Abstract
The human skin microbiome plays important roles in skin health and disease. However, bacterial population structure and diversity at the strain level is poorly understood. We compared the skin microbiome at the strain level and genome level of Propionibacterium acnes, a dominant skin commensal, between 49 acne patients and 52 healthy individuals by sampling the pilosebaceous units on their noses. Metagenomic analysis demonstrated that while the relative abundances of P. acnes were similar, the strain population structures were significantly different in the two cohorts. Certain strains were highly associated with acne and other strains were enriched in healthy skin. By sequencing 66 previously unreported P. acnes strains and comparing 71 P. acnes genomes, we identified potential genetic determinants of various P. acnes strains in association with acne or health. Our analysis suggests that acquired DNA sequences and bacterial immune elements may play roles in determining virulence properties of P. acnes strains and some could be future targets for therapeutic interventions. This study demonstrates a previously unreported paradigm of commensal strain populations that could explain the pathogenesis of human diseases. It underscores the importance of strain level analysis of the human microbiome to define the role of commensals in health and disease.
INTRODUCTION
The diversity of the human microbiota at the strain level and its association with human health and disease is largely unknown. However, many studies have shown that microbe-related human diseases are often caused by certain strains of a species, rather than the entire species being pathogenic. Examples include methicillin-resistant Staphylococcus aureus (MRSA) (Chambers and Deleo, 2009; Chen et al., 2010; Hansra and Shinkai) and Escherichia coli O157 (Chase-Topping et al., 2008; Tarr et al., 2005). Acne vulgaris (commonly called acne) is one of the most common skin diseases with a prevalence of up to 85% of teenagers and 11% of adults (White, 1998). Although the etiology and pathogenesis of acne are still unclear, microbial involvement is considered one of the main mechanisms contributing to the development of acne (Bojar and Holland, 2004; Cunliffe, 2002). In particular, Propionibacterium acnes has been hypothesized to be an important pathogenic factor (Webster, 1995). Antibiotic therapy targeting P. acnes has been a mainstay treatment for more than 30 years (Leyden, 2001). However, despite decades of study, it is still not clear how P. acnes contributes to acne pathogenesis while being a major commensal of the normal skin flora (Bek-Thomsen et al., 2008; Cogen et al., 2008; Costello et al., 2009; Dominguez-Bello et al., 2010; Fierer et al., 2008; Gao et al., 2007; Grice et al., 2009). Whether P. acnes protects the human skin as a commensal bacterium or functions as a pathogenic factor in acne, or both, remains to be elucidated.
Here we compared the skin microbiome at the strain level and genome level in 49 acne patients and 52 normal individuals using a combination of metagenomics and genome sequencing. First, for each sample, 16S ribosomal DNA (rDNA) was amplified, approximately 400 clones were sequenced, and an average of 311 nearly full length 16S rDNA sequences were analyzed. The population structure of P. acnes strains was determined in each sample. Second, each P. acnes strain was assigned an “acne index” by calculating its prevalence in acne patients based on the 16S rDNA metagenomic data. The P. acnes strains associated with the acne patient group were identified, as well as the strains enriched in the individuals with normal skin. This metagenomic approach is fundamentally different than prior approaches in determining disease associations; it is more powerful and less biased than traditional methods by bypassing the biases and selection in strain isolation and culturing. To our knowledge this study has the largest number of individual skin microbiomes reported at the strain level to date. Lastly, we sequenced 66 previously unreported P. acnes strains and compared 71 P. acnes genomes covering the major lineages of P. acnes found in the skin microbiota. By combining a metagenomic study of the skin microbiome and genome sequencing of this major skin commensal, this study provides insight into potential bacterial genetic determinants in acne pathogenesis and emphasizes the importance of strain level analysis of the human microbiome to understand the role of commensals in health and disease.
RESULTS
P. acnes dominates the pilosebaceous unit
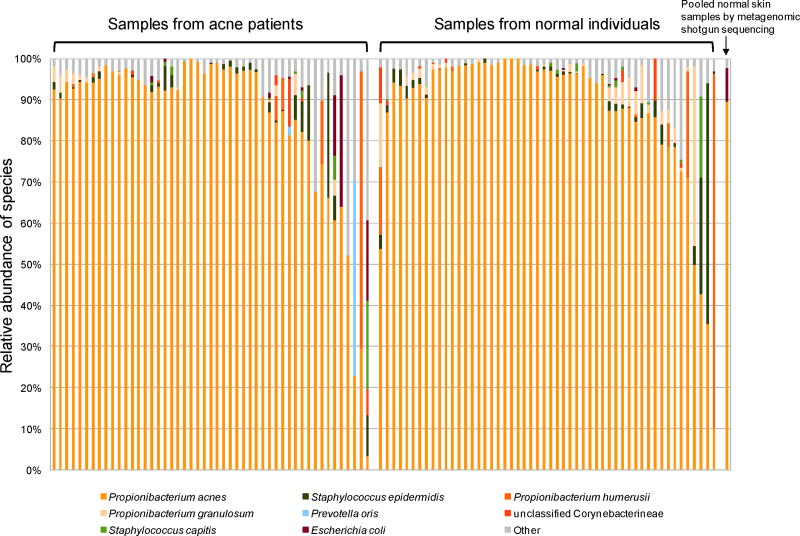
We characterized the microbiome in pilosebaceous units (“pores”) on the nose collected from 49 acne patients and 52 individuals with normal skin. Nearly full length 16S rDNA sequences were obtained using Sanger method, which allowed us to analyze the P. acnes at the strain level. After quality filtering, our final dataset contained 31,461 16S rDNA sequences ranging from position 29 to position 1483. 27,358 of the sequences matched to P. acnes with greater than 99% identity. Our data demonstrated that P. acnes dominates the microbiota of pilosebaceous units, accounting for 87% of the clones (Figure 1). Other commonly found species in pilosebaceous units included Staphylococcus epidermidis, Propionibacterium humerusii, and Propionibacterium granulosum, each representing 1% – 2.3% of the total clones. A total of 536 species level operational taxonomic units (SLOTUs) belonging to 42 genera and six phyla were identified in the samples (Table S1).
