1996). Also our previous transcriptome study of complete galls showed evidence of photosynthetic activity at 7 dai, but again these results could have been biased because the tissue had not been covered from indirect light (Kyndt et al., 2012a ). Up till now, this phenomenon had not been investigated in dark-grown giant cells. That is why, for the here-described experiments, the material was completely protected from light. To our surprise, a strong induction of photosynthesis-related transcripts and transcripts involved in the biogenesis of chloroplasts was consistently found in the giant cells. Genes related to the biosynthesis of tetrapyrroles, which mainly occurs inside chloroplasts, are induced in the giant cells (Fig. 3). Both transcriptome data and microscopical analysis (Fig. 4) confirmed that the 7 dai giant cells, even without light stimulation, hold chloroplast-like structures, which contain metabolites with a similar fluorescence emission spectrum to rice leaf tissue.
Sugar depletion due to nutrient sipping by the feeding nematode might be the trigger that activates photosynthetic gene expression (Oswald et al., 2001). Furthermore, recent findings suggest that phytohormones and environmental (stress) signals regulate the expression of genes that are related to tetrapyrrole metabolism. Whether there is a causal relationship between this disturbance of the hormone homeostasis and chlorophyll accumulation inside giant cells remains to be further studied.
Modulation of plant-hormone pathways in giant cells
Hormones act as signalling molecules in plants by mediating physiological responses, thereby coordinating growth and differentiation of cells as well as innate immunity.
Consistent with recent findings demonstrating a general induction of GA biosynthesis in galls on tomato and rice (Bar-Or et al., 2005; Kyndt et al., 2012a ) and syncytia in soybean (Klink et al., 2007), giant cells accumulate high levels of transcripts coding for enzymes involved in GA production and response (such as catabolism and DELLA proteins) (Supplementary Table S5). Gibberellins are important stimulators of cell division and elongation (Richards et al., 2001). These observations suggest that gibberellins are important players in the development, maintenance, and maturation of giant cells. GA plays a critical role in controlling and coordinating cell division, cell expansion, and, interestingly, also chloroplast biogenesis through influencing the DELLA protein family in leaf tissue of both dicotyledonous and monocotyledonous plant species. (Jiang et al., 2012).
Transcripts necessary for jasmonate biosynthesis are suppressed in the 7 and 14 dai giant cells, although this was not observed in whole-gall tissue (Nahar et al., 2011; Kyndt et al., 2012a ). Also Ithal et al. (2007b ) detected a strong and consistent, but very local, suppression of the jasmonic acid pathway in isolated syncytia after cyst nematode infection in soybean. It is important to note that activation of the jasmonic acid pathway by external methyl jasmonate application is an effective way to protect potato, tomato, and rice from RKN infection (Cooper et al., 2005; Nahar et al., 2011; Vieira dos Santos et al., 2013).
Although application of the SA analogue BTH only resulted in slightly less gall information in rice (Nahar et al., 2011), it has recently been shown to have a strong negative effect on Meloidogyne chitwoodi development in tomato and potato (Vieira dos Santos et al., 2013). SA is derived from the phenylpropanoid pathway, and in line with a general local and systemic repression of this pathway at 3 dai in galls and systemic tissues in rice (Kyndt et al., 2012a , 2012b ) and giant cells formed in Arabidopsis (Barcala et al., 2010), transcripts involved in the phenylpropanoid pathway are also strongly downregulated in the isolated 7 and 14 dai giant cells formed in rice roots. Ethylene (ET) is known to play a synergistic role with jasmonic acid in plant innate immunity (Pieterse et al., 2009). Nahar et al. (2011) showed that ET-insensitive mutants and pharmacological inhibition of ET biosynthesis in rice leads to significantly higher susceptibility for RKN, demonstrating that ET plays a role in defence against RKN in these plants. In addition, Fudali et al. (2013) showed that ET-overproducing Arabidopsis plants are less attractive to RKN. Nevertheless, ET has been suggested to be critical for syncytium formation during cyst–nematode infections in Arabidopsis (Goverse et al., 2000). Its ambiguous role in plant defence versus development of the feeding site might explain why no clear trend concerning the ET pathway was observed in the here-studied 7 and 14 dai giant cells (Supplementary Table S5).
A role for epigenetic processes in transcriptional reprogramming of the root cells
Different epigenetic processes have been described to affect the transcriptome of plant cells: (1) cytosine methylation influences gene expression by altering transcription and chromatin structure, (2) histone modifications have an important impact on the structure of chromatin and can make DNA more or less accessible for transcription, (3) small RNAs, like miRNA and siRNAs, influence gene expression through targeted degradation of mRNA (post-transcriptional gene silencing) or induction of methylation at complementary DNA sequences (transcriptional gene silencing).
