Abstract
Motivated from a colorectal cancer study, we propose a class of frailty semi-competing risks survival models to account for the dependence between disease progression time, survival time, and treatment switching. Properties of the proposed models are examined and an efficient Gibbs sampling algorithm using the collapsed Gibbs technique is developed. A Bayesian procedure for assessing the treatment effect is also proposed. The Deviance Information Criterion (DIC) with an appropriate deviance function and Logarithm of the Pseudomarginal Likelihood (LPML) are constructed for model comparison. A simulation study is conducted to examine the empirical performance of DIC and LPML and as well as the posterior estimates. The proposed method is further applied to analyze data from a colorectal cancer study.
Keywords: Competing risks, Panitumumab, Partial treatment switching, Posterior propriety, Semi-Markov model
1 Introduction
In chronic disease or cancer studies and clinical trials, it is very common to have both terminating events and nonterminating events in the data. This type of situation is referred as semi-competing risks, in which an event time can be censored by another event time but not vice versa. A terminating event potentially censors a nonterminating event, but the nonterminating event does not prevent subsequent observation of the terminating event. An example of this is the colorectal cancer clinical trial that we examine here, called the panitumumab 408 study, which was conducted by Amgen Inc. (see Section 5). In this study, disease progression is a nonterminating event, death is a terminating event, and disease progression can be censored by death but not vice-versa. In addition to semi-competing risks, treatment switching may also occur in clinical trials. In such trials, patients in the control arm who experience an intermediate event, such as disease progression, may begin taking the experimental treatment. In the panitumumab 408 study, there were a substantial proportion of patients in the control arm who switched treatment after disease progression (see Section 5). As discussed in Marcus and Gibbons (2001), an intent-to-treat (ITT) analysis leads to attenuated treatment effect estimates, and thus one must properly model the data accommodating this switching effect and then appropriately estimate the treatment effect.
In semi-competing risks data, there are two major issues: dependent censoring and identifiability. In order to deal with these issues, several modeling and inference approaches have been developed. One major approach is to model the joint distribution of TD and TE, where TD denotes the time to terminating event and TE denotes the time to nonterminating event. Day and Bryant (1997) used frailty models for the joint survival function using a relevant censoring process. Later, Fine et al. (2001) adopted this model and proposed a novel estimator for the marginal distribution of TE based on a bivariate location-shift model with a completely unspecified underlying distribution for TD and TE. Although this method is appropriate for modeling one recurrent event taking into account dependent censoring, it cannot be applied to more than two recurrent events. Furthermore, various types of copula models have been applied for modeling the joint distribution of (TE, TD) (Wang, 2003; Ghosh, 2006; Peng and Fine, 2007). Another approach is to model the gap time TG between TD and TE (Mandel, 2010). Nonparametric estimation of the gap time distribution and regression methods for gap time hazard functions have been developed. A third approach is similar to the above gap time model. In addition to modeling TE and TG, another event time is introduced, which denotes the terminating event that happens without the nonterminating event TE. Shen and Thall (1998) used such a model for obtaining the marginal distributions of TE, TG and . They assumed that the distributions of TE and are mutually independent. For the bivariate distribution of TE and TG, they used a bivariate generalized von Morgenstern distribution, which characterizes the positive or negative association between these two times using a single parameter. A conditional model is also developed (Zeng et al., 2012). Instead of modeling the joint distribution of TE and TG, a conditional model of TG given TE is used. Multistate modeling is another approach for survival data with semi-competing risks, in which no event, nonterminating event, and terminating event can be viewed as the three states in a multistate process. The focus of multistate modeling is mainly on the transition probabilities between different states. Aalen-Johansen estimators (Aalen et al., 1978; Andersen et al., 1993) can be used to estimate these transition probabilities. However, this approach does not provide much information on the dependence structure between the time to nonterminating event and the time to terminating event. Except for Zeng et al. (2012), most of the aforementioned articles do not directly deal with both semi-competing risks and treatment switching.