Figure 1.
P. acnes was dominant in pilosebaceous units in both acne patients and individuals with normal skin. By 16S rDNA sequencing, P. acnes sequences accounted for 87% of all the clones. Species with a relative abundance greater than 0.35% are listed in order of relative abundance. Species distribution from a metagenomic shotgun sequencing of pooled samples from normal individuals confirmed the high abundance of P. acnes in pilosebaceous units, as shown on the far right column.
To bypass the potential biases due to PCR amplification and due to uneven numbers of 16S rDNA gene copies among different species, we performed a metagenomic shotgun sequencing of the total DNA pooled from the pilosebaceous unit samples of 22 additional normal individuals. Microbial species were identified by mapping metagenomic sequences to reference genomes. The results confirmed that P. acnes was the most abundant species (89%) (Figure 1). This is consistent with the results obtained from 16S rDNA sequencing (87%).
Different P. acnes strain populations in acne
There was no statistically significant difference in the relative abundance of P. acnes when comparing acne patients and normal individuals. We next examined whether there were differences at the strain level of P. acnes by extensively analyzing the P. acnes 16S rDNA sequences. We define each unique 16S rDNA sequence as a 16S rDNA allele type, called a ribotype (RT). The most abundant P. acnes sequence was defined as ribotype 1 (RT1); all other defined ribotypes have 99% or greater sequence identity to RT1. Similar to the distributions seen at higher taxonomical levels (Bik et al.), at the strain level a few ribotypes were highly abundant in the samples with a significant number of rare ribotypes (Figure S1). After careful examination of the sequence chromatograms and manual correction of the sequences, a total of 11,009 ribotypes were assigned to the P. acnes 16S rDNA sequences. Most of the minor ribotypes were singletons. On average, each individual harbored 3±2 P. acnes ribotypes with three or more clones. Based on the genome sequences described below, all the sequenced P. acnes strains have three identical copies of 16S rDNA genes (note in Supplementary Information). This allowed us to compare the P. acnes strain populations in individuals based on the 16S rDNA sequences. The top ten major ribotypes with more than 60 clones and found in multiple subjects are shown in Table 1.
Table 1.
Top ten most abundant ribotypes found in pilosebaceous units
| Ribotype | Nucleotide changes from RT1 | Number of subjects | Number of clones | Percentage of clones from acne patientsa | Percentage of clones from normal individualsb | p-valuec |
|---|---|---|---|---|---|---|
| RT1 | - | 90 | 5536 | 48% | 52% | 0.84 |
| RT2 | T854C | 48 | 1213 | 51% | 49% | 0.36 |
| RT3 | T1007C | 60 | 2104 | 40% | 60% | 0.092 |
| RT4 | G1058C, A1201C | 23 | 275 | 84% | 16% | 0.049 |
| RT5 | G1058C | 15 | 205 | 99% | 1% | 0.00050 |
| RT6 | T854C, C1336T | 11 | 262 | 1% | 99% | 0.025 |
| RT7 | G529A | 10 | 188 | 99% | 1% | 0.12 |
| RT8 | G1004A, T1007C | 5 | 239 | 100% | 0% | 0.024 |
| RT9 | G1268A | 4 | 68 | 99% | 1% | 0.29 |
| RT10 | T554C, G1058C | 5 | 61 | 100% | 0% | 0.024 |
The percentage was calculated after the number of clones of each ribotype was normalized by the total number of clones in acne patients (acne index).
The percentage was calculated after the number of clones of each ribotype was normalized by the total number of clones in normal individuals.
Mann-Whitney-Wilcoxon rank sum test.