Recent studies have shown the importance of small RNAs during cyst nematode feeding site (syncytium) formation. Sequences of known miRNAs as well as siRNAs were identified by sequencing small RNA libraries isolated from feeding sites induced by the cyst nematode Heterodera schachtii in Arabidopsis roots (Hewezi et al., 2008), and the data suggested a role for small RNAs mediating gene-regulation processes during the plant–nematode interaction. Hewezi et al. (2012) reported a strong downregulation of miR396 in early syncytial cells in comparison with the surrounding root tissue, when nematodes were at the J2 or early J3 stage. At later time points, when nematodes reached the J3/J4 stage, a specific miR396 upregulation was observed in the developed feeding site. miR396 targets a set of growth-regulation factor genes (Hewezi et al., 2012).
In tomato giant cells, Portillo et al. (2013) reported that genes involved in epigenetic processes are induced from 3 dai and increasing at 7 dai. Histone acetylation/deacetylation and methylation are important mechanisms regulating gene expression in plants (Zhou et al., 2010), and may be largely involved in responses to environmental stimuli (Chen and Tian, 2007; Servet et al., 2010). The here-reported transcriptional changes at 7 dai in genes encoding histone-modifying and small-RNA-processing enzymes (Table 2) confirm a role for epigenetic processes in transcriptional reprogramming of the root cells to form nematode feeding sites. Which transcriptional changes are specifically targeted by these enzymes, and what is their functional relevance, remains to be elucidated.
Supplementary material
Supplementary material is available at JXB online.
Supplementary Table S1. Locus number, annotation, primer pairs and amplification efficiency of qRT-PCR analyses.
Supplementary Table S2. List of transcripts that are significantly differentially expressed in 7 dai giant cells induced by M. graminicola. The list shows the chromosomal location and the log2FC between giant cells and control vascular cells of each transcript.
Supplementary Table S3. qRT-PCR validation of differential expression patterns (log2FC) in 7 dai giant cell tissue versus control cells from the vascular root tissue.
Supplementary Table S4. List of transcripts that are significantly differentially expressed in 14 dai giant cells induced by M. graminicola. The list shows the chromosomal location and the log2FC between giant cells and control vascular cells of each transcript.
Supplementary Table S5. Expression patterns of genes involved in different hormone pathways as determined by mRNA-Seq analysis. Log2FC of expression in 7 dai complete galls (versus control root tips; data extracted from Kyndt et al., 2012a ), and the here-generated data of 7 dai giant cells and 14 dai giant cells versus the uninfected control cells.
Supplementary Table S6. List of putative novel transcriptionally active regions and novel splicing variants identified in giant cells, with their genomic location, BLASTn results against cDNA and genomic DNA of O. sativa cv. ‘Japonica’, BLASTx results against rice proteins, SwissProt/TrEMBL, and their differential expression pattern in comparison with the control cells. Y: yes; N: no.
Acknowledgements
We acknowledge the financial and infrastructural support of Ghent University (“Bioinformatics: from nucleotides to networks”, BOF_01GA0805, the Hercules Foundation (AUGE/013), the Belgian Research Fund (B/11599/17), and Stevin Supercomputer Infrastructure). We thank Sandra Soetaert, Jean-Pierre Renard, Sarah De Keulenaer and Lien De Smet for excellent technical assistance. Tina Kyndt, Annelies Haegeman and Tim De Meyer (in part) are supported by an FWO postdoctoral fellowship. Simon Denil is supported by an IWT doctoral grant. Kamrun Nahar and Hongli Ji are supported by a BOF Phd-scholarship (Bijzonder Onderzoeksfonds, Ghent University).
Glossary
Abbreviation:
- EST
expressed sequence tag
- ET
ethylene
- GA
gibberellic acid
- J
juvenile
- LCM
laser-capture microdissection
- mRNA-Seq
mRNA sequencing
- nTAR
novel transcriptionally active region
- qRT-PCR
quantitative reverse transcriptase-PCR
- RKN
root knot nematode
- SA
salicylic acid.
