To the Editor:
Proteases of the ADAM (A disintegrin and metalloproteinase) family form a large group of metalloenzymes that function primarily to cleave (or shed) extracellular domains from plasma membrane bound signaling molecules. Whereas ADAM proteases are not thought to have strict cleavage site specificities, ADAM17 and ADAM10 have evolved distinct substrate preferences despite being the most closely related members of the family. ADAM17 is the principal sheddase of TNF-α and the majority of the EGFR-ligands (Blobel, 2005; Peschon et al., 1998), whereas ADAM10 is required for Notch and amyloid precursor protein (APP) shedding (Kuhn et al., 2010; van Tetering et al., 2009). The physiological importance of ADAM17 in EGFR-signaling is based on the striking phenotypic similarities between Adam17−/− and Egfr−/− mutant mice. Both die at birth due to multiple developmental defects involving the heart, lung, and skin (Blobel, 2005; Miettinen et al., 1995; Peschon et al., 1998).
Notch receptors are conserved membrane-anchored transcription regulators activated by proteolysis (Kopan and Ilagan, 2009; van Tetering and Vooijs, 2011). During maturation in the Golgi, Notch receptors are cleaved by furin-like convertases at site 1 (S1) located in a loop protruding from the negative regulatory region (NRR). When denatured for western blot analysis, the membrane–bound form of Notch1 (TMIC) migrates at ~120 kDa. Ligand binding unfolds the Lin12-Notch repeat (LNR) module to expose and allow cleavage at Val1711 (“S2”) creating “NEXT”, which is 55 amino acids shorter than TMIC. S2 cleavage is followed by γ-secretase cleavage at Val1744 (“S3”), releasing the ~110 kDa Notch intracellular domain (NICD) that acts as a transcriptional regulator (Figure S1). In all metazoan, the essential role of ADAM10 and its orthologs (Kuzbanian and Sup-17) in executing the Notch S2 cleavage is evident by the phenotypic similarities between Notch and Adam10/Kuz/Sup-17 mutants in mice, flies and worms, respectively (van Tetering and Vooijs, 2011).
ADAM17 can mediate S2 cleavage under special, non-physiological circumstances. It cleaves T-ALL-associated, mutant forms of Notch that are ligand independent in tissue culture cells (Bozkulak and Weinmaster, 2009). It also cleaves Notch proteins unfolded in vitro with Ca2+ chelators (Rand et al., 2000); however, a physiological activity for ADAM17 in S2 cleavage should be ruled out for two reasons. First, most Adam10 loss of function phenotypes examined to date (including skin phenotypes) phenocopied Notch loss (Demehri et al., 2008; Dumortier et al., 2010; Weber et al., 2011) indicating that ADAM17 cannot rescue S2 cleavage in Adam10 deficient tissues. Second, in cell-based assays, ligand-induced Notch signaling depends on ADAM10 (Bozkulak and Weinmaster, 2009; van Tetering et al., 2009) and occurs at Val1711 S2 cleavage site during ligand dependent Notch1 signaling (van Tetering et al., 2009).
Recently, Murthy and colleagues (Murthy et al., 2012) suggested a role for ADAM17 in maintaining a baseline level of Notch1 activity (“tonic” Notch signal) in epidermal keratinocytes in a ligand-independent manner. We examined the data supporting this function in the same genetic model used by Murthy et al, arriving at different conclusions. Both Murthy et al. (Murthy et al., 2012) and Franzke et al. (Franzke et al., 2012) characterized the same keratinocyte-specific Adam17 knockout mouse model (Krt14-A17ΔKC), and observed altered epidermal differentiation and the onset of chronic dermatitis and severe myeloproliferative disease. Although both studies concur that ADAM10 is required for ligand-dependent Notch signaling during skin development, Murthy et al., concluded that Adam17 loss alleviated a Notch-dependent suppression of pro-inflammatory cytokine thymic stromal lymphopoietin (TSLP). According to Murthy and colleagues, ADAM17 maintains a ligand-independent (“tonic”) mode of Notch signaling in skin, which in turn suppressed TSLP production. Measurements of TSLP by ELISA demonstrate that γ-secretase-deficient keratinocytes produced indistinguishable amounts of TSLP from the wild type cells (Demehri et al., 2008) however are inconsistent with the idea that a “tonic” Notch signal regulates TSLP in skin.