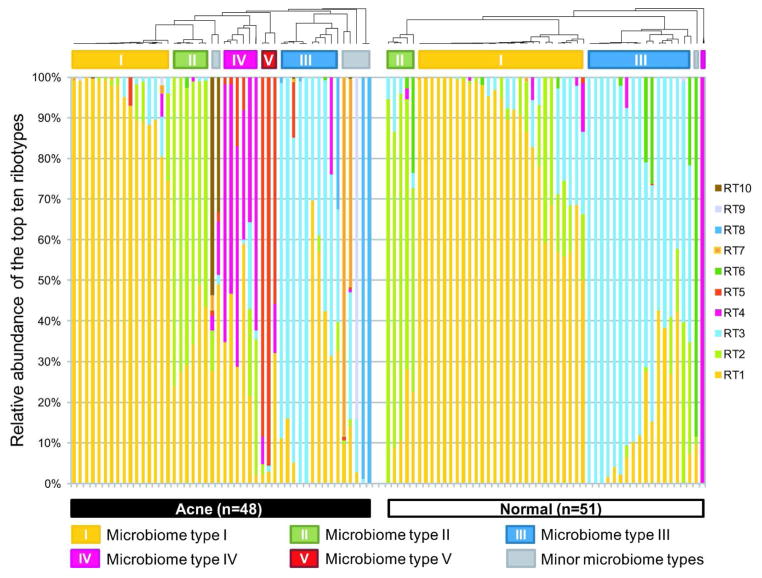
Analysis of the top ten ribotypes showed both disease-specific and health-specific associations. The three most abundant ribotypes (RT1, RT2 and RT3) were fairly evenly distributed among acne and normal individuals. However, the next seven major ribotypes were significantly skewed in their distribution (Table 1). Ribotypes 4, 5, 7, 8, 9 and 10 were found predominantly in acne patients, with four of these six statistically significantly enriched in acne (p<0.05, Wilcoxon test). Ribotypes 4, 5 and 10 contain a nucleotide substitution G1058C in the 16S rDNA sequences, which has previously been shown to confer increased resistance to tetracycline (Ross et al., 1998a; Ross et al., 2001). However, only a small percentage of the subjects in our study harboring these ribotypes had been treated with antibiotics (Table S2), therefore enrichment of these three ribotypes in the acne group was not correlated with antibiotic treatment. This is consistent with previous studies, which showed that previous use of antibiotics was not always associated with the presence of antibiotic resistant strains and that some patients who were not previously treated with antibiotics harbored strains already resistant to antibiotics (Coates et al., 2002; Dreno et al., 2001). On the other hand, one ribotype, RT6, although detected in only 11 subjects, was strongly associated with normal skin (p=0.025, Wilcoxon test) (Table 1). Its relative abundance in our normal group was similar to that found in the healthy cohort data from the Human Microbiome Project (HMP) (Supplementary Information, Figure S2). The percentage of positive subjects (11/52) was similar as well. Three of the 14 HMP subjects had RT6 found in the anterior nares, and one additional subject had RT6 in the left retroauricular crease.
Based on the distributions of the top ten ribotypes, statistical analysis using several different tests showed significant differences in P. acnes population structure between acne and normal skin (Table S3). This is consistent with a principal coordinate analysis, where acne samples and normal skin samples were separated by mostly principal coordinates 1 and 2 (Figure S3), explaining 44% and 20% of the variation, respectively.
To examine whether different individuals share similar P. acnes population structures, we clustered the samples based on the relative abundance of the top ten ribotypes. Five main microbiome types were observed at the P. acnes strain level (microbiome types I to V). Types IV and V, which are dominated by P. acnes RT4 and RT5, respectively, were mainly found in acne patients (Figures 2 and S4). The same five main microbiome types were observed in the HMP data and the data from Grice et al. (Grice et al., 2009) (Supplementary Information, Figure S5).
Figure 2.
Distribution of the top ten most abundant P. acnes ribotypes in acne patients and individuals with normal skin. Each column represents the percentage of the top ten ribotypes identified in each subject. The average P. acnes clone number per subject was 262 and the average clone number of top ten ribotypes was 100. Five major microbiome types at the P. acnes strain level were observed in the data. Types IV and V were mostly found in acne patients. Two samples (one from acne, one from normal skin) with fewer than 50 P. acnes 16S rDNA sequences are not displayed.
Genome sequence analysis of 71 P. acnes strains
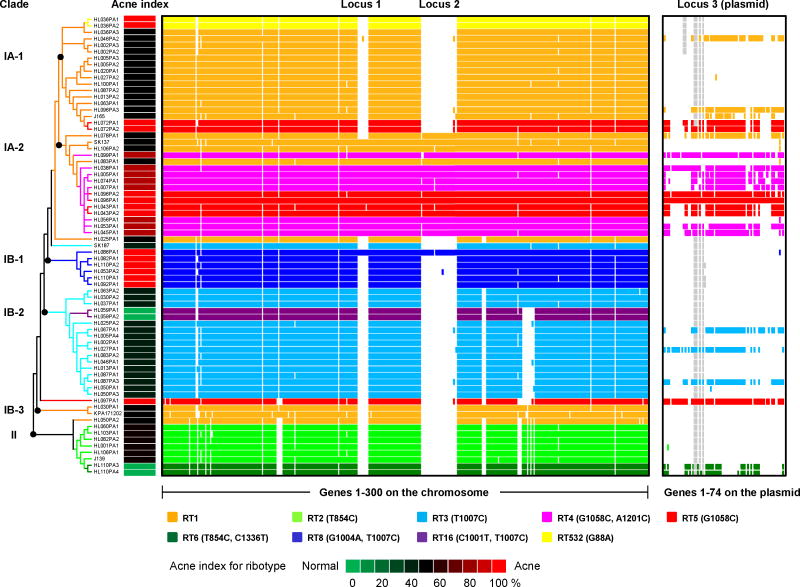
All of the top ten most abundant ribotypes differ from RT1 by only one or two nucleotide changes in the 16S rDNA sequence (Table 1). To determine whether such small changes in the 16S rDNA sequence reflect the lineages and evolutionary history at the genome level, we selected 66 P. acnes isolates representing major ribotypes 1, 2, 3, 4, 5, 6, and 8 as well as two minor ribotypes, 16 and 532, for genome sequencing. The genomes of these 66 isolates were fully sequenced and assembled to high quality drafts or complete genomes with 50X coverage or more. Five other P. acnes genomes, KPA171202 (Bruggemann et al., 2004), J165, J139, SK137, and SK187, were publicly available and were included in our analysis. We constructed a phylogenetic tree based on 96,887 unique single nucleotide polymorphism (SNP) positions in the core genome obtained from these 71 P. acnes genomes. Most of the genomes with the same ribotypes clustered together. The tree suggests that the 16S rDNA ribotypes do represent the relationship of the lineages to a large extent and that16S rDNA sequence is a useful molecular marker to distinguish major P. acnes lineages (Figures 3 and S6).