References
- Bae HH, Herman E, Bailey B, Bae HJ, Sicher R. 2005. Exogenous trehalose alters Arabidopsis transcripts involved in cell wall modification, abiotic stress, nitrogen metabolism, and plant defense. Physiologia Plantarum 125, 114–126. [Google Scholar]
- Bar-Or C, Kapulnik Y, Koltai H. 2005. A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root knot nematode Meloidogyne javanica. European Journal of Plant Pathology 111, 181–192. [Google Scholar]
- Barcala M, Garcia A, Cabrera J, Casson S, Lindsey K, Favery B, Garcia-Casado G, Solano R, Fenoll C, Escobar C. 2010. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. The Plant Journal 61, 698–712. [DOI] [PubMed] [Google Scholar]
- Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447, 407–412. [DOI] [PubMed] [Google Scholar]
- Boudet AM. 2000. Lignins and lignification: selected issues. Plant Physiology and Biochemistry 38, 81–96. [Google Scholar]
- Bridge J, Plowright RA, Peng D. 2005. Nematode parasites of rice. In: Luc M, Sikora RA, Bridge J, eds. Plant-parasitic nematodes in subtropical and tropical agriculture. Wallingford: CAB International, 87–130. [Google Scholar]
- Chen ZJ, Tian L. 2007. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochimica et Biophysica Acta Gene Structure and Expression 1769, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WR, Jia L, Goggin L. 2005. Effects of jasmonate-induced defenses on root-knot nematode infection of resistant and susceptible tomato cultivars. Journal of Chemical Ecology 31, 1953–1967. [DOI] [PubMed] [Google Scholar]
- De Almeida Engler J, Van Poucke K, Karimi M, De Groodt R, Gheysen G, Engler G. 2004. Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode-infected roots. The Plant Journal 38, 12–26. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo BY. 1987. Histopathology and ultrastructure of crops invaded by certain sedentary endoparasitic nematodes. Vistas on Nematology: A Commemoration of the Twenty fifth Anniversary of the Society of Nematologists. Hyattsville, MD: Society of Nematologists, 196–210. [Google Scholar]
- Fernandez O, Bethencourt L, Quero A, Sangwan RS, Clement C. 2010. Trehalose and plant stress responses: friend or foe? Trends in Plant Science 15, 409–417. [DOI] [PubMed] [Google Scholar]
- Flores HE, Dai YR, Cuello JL, Maldonadomendoza IE, Loyolavargas VM. 1993. Green roots – Photosynthesis and photoautotrophy in an underground plant organ. Plant Physiology 101, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosu-Nyarko J, Jones MGK, Wang ZH. 2009. Functional characterization of transcripts expressed in early-stage Meloidogyne javanica-induced giant cells isolated by laser microdissection. Molecular Plant Pathology 10, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudali SL, Wang CL, Williamson VM. 2013. Ethylene signaling pathway modulates attractiveness of host roots to the root-knot nematode Meloidogyne hapla. Molecular Plant-Microbe Interactions 26, 75–86. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG. 2011. How nematodes manipulate plant development pathways for infection. Current Opinion in Plant Biology 14, 415–421. [DOI] [PubMed] [Google Scholar]
- Golinowski W, Grundler FMW, Sobczak M. 1996. Changes in the structure of Arabidopsis thaliana during female development of the plant-parasitic nematode Heterodera schachtii. Protoplasma 194, 103–116. [Google Scholar]
- Goverse A, Overmars H, Engelbertink J, Schots A, Bakker J, Helder J. 2000. Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Molecular Plant-Microbe Interactions 13, 1121–1129. [DOI] [PubMed] [Google Scholar]
- Hardcastle TJ, Kelly KA. 2010. baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Howe P, Maier TR, Baum TJ. 2008. Arabidopsis small RNAs and their targets during cyst nematode parasitism. Molecular Plant-Microbe Interactions 21, 1622–1634. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ. 2012. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiology 159, 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J, Szakasits D, Blochl A, Sobczak M, Daxbock-Horvath S, Golinowski W, Bohlmann H, Grundler FMW. 2008. Starch serves as carbohydrate storage in nematode-induced syncytia. Plant Physiology 146, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J, El Ashry A, Anwar S, Erban A, Kopka J, Grundler F. 2010. Metabolic profiling reveals local and systemic responses of host plants to nematode parasitism. The Plant Journal 62, 1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutangura P, Mathesius U, Jones MGK, Rolfe BG. 1999. Auxin induction is a trigger for root gall formation caused by root-knot nematodes in white clover and is associated with the activation of the flavonoid pathway. Australian Journal of Plant Physiology 26, 221–231. [Google Scholar]
- Ibrahim HMM, Hosseini P, Alkharouf NW, Hussein EHA, El-Din AYG, Aly MAM, Matthews BF. 2011. Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Hearne L, Maier T, Baum TJ, Mitchum MG. 2007a. Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Molecular Plant-Microbe Interactions 20, 293–305. [DOI] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, Mitchum MG. 2007b. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Molecular Plant-Microbe Interactions 20, 510–525. [DOI] [PubMed] [Google Scholar]
- Jammes F, Lecomte P, Almeida-Engler J, Bitton F, Martin-Magniette ML, Renou JP, Abad P, Favery B. 2005. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. The Plant Journal 44, 447–458. [DOI] [PubMed] [Google Scholar]