We have provided an alternative explanation by showing that loss of ADAM17-dependent EGFR signaling impaired the maintenance of epidermal differentiation and barrier integrity in Krt14-A17ΔKC mice (Franzke et al., 2012), which would be sufficient for TSLP induction as demonstrated in several animal models with epidermal barrier defects, including Gata3−/− (de Guzman Strong et al., 2006), epidermis-specific loss of Notch (Demehri et al., 2008), or mice lacking the lipid transporter FATP4 (Demehri et al., 2008). Mice with a persistent defect in epidermal barrier maintenance have chronically elevated TSLP and GCSF expression, which together with additional cytokines drive leukocyte infiltration and promote development of atopic dermatitis, myeloproliferative disease, and asthma. Because this TSLP production also occurs in mice with a competent Notch signaling system but a defective epidermal differentiation program, TSLP expression is triggered by a mechanism that does not involve a cell autonomous Notch function (Demehri et al., 2008).
In addition to the genetic and cell-based data summarized above, both themethods and the interpretation of the data, which supported a physiological role for ADAM17 in tonic Notch signaling, raise several concerns. To support the conclusion that ADAM17 regulates ligand-independent Notch signaling, Murthy and colleagues activated Notch with EDTA, an in vitro assay with no physiological correlate rendering Notch permissive to cleavage by either ADAM10 or ADAM17. To provide physiological evidence, they presented data suggesting that NICD amounts were reduced in Krt14-A17ΔKC epidermis in vivo at postnatal day 56 (P56) versus control mice. A 72 kDa band labeled as NICD (Val1744) is greatly diminished in Adam17-deficient epidermis (Murthy et al., 2012). Based on the molecular weight of an intact NICD (~110kDa in SDS PAGE), this fragment is most likely a metabolic product of NICD that retained the VLLS epitope but lost a fraction of its C-terminus. Since ADAM10 is present, it was unclear to us why loss of “tonic” signal will cause such a pronounced change in NICD levels.
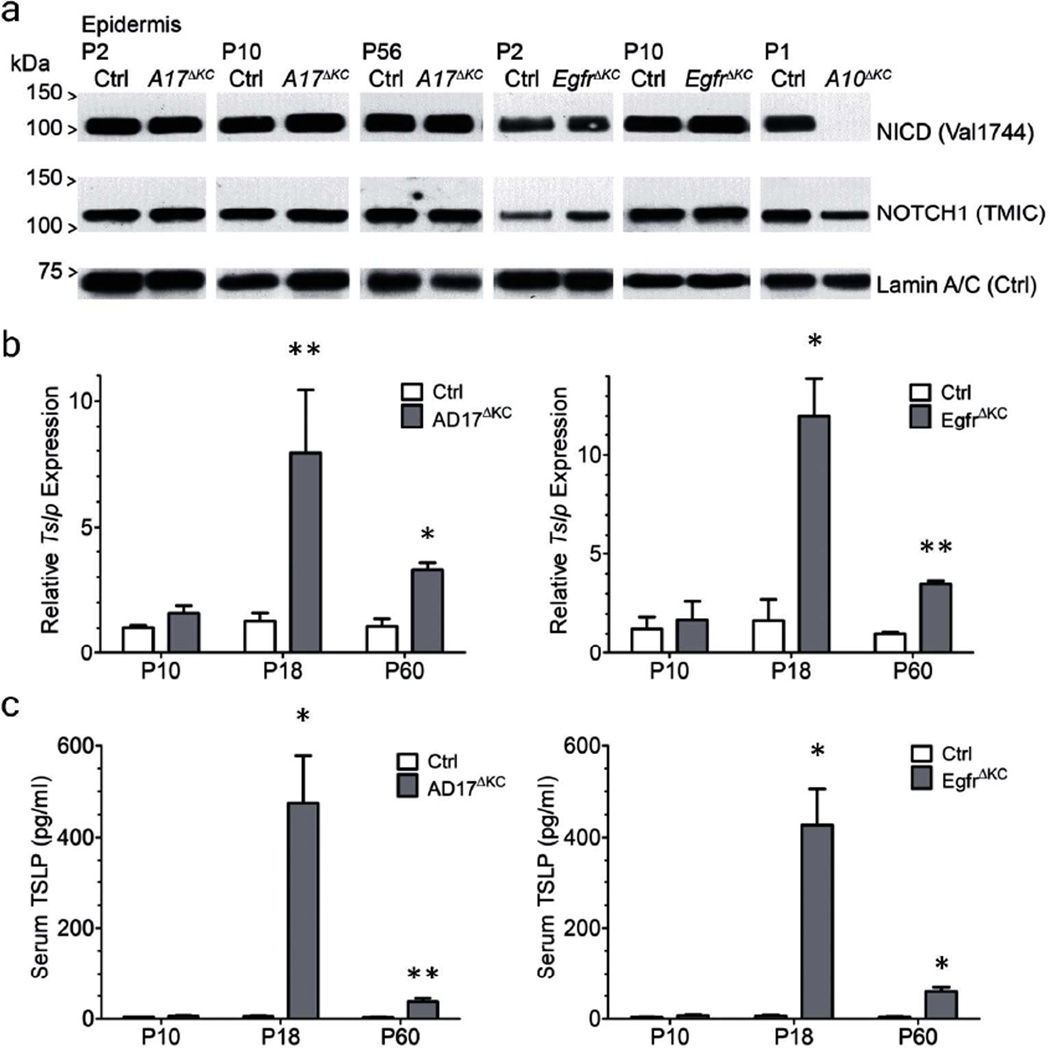