Figure 3.
Genome comparison of 71 P. acnes strains showed that the genomes of RT4 and RT5 are distinct from others. Two chromosomal regions, loci 1 and 2, are unique to clade IA-2 and one other genome HL086PA1. Clade IA-2 consists of mainly RT4 and RT5 that were highly enriched in acne. The presence of a plasmid (locus 3) is also characteristic of RT4 and RT5. Each row represents a P. acnes genome colored according to the ribotypes. Rows are ordered by the phylogeny calculated based on the SNPs in the P. acnes core genome. Only the topology is shown. The clades were named based on their recA types (IA, IB and II). Columns represent predicted open reading frames (ORFs) in the genomes and are ordered by ORF positions along the finished genome HL096PA1, which encodes a 55 Kb plasmid. Only the first 300 ORFs on the chromosome (on the left) and all the ORFs on the plasmid (on the right) are shown. The colored plasmid regions represent genes on contigs that match exclusively to the HL096PA1 plasmid region. The genes that fall on contigs that clearly extend beyond the plasmid region are likely to be chromosomally located and are colored in grey. Acne index for the ribotypes was calculated based on the percentage of clones of each ribotype found in acne as shown in column 5 in Table 1.
Genetic elements detected in P. acnes
We further performed comparative genome analysis among all 71 genomes grouped by ribotypes. Our analysis revealed potential genetic elements by which acne-associated strains could contribute to acne pathogenesis and the elements by which health-associated strains could contribute to maintaining skin health. Specifically, we describe here the unique genome regions of RT4 and RT5, which had a strong association with acne, and RT6, which was found enriched in normal skin. Three distinct regions, loci 1, 2 and 3, were found almost exclusively in strains that belong to clade IA-2 in the phylogenetic tree. Clade IA-2 consists of mainly RT4 and RT5 (Figures 3 and S7). Loci 1 and 2 are located on the chromosome. Locus 1 contains prophage-related genes and appears to be a genomic island. Locus 2 has plasmid integration sites and thus could be derived from a plasmid sequence. Locus 3 appears to be on a large mobile genetic element, likely a plasmid. The plasmid is approximately 55 Kb long and has inverted terminal repeats according to our finished genome HL096PA1 (Supplementary Information). The sequence data suggest that the plasmid is linear and possibly originated from a phage (Hinnebusch and Tilly, 1993). All but one of the fifteen genomes of RT4 and RT5 have at least 60% of the genes of the plasmid represented, and all of them have regions homologous to the inverted terminal repeat in the plasmid, suggesting that they harbor the same or a similar linear plasmid (Figure 3). The copy number of the plasmid in the genomes ranges from 1 to 3 based on genome sequencing coverage, which was confirmed by quantitative PCR (Figures S8 and S9).
The fact that acne-enriched RT4 and RT5 carry a linear plasmid and two unique loci of genomic islands suggests that these plasmid and chromosomal regions may play a role in acne pathogenesis. In fact, the linear plasmid encodes a tight adhesion (Tad) locus, which has been suggested to play a role in virulence in other organisms (Kachlany et al., 2000; Schreiner et al., 2003). The complete Tad locus is found in all but one of the fifteen genomes of RT4 and RT5, and is only occasionally found in other ribotypes. Additionally, in locus 2, a Sag gene cluster is encoded, which has been shown to contribute to hemolytic activity in pathogens (Fuller et al., 2002; Humar et al., 2002; Nizet et al., 2000). Table S4 summarizes the genes that are mostly unique to RT4 and RT5, several of which play essential roles in virulence in other organisms. We speculate that some of these genes encoded in RT4 and RT5 may increase virulence, promote stronger adherence to the human host, or induce a pathogenic host immune response.
In genome comparison analysis, we found that all the genomes of RT2 and RT6 encode Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR). Among the sequenced genomes, RT2 and RT6 are the only ribotypes encoding CRISPR. CRISPR have been shown to confer protective “immunity” against viruses, phage and plasmids (Horvath and Barrangou, 2010; Makarova et al., 2011). The CRISPR locus encoded in P. acnes consists of a series of cas genes - cas3, cse1, cse2, cse4, cas5e, cse3, cas1 and cas2, which are homologous to the CRISPR locus reported in E. coli (Figure S10) and the CRISPR4 locus in Streptococcus thermophilus (Horvath and Barrangou, 2010).