- Jiang XS, Li HY, Wang T, Peng CL, Wang HY, Wu H, Wang XJ. 2012. Gibberellin indirectly promotes chloroplast biogenesis as a means to maintain the chloroplast population of expanded cells. The Plant Journal 72, 768–780. [DOI] [PubMed] [Google Scholar]
- Jones JT, Furlanetto C, Phillips MS. 2007. The role of flavonoids produced in response to cyst nematode infection of Arabidopsis thaliana. Nematology 9, 671–677. [Google Scholar]
- Kim SY, Volsky DJ. 2005. PAGE: Parametric analysis of gene set enrichment. BMC Bioinformatics 6, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF. 2007. Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 226, 1389–1409. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Denil S, Haegeman A, Trooskens G, Bauters L, Van Criekinge W, De Meyer T, Gheysen G. 2012a. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytologist 196, 887–900. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Nahar K, Haegeman A, De Vleesschauwer D, Hofte M, Gheysen G. 2012b. Comparing systemic defence-related gene expression changes upon migratory and sedentary nematode attack in rice. Plant Biology 14, 73–82. [DOI] [PubMed] [Google Scholar]
- Lu TT, Lu GJ, Fan DL, Zhu CR, Li W, Zhao QA, Feng Q, Zhao Y, Guo YL, Li WJ, Huang XH, Han B. 2010. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Research 20, 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, De Vleesschauwer D, Hofte M, Gheysen G. 2011. The Jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiology 157, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HS, Collmer A. 2005. Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. The Plant Journal 44, 348–359. [DOI] [PubMed] [Google Scholar]
- Orion D, Wergin WP. 1982. Chloroplast differentiation in tomato root galls induced by the root-knot nematode Meloidogyne incognita . Journal of Nematology 14, 77–83. [PMC free article] [PubMed] [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA. 2001. Plastid redox state and sugars: Interactive regulators of nuclear-encoded photosynthetic gene expression. Proceedings of the National Academy of Sciences, United States of America 98, 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrós R, Moya I, Goulas Y, Jacquemoud S. 2008. Chlorophyll fluorescence emission spectrum inside a leaf. Photochemical & Photobiological Sciences 7, 498–502. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. 2009. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Portillo M, Lindsey K, Casson S, Garcia-Casado G, Solano R, Fenoll C, Escobar C. 2009. Isolation of RNA from laser-capture-microdissected giant cells at early differentiation stages suitable for differential transcriptome analysis. Molecular Plant Pathology 10, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo M, Cabrera J, Lindsey K, et al. 2013. Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytologist 197, 1276–1290. [DOI] [PubMed] [Google Scholar]
- Reversat G, Boyer J, Sannier C, Pando-Bahuon A. 1999. Use of a mixture of sand and water-absorbent synthetic polymer as substrate for the xenic culturing of plant-parasitic nematodes in the laboratory. Nematology 1, 209–212. [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd NP. 2001. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annual Review of Plant Physiology and Plant Molecular Biology 52, 67–88. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servet C, Silva NCE, Zhou DX. 2010. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Molecular Plant 3, 670–677. [DOI] [PubMed] [Google Scholar]
- Sijmons PC, Grundler FMW, Vonmende N, Burrows PR, Wyss U. 1991. Arabidopsis thaliana as a new model host for plant-parasitic nematodes. The Plant Journal 1, 245–254. [Google Scholar]
- Sultan M, Schulz MH, Richard H, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321, 956–960. [DOI] [PubMed] [Google Scholar]
- Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FMW, Bohlmann H. 2009. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. The Plant Journal 57, 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. 2007. Tetrapyrrole biosynthesis in higher plants. Annual Review of Plant Biology 58, 321–346. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, et al. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira dos Santos MC, Curtis RHC, Abrantes I. 2013. Effect of plant elicitors on the reproduction of the root-knot nematode Meloidogyne chitwoodi on susceptible hosts. European Journal of Plant Pathology 136, 193–202. [Google Scholar]
- Wang ZH, Potter RH, Jones MGK. 2003. Differential display analysis of gene expression in the cytoplasm of giant cells induced in tomato roots by Meloidogyne javanica . Molecular Plant Pathology 4, 361–371. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243. [DOI] [PubMed] [Google Scholar]
- Wuyts N, Swennen R, De Waele D. 2006. Effects of plant phenylpropanoid pathway products and selected terpenoids and alkaloids on the behaviour of the plant-parasitic nematodes Radopholus similis, Pratylenchus penetrans and Meloidogyne incognita. Nematology 8, 89–101. [Google Scholar]
- Zhang GJ, Guo GW, Hu XD, et al. 2010. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Research 20, 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JL, Wang XF, He K, Charron JBF, Elling AA, Deng XW. 2010. Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Molecular Biology 72, 585–595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.