In this paper, we introduce a Bayesian frailty model for survival data with semi-competing risks in the presence of partial treatment switching (i.e., not every subject in the control arm switched to active treatment). In the frequentist inference, the Monte Carlo EM (MCEM) algorithm is often used to obtain the maximum likelihood estimates in the presence of the unobserved frailty variables. However, the MCEM algorithm may fail to converge when fitting a semi-competing risks frailty model with unknown parameters in the frailty distribution since the estimates of these unknown parameters are unstable. To overcome this challenging computational issue, we develop an efficient Gibbs sampling algorithm via the introduction of latent variables, reparameterization, and the collopsed Gibbs sampler. The Bayesian framework also allows us to characterize the conditions for model identifiability by examining posterior propriety. In addition, to appropriately estimate the treatment effect, we extend the method of Zeng et al. (2012) to derive the predictive survival function with partial treatment switching under the semi-competing risks frailty model and carry out Bayesian inference on this quantity without resorting to asymptotics.
The rest of the paper is organized as follows. Section 2 presents a detailed development of the semi-competing risks model via a gamma frailty including explicit expressions for the likelihood function based on the observed data. In Section 3, we characterize posterior propriety conditions under this complex model, provide the Bayesian formulation of the predictive survival function with partial treatment switching, develop an efficient Gibbs sampling algorithm, and introduce two Bayesian model comparison criteria. A simulation study is carried out to examine the empirical performance of the posterior estimates and Bayesian model criteria in Section 4, and a detailed analysis of a subset of the data from the panitumumab 408 study is presented in Section 5. We conclude the paper with a brief discussion in Section 6. The proofs of all theorems and detailed derivations of the computational development are given in the Appendices.
2 The Semi-Competing Risks Frailty Models
2.1 Models
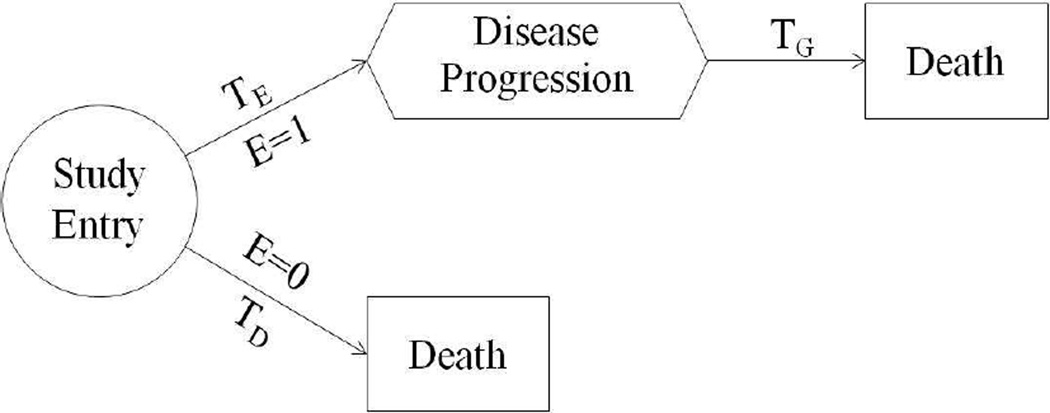
To introduce the proposed model, we use the following notation. As motivated from the panitumumab 408 study, we consider disease progression as a nonterminating event. However, the proposed model can be applied to any other type of nonterminating event. Let E be a dichotomous variable to denote the disease progression status of subjects, where E = 1 if the subject is in the disease progression population, which include subjects who eventually develop disease progression before death, and E = 0 if otherwise. Also let TD denote the time from study entry to death for subjects with E = 0. For the disease progression population (E = 1), we further let TE denote the time from study entry to disease progression and let TG denote the time from disease progression to death. A graphical illustration of these variables is shown in Fig. 1.
Fig. 1.
A graphical illustration of key random variables in the semi-competing risks model.
The proposed statistical model consists of the following three components. The first component is to model the disease progression status E given the baseline covariates x and the treatment indicator A (A = 1 if the subject is on the treatment arm and A = 0 if the subject is on the placebo or control arm). To this end, we assume
| (2.1) |
where α0, α1, and α2 are unknown coefficients and . The second component models the survival distribution of the non-progression population given x and A, which is defined by
| (2.2) |
where hD(t|A, x, E = 0) is the conditional hazard function of TD given the covariates, h0(t) is an unknown baseline hazard function, and (β0, γ0) are unknown regression coefficients.