To evaluate the contribution of ADAM17 to NICD production in vivo, we repeated this experiment using epidermal lysates from the same Krt14-17ΔKC mice (named Adam17Δep in (Murthy et al., 2012)) and analyzed these at the same and more stages for Notch proteolysis. Due to the small differences in molecular weight between NEXT and NICD (33 amino acid residues), we visualized NICD with cleavage-specific antibodies (Figure 1A). We analyzed the epidermis from newborn, P10 and P56 Krt14-A17ΔKC epidermis using an antibody recognizing the C-terminus to identify total Notch1 (TMIC) and a Val1744 neo-epitope antibody to detect NICD unequivocally by western blot. As expected, NICD production in Krt14-A17ΔKC, Krt14-EgfrΔKC and control epidermis is indistinguishable in all postnatal ages, consistent with an intact, ADAM10-dependent, canonical signaling pathway. In contrast NICD production in the Krt14-A10ΔKC (named Adam10epi−/− in (Weber et al., 2011)) epidermis at P1 was impaired indicating ADAM10 is the sheddase of Notch1 in the epidermis (Figure 1A) (Weber et al., 2011).
Figure 1. Keratinocyte-derived ADAM17 is dispensable for Notch1 activation and the upregulation of the epidermal alarmin TSLP in Krt14-A17ΔKC mice depends on skin barrier defects.
(A) Western blot analysis of epidermal lysates for activated S3 cleaved NOTCH1; NICD (Val1744, upper panels) in Krt14-A17ΔKC and Krt14-EgfrΔKC mice revealed no differences to their littermate controls at different postnatal ages, while NICD was completely lost in the epidermis of Krt14-A10ΔKC animals. NOTCH1 protein (TMIC, middle panel) and Lamin A/C (Ctrl, bottom panels) were used as loading controls. Data from each postnatal age are representative of at least three mice (for each genotype). (B) Quantitative Tslp gene expression analysis of Krt14-A17ΔKC or Krt14-EgfrΔKC skin revealed no changes in expression on P10, but significantly elevated expression at P19 and P56, when the skin barrier defects are prominent. Mice analyzed for Krt14-A17KC and controls: P10, n=5; P19, n=5; P56, n=4; and for Krt14-EgfrΔKC and control littermates n=3 for all postnatal ages. (C) Analysis of serum TSLP levels of Krt14-A17ΔKC or Krt14-EgfrΔKC animals at P10, P19, and P56 revealed increased TSLP levels due to severe skin barrier defects starting at P19. The data derived from three different mice for each genotype. Data in B and C are shown as mean ± SEM, *p<0.05; **p<0.01.