CRISPR arrays are composed of a cluster of identical repetitive sequences separated by spacer sequences of similar length but with different nucleotide sequences. Spacer sequences have been found identical or with one or two mismatches to phage or plasmid DNA sequences. A total of 39 spacer sequences were found in eight P. acnes strains, 25 of which were unique as shown in Table 2. As expected, most of the identifiable spacers target to known P. acnes phage sequences. However, among the unique CRISPR spacer sequences, one matched locus 2 on the chromosome and three matched the plasmid region (locus 3) in P. acnes genomes of mainly RT4 and RT5. This suggests that these loci may have been acquired by RT4 and RT5, while the genomes of RT2 and RT6 may be capable of protecting against the invasion of the plasmids or other foreign DNA through the CRISPR mechanism.
Table 2.
CRISPR spacer sequences found in the genomes of RT2 and RT6
| Ribotype | Strain | Spacer number | Spacer sequence | BLAST result | Match found |
|---|---|---|---|---|---|
| RT2 | HL001PA1 | 1 | CATGGCCTGCACACCAGGCGCTTTTAGCACCT | No hits | |
| 2 | CATGGCCTGCACACCAGGCGCTTTTAGCACCT | No hits | |||
| 3 | CATGGCCTGCACACCAGGCGCTTTTAGCACCT | No hits | |||
| 4 | GGCGTATGACGAGTTGTGGTCGGCGTTTCCTC | P. acnes phage PA6 gp15 (minor tail protein) | |||
| 5 | CGGTGTTAACGGCTTGCCTGGCTTGGATGGAG | No hits | |||
| RT2 | HL060PA1 | 1 | CGCCTACCGTCAGCTGACTCACGCCTCCGCGTT | No hits | |
| 2 | TCACACCAGTCATCAGCGTCATAGTCCTCTCGG | No hits | |||
| RT2 | HL082PA2 | 1 | GGCTCAGCCCTGCCCGATGCCTACGCCAAATGG | C. leptum DSM 753 CLOLEP_00129 (cell wall-associated hydrolases (invasion-associated proteins)) | Locus 3 |
| 2 | TCACACCAGTCATCAGCGTCATAGTCCTCTCGG | No hits | |||
| RT2 | HL103PA1 | 1 | CACCGGGCCCATCCCGGTCGGCCTCCTGAAAGG | C. leptum DSM 753 CLOLEP_00135 | Locus 3 |
| RT2 | HL106PA1 | 1 | GATCGAGTTGGCTGAGTCGAAGGTGTTGCGGTT |
P. acnes phage PA6 gp16
(conserved protein) P. acnes phage PAD20 gp16 |
|
| 2 | CTGCTCATCGCTCAGCTCCTGCGCCTCATCACA | No hits | |||
| 3 | CTGCGCCAACAGCCGCATCTGATCCGAATACGG | P. acnes phage PA6 gp3 (phage portal protein) | |||
| 4 | CGCAGCAATCTCAGAAGGCCACAACAAGTTCGT |
P. acnes phage PA6 gp7
(conserved protein) P. acnes phage PAD20 gp7 P. acnes phage PAS50 gp7 |
|||
| 5 | CAAATCACCCAAGCCCAACACGCCGCCACCACC | No hits | |||
| 6 | TGTCACCGATTCAATGTATCTATGAGTGGTGTA | No hits | |||
| 7 | TTGGGTGGGTGAGGTCGGGTCGTCAGTCATGAG | No hits | |||
| 8 | GTCGATGTCGAGATTGGCCTGGGGGTCCATGTC | Clostridium leptum DSM 753 CLOLEP_00142 | Locus 3 | ||
| 9 | ACGTCGTGAACGTACCCCTTGACGGAGACGGCA | No hits | |||
| RT2 | J139 | 1 | CGAGGGCTACCACGTGGTCGATTTGGACTGTCG |
C. leptum DSM 753
CLOLEP_00167 P. acnes SK137 HMPREF0675_3193 (domain of unknown function) |
Locus 2 |
| 2 | CAGGCGCTCCACTCCCTCGCCCTGGCCACCAAC | No hits | |||
| RT6 | HL110PA3 HL110PA4 |
1 | CTATGTGGACAGTGTTGGTTACTGTGGGGGGAA | P. acnes phage PA6 intergenic region between gp45 and gp46 | |
| 2 | GCACTGCACCGATATCGTCGTGGCTGTCACTTG | No hits | |||
| 3 | CCCAGACAACCTCGACAACCTGTTCAGGGGATG | P. acnes phage PAS50 gp25 | |||
| 4 | CATGGCTAGCCCGGATTTTTGGCTGCCTGAGCG |
P. acnes phage PA6 gp34
(mutidrug resistance protein-like transporters) P. acnes phage PAD20 gp34 (DNA helicase) |
|||
| 5 | CGGCCTGCGGCAGATTTTTGTTGCGTTGAATCC |
P. acnes phage PA6 gp14
(tape measure protein) P. acnes phage PAD20 gp14 (tape measure protein) P. acnes phage PAS50 gp14 (tape measure protein) |
|||
| 6 | CGGGCAGAGGATGTGTTGCTCGTTCCTGGATGG |
P. acnes phage PA6 gp32
(CHC2 zinc finger) P. acnes phage PAD20 gp32 (DNA primase) P. acnes phage PAS50 gp32 (DNA primase) |
|||
| 7 | GTTACGCTGGAACCCCCAATGAACACGCGAGAA |
P. acnes phage PAD42 major
head protein gene P. acnes phage PAD20 major head protein gene P. acnes phage PAD9 major head protein gene P. acnes phage PAS40 major head protein gene P. acnes phage PAS12 major head protein (gene P. acnes phage PAS10 major head protein gene P. acnes phage PAD21 major head protein gene P. acnes phage PAS2 major head protein gene P. acnes phage PA6 gp6 (Phage capsid family) P. acnes phage PAS50 gp6 major head protein gene |
|||
| 8 | CGAGGGCTACCACGTGGTCGATTTGGACTGTCG |
C. leptum DSM 753
CLOLEP_00167 P. acnes SK137 HMPREF0675_3193 (Domain of unknown function) |
Locus 2 | ||
| 9 | CAGGCGCTCCACTCCCTCGCCCTGGCCACCAAC | No hits |
Abbreviations: BLAST, Basic Local Alignment Search Tool; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeat; C. leptum, Clostridium leptum; P. acnes, Propionibacterium acnes; RT, ribotype.