As shown in Fig. 1, TE and TG are potentially dependent. To capture this dependence, we assume the frailty model
| (2.3) |
where hE(t|A, x, E = 1, ω) is the conditional hazard function for TE, hG(t|A, z, E = 1, ω) is the conditional hazard function for TG, both h1(t) and h2(t) are unknown baseline hazard functions, and the β’s and γ’s are regression coefficients. Here, V is the treatment switching indicator (1 = switching; 0 = no switching) and z reflects the covariates collected at baseline or at disease progression, which could be prognostic factors for the treatment switching decision. In (2.3), ω is a latent gamma-frailty, which is assumed to follow a Gamma distribution, Gamma(1/τ, 1/τ ), with mean one, variance τ (τ > 0), and density given by . Given ω, TE and TG are conditionally independent. Unconditionally, TE and TG are dependent and, moreover, the local measure of dependence (Oakes, 1989) between TE and TG is ϕFM = 1 + τ, indicating a positive association between TE and TG. When τ → 0, ϕFM → 1 and TE and TG become independent. The model defined by (2.1) – (2.3) is thus called the semi-competing risks frailty model abbreviated by FM. As an alternative to (2.3), we may consider the following models for TE and TG:
| (2.4) |
where γ21 is the regression coefficient corresponding to TE and γ22 is the corresponding vector of regression coefficients associated with z. The model defined by (2.1), (2.2), and (2.4) is called the conditional semi-competing risks model, denoted by CM. After some algebra, we can show that the local measure of dependence between TE and TG under CM is given by ϕCM =[∫tE{exp[−H2(tG) exp{Aβ21 + V (1 − A)β22 + γ21u + z′γ22}]h1(u) exp(Aβ1 + x′γ1) × exp{−H1(u) exp(Aβ1 + x′γ1)}}du exp(tE γ21)] × [∫tE {exp[−H2(tG) exp{Aβ21+V (1−A)β22+γ21u+z′γ22}]h1(u) exp(Aβ1+x′γ1)×exp{−H1(u) exp(Aβ1+x′γ1)} exp(uγ21)}du]−1 for tE > 0 and tG > 0, where for j = 1, 2. Unlike ϕFM, ϕCM depends on (tE, tG). It is easy to see that ϕCM > 1 when γ21 < 0, ϕCM = 1 when γ21 = 0, and ϕCM < 1 when γ21 > 0. This result implies that CM allows a positive or negative association between TE and TG. As discussed in Zhao (2009), FM is a homogeneous Markov model when h2(t) is constant while CM is a homogeneous semi-Markov model since the hazard function for TG in (2.4) depends on the progression time TE. On the other hand, the marginal distributions of TE and TG after integrating out the gamma frailty belong to the class of generalized odds-rate hazards (GORH) models (see Banerjee et al, 2007). As the GORH model is a non-proportional hazards model, FM is more robust to the proportional hazards assumption than CM.
We further assume piecewise exponential models for the baseline hazard functions h0(t), h1(t), and h2(t). For k = 0, 1, 2, let 0 < sk1 < sk2 < … < skJk be a finite partition of the time axis. Thus, we have the Jk intervals: (0, sk1], (sk1, sk2], … (sk,Jk−1, skJk], where skJk = ∞. In the jth interval, we assume a constant baseline hazard, hk(y|λk) = λkj for y ∈ (sk,j−1, skj]. Letting λk = (λk1, λk2, …λkJk)′, the cumulative baseline hazard function corresponding to hk(t) is given by
| (2.6) |
for k = 0, 1, 2.
2.2 Likelihood Function
Suppose we have n subjects. Let yi denote the observed death time or censoring time, xi is the vector of baseline covariates, Ai is the treatment indicator, yEi is the observed disease progression time, zi is the vector of covariates collected at baseline or at disease progression, and Vi is the indicator for treatment switching for the ith subject for i = 1,…, n. Also let νi be the censoring variable such that νi = 1 if yi is a death time and νi = 0 if yi is a right censoring time, and let di be the indicator variable such that di = 1 if yEi is a disease progression time and 0 if there is no disease progression for the ith individual. When di = 0, yEi is assumed to be equal to infinity. Finally, we use Ei to denote the disease progression indicator such that Ei = 1 if subject i is in the disease progression population and 0 otherwise. Let , where β2 = (β21, β22)′ and ωi is a latent frailty for the ith subject. Based on the nature of the semi-competing risks, the observations in the observed data can be classified into four different cases. Under FM, the likelihoods for these four cases are derived as follows.