If “tonic” Notch signaling exists, loss of tonic signals predicts that elevated TSLP production (released from Notch mediated inhibition) should precede (perhaps even drive) the defects in epidermal differentiation and barrier integrity, as seen in Notch and Fatp4 deficient animals (Demehri et al., 2008). Conversely, if EGFR deficiency caused barrier defects that then trigger Tslp expression, TSLP levels should rise only after the barrier defects are manifested. In order to analyze the time course of keratinocyte derived TSLP production in Krt14-A17ΔKC animals, we measured the Tslp mRNA in P10, P19, and P56 skin. Importantly, Tslp mRNA expression in P10 skin is identical to controls. Only when pronounced differentiation and barrier defects were noted (P19) (Franzke et al., 2012) did Tslp mRNA accumulate, and stayed consistently elevated in the skin of two-month-old animals (Figure 1B). Accordingly, TSLP serum levels were significantly increased in P19 and P56 animals, but unchanged compared to controls at P10 preceding barrier dysfunction (Figure 1C). In addition, we have seen the same time course for an increase in TSLP in Krt14-EgfrΔKC mice (Figure 1B, C). These results argue against a cell intrinsic upregulation of keratinocyte Tslp through loss of Notch-dependent suppression and support the null hypothesis that TSLP production follows barrier disruption (Demehri et al., 2008).
Since Adam17 disruption in these studies is achieved using the identical Krt14-A17ΔKC animal model, and since deletion of Adam17 in keratinocytes did not lead to elevation of Tslp mRNA until after a barrier defect was detected, Adam17 loss was unlikely to be associated with reduced epidermal Notch signaling in vivo. All the data can be explained by strongly diminished activation of EGFR in the epidermis of Krt14-A17ΔKC mice caused by reduced ADAM17 dependent EGFR-ligand shedding, which led to reduced expression of transglutaminases, reducing crosslinking of barrier proteins and thereby causing a differentiation and skin barrier defect (Franzke et al., 2012). Though it is possible that ADAM17 may cleave Notch proteins in vivo under some yet to be described circumstances, the data and analysis provided herein do not support the “tonic” Notch signaling inhibiting TSLP hypothesis.
Supplementary Material
Acknowledgments
We thank Mario Hartmann and Marlies Rusch for excellent technical assistance. We are grateful to Leena Bruckner-Tuderman for critically reading the manuscript. This work was supported by the Research Commission of the Medical Faculty, University of Freiburg (FRA725/09) and the German Research Foundation DFG (SFB 850/B6) to C.-W.F., SFB877 to P.S., GM64750 to C.P.B., and the European Research Council (FP7/2007-2013)/ ERC Grant 208259 to M.V.
Abbreviations
- ADAM
a disintegrin and metalloproteinase
- EGFR
epidermal growth factor receptor
- NICD
Notch intracellular domain
- TSLP
thymic stromal lymphopoietin
Footnotes
Conflict of interest
The authors state no conflict of interests.
References
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Molecular and cellular biology. 2009;29:5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guzman Strong C, Wertz PW, Wang C, Yang F, Meltzer PS, Andl T, et al. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. The Journal of cell biology. 2006;175:661–670. doi: 10.1083/jcb.200605057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A, Durham AD, Di Piazza M, Vauclair S, Koch U, Ferrand G, et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS One. 2010;5:e9258. doi: 10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke CW, Cobzaru C, Triantafyllopoulou A, Loffek S, Horiuchi K, Threadgill DW, et al. Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand-dependent terminal keratinocyte differentiation. J Exp Med. 2012;209:1105–1119. doi: 10.1084/jem.20112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, et al. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. The EMBO journal. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, et al. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- Murthy A, Shao YW, Narala SR, Molyneux SD, Zuniga-Pflucker JC, Khokha R. Notch Activation by the Metalloproteinase ADAM17 Regulates Myeloproliferation and Atopic Barrier Immunity by Suppressing Epithelial Cytokine Synthesis. Immunity. 2012;36:105–119. doi: 10.1016/j.immuni.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Molecular and cellular biology. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. The Journal of biological chemistry. 2009;284:31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tetering G, Vooijs M. Proteolytic cleavage of Notch: "HIT and RUN". Current molecular medicine. 2011;11:255–269. doi: 10.2174/156652411795677972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S, Niessen MT, Prox J, Lullmann-Rauch R, Schmitz A, Schwanbeck R, et al. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development. 2011;138:495–505. doi: 10.1242/dev.055210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.