DISCUSSION
Our study of the human skin microbiome associated with acne provides a previously unreported portrait of the microbiota of pilosebaceous units at the bacterial strain level. Since P. acnes is the major skin commensal bacterium found in both acne and healthy skin, this strain-level analysis is important to help understand the role of P. acnes in acne pathogenesis and in skin health. We demonstrate a strong association between strains of RT4 and RT5 with acne and a strong association between strains of RT6 and healthy skin, each with unique genetic elements. Other P. acnes strains, including ribotypes 7, 8, 9 and 10, or interactions among different strains, may also contribute to the development of the disease. In addition, host factors, such as hormone level, sebum production and physical changes in the pilosebaceous unit, may also play a role in acne pathogenesis. Further studies aimed at identifying the specific functions of these strains, host factors in the development of acne, as well as the associations of microbiome characteristics with the sub-types of acne (comedonal, pustular, inflammatory, cystic, etc.) with larger cohort sizes may improve our understanding of the molecular mechanisms of the disease. These studies may also help to develop a targeted therapeutic approach to treat this extremely common and sometimes disfiguring skin disease.
Our metagenomic approach in revealing the association of P. acnes strains with the disease or health is to our knowledge previously unreported, and is more powerful than previous studies using traditional methods (Lomholt and Kilian, 2010; McDowell et al., 2011). Because the skin microbiota of each individual and each skin site may harbor “good”, “neutral” and “bad” strains at the same time, which may have different growth rates under in vitro culturing conditions, culturing a few isolates from a disease lesion or healthy skin site may not provide an accurate and unbiased measurement of the association of the strains with the disease or health. The sampling technique and disease associations in this study do not depend on sampling locations, on the presence of lesions in the sampling field, or on inherently biased culture techniques. While sampling lesional skin intentionally may yield interesting results, these results would not be capable of defining the disease associations that unbiased sampling can. The metagenomic approach employed in this study to identify underlying strain differences in acne might also be applied to the study of other disease/health associations with commensal or pathogenic bacteria. Ultimately, these studies could lead to targeted therapeutics to restore natural commensal population structures, and could help determine if therapeutic modulations of the microbiota can return the host to a state of health.
MATERIALS AND METHODS
Subjects
Subjects with acne and subjects with normal skin were recruited from various clinics in Southern California including private practice, managed care, and public hospital settings, as well as outside of dermatology clinics, to best represent the diversity of populations and history of medical care. The subject data are available at dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000263.v1.p1). The diagnosis of acne was made by board-certified dermatologists. The presence of acne was graded on a scale of 0 to 5 relating closely to the Global Acne Severity Scale (Dreno et al., 2011). Grades were recorded for both the face and the nose separately where zero represents normal skin and 5 represents the most severe inflammatory cystic acne. In acne patients, the grades of the face ranged from 1 to 5 with an average of 2.1, and the grades of the nose ranged from 0 to 2 with an average of 0.3. The presence of scarring was also noted. Subjects with normal skin were determined by board-certified dermatologists and were defined as people who had no acneiform lesions on the face, chest, or back. They were also excluded if they had other skin problems that the investigators felt would affect sampling or the microbial population on the skin. Among the 101 subjects, 59 were female (31 acne patients and 28 normal subjects) and 42 were male (18 acne patients and 24 normal subjects). The average age of the acne cohort was 22.2 and the average age of the normal cohort was 29.6. There was no significant difference in ethnicity between the acne and normal populations. The subjects responded to a written questionnaire, administered by a physician or a well trained study coordinator who went over each question with the subjects. Most of the subjects had not been treated for acne in the past or were not being treated when samples were collected (Table S2). Only nine out of 78 subjects, who provided treatment information, were being treated for acne when samples were taken. Among the nine subjects, two were being treated with antibiotics, five were being treated with topical retinoids, one was being treated with both antibiotics and retinoids, and one did not list the treatment. We also asked subjects for acne treatment history in the past (anytime in their life). Eighteen out of 73 subjects, who provided treatment history, had been treated for acne in the past. Among them, seven had been treated with antibiotics, eight had been treated with retinoids, two had been treated with both antibiotics and retinoids, and one did not list the treatment. All subjects provided written informed consent. All protocols and consent forms were approved by both the UCLA and Los Angeles Biomedical Research Institute IRBs. The study was conducted in adherence to the Helsinki Guidelines.