Case 1
Subject died at time yi and no disease progression was observed. Then we have Ei = 0, di = 0 and νi = 1 and the observation is Di = (Ei = 0, yi, di = 0, νi = 1, xi, Ai). The likelihood function is given as follows:
| (2.5) |
Case 2
Subject was observed to have disease progression at yEi and died at yi. Then we have Ei = 1, di = 1, and νi = 1, and the observation is di = (Ei = 1, yEi, yGi = yi − yEi, di = 1, νi = 1, xi, Ai, Vi (1−Ai), zi) with the likelihood function given by:
| (2.6) |
where P(Ei = 1|Ai, xi, α) = 1 − P(Ei = 0|Ai, xi, α).
Case 3
Subject was observed to have disease progression at yEi and right censored at yi. Then we have Ei = 1, di = 1, and νi = 0, and the observation is Di = (Ei = 1, yEi, yGi = yi − yEi, di = 1, νi = 0, xi, Ai, Vi(1 − Ai), zi) with the likelihood function given by
| (2.7) |
Case 4
Subject was only observed to be right censored at yi and no disease progression occurred before yi. Then we have di = 0 and νi = 0 and the observation is Di = (yi, di = 0, νi = 0, xi, Ai) and for such a subject, it is possible that Ei = 1 or Ei = 0. The likelihood function is given by
| (2.8) |
Let Dobs = (Di, i = 1,…, n) denote the observed data, where Di is defined by (2.5) – (2.8). Then, the observed-data likelihood function under FM is given by
| (2.9) |
where 1{B} denotes the indicator function such that 1{B} = 1 if B is true and 0 otherwise, L1i(α, β0, γ0, λ0|Di) is defined by (2.5), L2i(α, β1, γ1,λ1, β2, γ2, λ2|Di) = P(Ei = 1|Ai, xi, α)[(1 + τ) h1(yEi|λ1)
The likelihood under CM can be derived in a similar way. Specifically, under CM, P(Ei = 0|Ai, xi, α), S0(yi|Ai, xi, β0, γ0,λ0), f0(yi|Ai, xi, β0, γ0,λ0), and (2.5) remain the same while (2.6) – (2.8) are obtained using
3 Posterior Inference and Computation
3.1 Prior and Posterior Distributions
Let denote the vector of all the model parameters. To carry out a Bayesian analysis, we need to specify a prior distribution for θ.We assume that α, (β0, γ0), (β1, γ1), (β21, β22, γ2), λ0, λ1, λ2 and τ are independent, a priori, and the following priors are specified for these parameters: , and τ ~ IG(aτ, bτ), which is an inverse Gamma distribution with mean bτ/(aτ −1) and variance (bτ)2/[(aτ − 1)2(aτ − 2)], where pa, p0, p1, and p2 are the dimensions corresponding to the respective vectors of the model parameters, and Σa, Σ0, Σ1, Σ2, and (aτ, bτ ) are pre-specified hyperparameters. Independently, we assume λkj ~ Gamma(akj, bkj) for j = 1,…, Jk and k = 0, 1, 2. Let πa(α), π0 (β0, γ0), π1 (β1, γ1), π2 (β2, γ2), π(τ |aτ, bτ), π0λ(λ0), π1λ (λ1), and π2λ(λ2) denote the above prior distributions, respectively. Then, the joint prior of θ is given by π(θ) ∝ πa(α)π0(β0, γ0)π1(β1, γ1)π2(β2, γ2)π(τ |aτ, bτ) π0λ(λ0) π1λ(λ1)π2λ(λ2). In the simulation study in Section 4 and the analysis of the real data from a colorectal cancer study in Section 5, these hyperparameters were specified as Σa = 1000Ipa, Σ0 = 1000Ip0, Σ1 = 1000Ip1, Σ2 = 1000Ip2, aτ = bτ = 0.01, and akj = bkj = 0 for j = 1,…, Jk and k = 0, 1, 2, where Ipa, Ip0, Ip1, and Ip2 are the identity matrices. Using (2.9) and π(θ), the posterior distribution of θ given the observed data Dobs under FM is of the form
| (3.1) |
When π(θ) is proper, the posterior distribution π(θ|Dobs) is also proper. However, even when π(θ) is improper, the posterior distribution can still be proper under certain mild conditions. To formally establish posterior propriety in this case, let 𝒩j denote the set which consists of subjects who were in Case j and nj = |𝒩j|, which is the total number of subjects in Case j for j = 1, …, 4, respectively. Write , which is an (n1 +n2 +n3)×pa matrix with rows , where . Let δi0j = 1 if yi ∈ (s0,j−1, s0j] and 0 otherwise for j = 1, 2, …J0 for i ∈ 𝒚1; δi1j = 1 if yEi ∈ (s1,j−1, s1j] and 0 otherwise for j = 1, 2, …J1 and i ∈ 𝒩2∪𝒩3; and δi2j = 1 if yi−yEi ∈ (s2,j−1, s2j] and 0 otherwise for j = 1, 2, …J2 and i ∈ 𝒩2 ∪ 𝒩3. Define X0 to be an n1 × (J0 + p0) matrix with rows for i ∈ 𝒩1, X1 an (n2 +n3) ×(J1 + p1) matrix with rows() for i ∈ 𝒩2, ∪ 𝒩3 X2 an n2 × (J2 + p3) matrix with rows for i ∈ 𝒩2 We are led to the following theorem.
Theorem 1
Assume πa (α) ∝ 1, π0 (β0, γ0) ∝ 1, π1(β1, γ1) ∝ 1, π2(β2, γ2) ∝ 1, and akj = bkj = 0 for j = 1, …, Jk and k = 0, 1, 2. If the following conditions are satisfied: (i) Xa is of full rank; (ii) there exists a positive vector c = (c1, …, cn* )′ ∈ Rn1+n2+n3, i.e., each component ci > 0, such that ; (iii) X0, X1, and X2 are of full rank; and (iv) aτ > 0 and bτ > 0, then the joint posterior π(θ|Dobs) in (3.1) is proper, i.e., ∫ L(θ|Dobs)π(θ)dθ < ∞.
The proof of Theorem 1 is given in Appendix A. When akj = bkj = 0 for j = 1, …, Jk and k = 0, 1, 2, we specify improper (Jeffreys’s) priors for all the λjk’s, namely, for j= 1,… Jk and k = 0, 1, 2. Conditions (i) and (ii) ensure posterior propriety for α, Condition (iii) leads to the posterior propriety of (λ0, β0, γ0) and Conditions (iii) and (iv) are required for the posterior propriety of (λ1, β1, γ1, λ2, β2, γ2, τ ). Condition (iii) is quite mild and essentially requires that at least one event (death or disease progression) occurs in each interval (sk,j−1, skj], and the corresponding covariate matrix is of full rank. These conditions are easily satisfied in most applications and are quite easy-to-check.
3.2 The Predictive Survival Function with Partial Treatment Switching
An inferential research goal in this research is to compare the survival function of the death time in the setting when no subjects have switched treatment. Let (a) denote a potential survival time when a subject receives treatment a at the time of randomization and stays on the same treatment over the entire study duration. Let . Following Zeng et al. (2012), we state the following two assumptions: (i) Treatment A is completely randomized and if a subject never switches treatment; and (ii) Given (A = 0, z, TE = u) or (A = 1, z, TE = u), V is independent of the potential outcomes . We note that these two assumptions are only used to compute Sa(t|θ). Similar to Zeng et al. (2012), under Assumptions (i) and (ii), we have
| (3.2) |
where fX(x | A = a) is the conditional density of X given A = a, and fZ (z | A = a, x, E = 1) is the conditional density of Z given A = a, x, and E = 1. When J0 = J1 = J2 = 1, after some algebra, we obtain
| (3.3) |
A detailed derivation of (3.3) is given in Appendix B. We assume nonparametric distributions for fX(x | A = a) and fZ(z | X, A = a, E = 1) as follows: and . Since Sa(t|θ) is a function of θ, the posterior estimates of Sa(t|θ) can be easily obtained using the MCMC samples from the posterior distribution of θ.