Samples
Skin microcomedone (white head or black head) samples were taken from the nose of the subjects using Bioré Deep Cleansing Pore Strips (Kao Brands Company, Cincinnati, OH) following the instruction of the manufacturer. Clean gloves were used for each sampling. After being removed from the nose, the strip was immediately placed into a 50 mL sterile tube and kept on ice or at 4 °C. The cells were lysed within four hours in most of the cases.
Metagenomic DNA extraction, 16S rDNA amplification, cloning and sequencing
Individual microcomedones were isolated from the adhesive nose strip using sterile forceps. Genomic DNA was extracted using QIAamp DNA Micro Kit (Qiagen). 16S rDNA was amplified and cloned according to the protocol by HMP, which is described in detail in Supplementary Information. Nearly full length sequences were obtained by Sanger method.
16S rDNA sequence analysis
Base calling and quality was determined with Phred (Ewing and Green, 1998; Ewing et al., 1998). Bidirectional reads were assembled and aligned to a core set of NAST-formatted sequences (rRNA16S.gold) using AmosCmp16Spipeline and NAST-ier. Suspected chimeras were identified using ChimeraSlayer and WigeoN (Haas et al., 2011). 16S rDNA sequences were extensively manually examined. Chromatograms were visually inspected at all bases with a Phred quality score < 30. Appropriate corrections were applied. QIIME (Caporaso et al., 2010b) was used to cluster the sequences into OTUs.
P. acnes isolation and genotyping
Colonies with the macroscopic characteristics of P. acnes were picked from each sample plate and were passed twice. The ribotype of each isolate was determined by PCR amplification and sequencing of the full length of the 16S rDNA gene by Sanger method.
Whole genome shotgun sequencing, assembly and annotation
Genome HL096PA1 was sequenced using Roche/454 FLX and was assembled using a combination of PHRAP/CONSED (Gordon et al., 1998) and GSMAPPER (Roche) with extensive manual editing in CONSED. The remaining 65 genomes were sequenced using Illumina/Solexa GAIIx. Sequence datasets were processed by quality trimming and were assembled using Velvet (Zerbino and Birney, 2008). Coding sequences were predicted using GeneMark (Borodovsky and McIninch, 1993) and GLIMMER (Salzberg et al., 1998). The final gene set was processed through a suite of protein categorization tools consisting of Interpro, psort-b and KEGG. A more detailed protocol can be found at http://hmpdacc.org/doc/sops/reference_genomes/annotation/WUGC_SOP_DACC.pdf.
Comparative genome analysis
Seventy-one P. acnes genome sequences were compared using Nucmer (Kurtz et al., 2004). Phylogenetic analysis was performed using MEGA5 (Tamura et al., 2007). CRISPRFinder (Grissa et al., 2007) was used to identify the CRISPR repeat-spacer sequences.
Supplementary Material
Acknowledgments
We thank G. Kasimatis, B. Shi, E. E. Curd, R. Yan, M. Wong and J. Liu for comments and technical support. We thank C. Lee for performing statistical analyses in the initial phase. We also thank Z. Guo and C. S. Miller for critical reading of the manuscript. This research was funded as one of the Demonstration Projects by the NIH Human Microbiome Project (HMP). It was supported by Grant Number 1UH2AR057503 from NIH/NIAMS and U54HG004968 from NIH.
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
The data reported in this paper are tabulated in Supplementary Information and archived at GenBank.
The authors declare no competing financial interests.
S.F. and S.T. analyzed the data. B.C., L.N. and C.D. performed experiments. D.E. performed some of the initial statistical analyses. M.C.E., A.L., J.K., R.L.M. and N.C. collected samples. E.S. and G.M.W. directed sequencing, genome assembly and annotation. H.L., M.L., R.L.M. and J.F.M. conceived the demonstration project in the initial phase. H.L. designed and directed the project, analyzed the data and wrote the paper. S.T., S.F. and N.C. co-wrote the paper.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bek-Thomsen M, Lomholt HB, Kilian M. Acne is not associated with yet-uncultured bacteria. J Clin Microbiol. 2008;46:3355–60. doi: 10.1128/JCM.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. Isme J. 4:962–74. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojar RA, Holland KT. Acne and Propionibacterium acnes. Clin Dermatol. 2004;22:375–9. doi: 10.1016/j.clindermatol.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Borodovsky M, McIninch J. Recognition of genes in DNA sequence with ambiguities. Bio Systems. 1993;30:161–71. doi: 10.1016/0303-2647(93)90068-n. [DOI] [PubMed] [Google Scholar]
- Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978;75:4801–5. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, et al. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science. 2004;305:671–3. doi: 10.1126/science.1100330. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010a;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010b;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat Rev Microbiol. 2008;6:904–12. doi: 10.1038/nrmicro2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Su LH, Lin TY, Huang YC. Molecular analysis of repeated methicillin-resistant Staphylococcus aureus infections in children. PLoS One. 2010;5:e14431. doi: 10.1371/journal.pone.0014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Coates P, Vyakrnam S, Eady EA, Jones CE, Cove JH, Cunliffe WJ. Prevalence of antibiotic-resistant propionibacteria on the skin of acne patients: 10-year surveillance data and snapshot distribution study. Br J Dermatol. 2002;146:840–8. doi: 10.1046/j.1365-2133.2002.04690.x. [DOI] [PubMed] [Google Scholar]
- Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence? Br J Dermatol. 2008;158:442–55. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe WJ. Looking back to the future - acne. Dermatology. 2002;204:167–72. doi: 10.1159/000057876. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreno B, Poli F, Pawin H, Beylot C, Faure M, Chivot M, et al. Development and evaluation of a Global Acne Severity Scale (GEA Scale) suitable for France and Europe. Journal of the European Academy of Dermatology and Venereology : JEADV. 2011;25:43–8. doi: 10.1111/j.1468-3083.2010.03685.x. [DOI] [PubMed] [Google Scholar]
- Dreno B, Reynaud A, Moyse D, Habert H, Richet H. Erythromycin-resistance of cutaneous bacterial flora in acne. Eur J Dermatol. 2001;11:549–53. [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–94. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–85. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–9. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller JD, Camus AC, Duncan CL, Nizet V, Bast DJ, Thune RL, et al. Identification of a streptolysin S-associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect Immun. 2002;70:5730–9. doi: 10.1128/IAI.70.10.5730-5739.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104:2927–32. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. Viewing and editing assembled sequences using Consed. Curr Protoc Bioinformatics. 2003;Chapter 11(Unit11):2. doi: 10.1002/0471250953.bi1102s02. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome research. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansra NK, Shinkai K. Cutaneous community-acquired and hospital-acquired methicillin-resistant Staphylococcus aureus. Dermatol Ther. 24:263–72. doi: 10.1111/j.1529-8019.2011.01402.x. [DOI] [PubMed] [Google Scholar]
- Hinnebusch J, Tilly K. Linear plasmids and chromosomes in bacteria. Mol Microbiol. 1993;10:917–22. doi: 10.1111/j.1365-2958.1993.tb00963.x. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Humar D, Datta V, Bast DJ, Beall B, De Azavedo JC, Nizet V. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet. 2002;359:124–9. doi: 10.1016/S0140-6736(02)07371-3. [DOI] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, et al. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–76. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome biology. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyden JJ. The evolving role of Propionibacterium acnes in acne. Semin Cutan Med Surg. 2001;20:139–43. doi: 10.1053/sder.2001.28207. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One. 2010;5:e12277. doi: 10.1371/journal.pone.0012277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell A, Gao A, Barnard E, Fink C, Murray PI, Dowson CG, et al. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology. 2011;157:1990–2003. doi: 10.1099/mic.0.049676-0. [DOI] [PubMed] [Google Scholar]
- McDowell A, Perry AL, Lambert PA, Patrick S. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol. 2008;57:218–24. doi: 10.1099/jmm.0.47489-0. [DOI] [PubMed] [Google Scholar]
- McDowell A, Valanne S, Ramage G, Tunney MM, Glenn JV, McLorinan GC, et al. Propionibacterium acnes types I and II represent phylogenetically distinct groups. J Clin Microbiol. 2005;43:326–34. doi: 10.1128/JCM.43.1.326-334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, et al. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun. 2000;68:4245–54. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M, Phillippy A, Delcher AL, Salzberg SL. Comparative genome assembly. Brief Bioinform. 2004;5:237–48. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JI, Eady EA, Cove JH, Cunliffe WJ. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob Agents Chemother. 1998a;42:1702–5. doi: 10.1128/aac.42.7.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JI, Eady EA, Cove JH, Jones CE, Ratyal AH, Miller YW, et al. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob Agents Chemother. 1997;41:1162–5. doi: 10.1128/aac.41.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JI, Eady EA, Cove JH, Ratyal AH, Cunliffe WJ. Resistance to erythromycin and clindamycin in cutaneous propionibacteria is associated with mutations in 23S rRNA. Dermatology. 1998b;196:69–70. doi: 10.1159/000017871. [DOI] [PubMed] [Google Scholar]
- Ross JI, Snelling AM, Eady EA, Cove JH, Cunliffe WJ, Leyden JJ, et al. Phenotypic and genotypic characterization of antibiotic-resistant Propionibacterium acnes isolated from acne patients attending dermatology clinics in Europe, the U.S.A., Japan and Australia. The British journal of dermatology. 2001;144:339–46. doi: 10.1046/j.1365-2133.2001.03956.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic acids research. 1998;26:544–8. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci U S A. 2003;100:7295–300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular biology and evolution. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–86. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- Webster GF. Inflammation in acne vulgaris. J Am Acad Dermatol. 1995;33:247–53. doi: 10.1016/0190-9622(95)90243-0. [DOI] [PubMed] [Google Scholar]
- White GM. Recent findings in the epidemiologic evidence, classification, and subtypes of acne vulgaris. J Am Acad Dermatol. 1998;39:S34–7. doi: 10.1016/s0190-9622(98)70442-6. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome research. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.