3.3 Posterior Computation
Due to the complexity of the likelihood structure for the proposed frailty model, an analytical evaluation of the posterior distribution is not possible. In order to carry out posterior inference, we develop an efficient Gibbs sampling algorithm to sample θ from the posterior distribution in (3.1). We first consider the transformation . The Jacobian of this transformation is for k = 1, 2. Write . After the transformation, the posterior distribution of θ* is given by
| (3.4) |
where L(α, β0, γ0, λ0, β1, γ1, λ1, β2, γ2, λ2, τ|Dobs) is defined in (2.9).
To facilitate the posterior computation, we introduce two sets of latent variables and w = (w1, w2, …, wn) so that the augmented posterior distribution of (θ*,E*,w) is given by
| (3.5) |
Where L1i(α, β0, γ0, λ0|di) is defined by (2.5), . It can be shown that ∑E* ∫ π(θ*, w, E*|Dobs)dw = π(θ*|Dobs), which is given by (3.4). We note that the latent variables (the wi’s) in (3.5) are different than those ωi’s in (2.6) – (2.8).
Let [A|B] denote the conditional distribution of A given B. To run the Gibbs sampling algorithm, we sample from the following conditional distributions in turn: (i) ; (ii) ; and (iii) [α|E*,Dobs]. For (ii), we use the modified collapsed Gibbs technique (Liu, 1994; Chen et al., 2000). It is easy to show that
| (3.6) |
For (ii), following Chen et al. (2000) and using (3.6), we run a sub-Gibbs sampling algorithm to draw from the following conditional distributions: (iia) ; (iib) ; (iic) ; and (iid) . Next, we will only discuss the properties of the conditional distribution and how to sample τ from this conditional distribution. All other conditional distributions are discussed in detail in Appendix B. We first consider the transformation τ* = 1/τ . Then, the conditional posterior density of τ* is given by
| (3.7) |
We are led to the following theorem.
Theorem 2
Assume that Then we have (i) the conditional density of τ* given by (3.7) is log-concave; and (ii) the mode of (3.7) is analytically available and given by
| (3.8) |
where
The proof of Theorem 2 is given in Appendix A. The assumption ensures the log-concavity and the existence of the mode. This assumption is quite mild. As long as there are more than three patients with disease progression, this assumption still holds even when the improper priors with aτ = 0 and akj = 0 for all k and j are specified for τ and the λjk’s. With the log-concavity property, τ* can be exactly drawn from the conditional distribution in (3.7) using the adaptive rejection algorithm of Gilks and Wild (1992). After τ* is generated, we let τ = 1/τ* and then the value of τ is a sample from the conditional distribution in (iic). With the analytical form of the mode, the performance of the rejection algorithm can be improved substantially as the algorithm does not need to search for the mode.
3.4 Model Comparison
To carry out Bayesian model comparison, we consider the deviance information criterion (DIC) and the Logarithm of the PseudoMarginal Likelihood (LPML).We define the deviance Dev(θ) = −2 log L(θ|Dobs), where L(θ|Dobs) is the observed-data likelihood defined in (2.9). Let θ̄ and denote the posterior mean of θ and Dev(θ), respectively. According to Spiegelhalter et al. (2002), the DIC measure is defined as DIC = Dev(θ̄) + 2pD, where is the effective number of model parameters. The smaller the DIC value, the better the model fits the data. LPML is another useful Bayesian measure of goodness-of-fit statistic, which is defined based on the Conditional Predictive Ordinate (CPO). For the ith observation, we define CPO as CPOi = ∫ L(θ|Di)π(θ|D(−i))dθ, where Di is the observed data defined in Section 2.2, L(θ|Di) is the observed likelihood for the ith subject, which is the term inside the product in (2.9), D(−i) is the data with Di deleted, and π(θ|D(−i)) is the posterior density of θ based on the data D(−i). According to Ibrahim et al. (2001), log (CPOi). The larger the LPML value, the better the model fits the data.
4 A Simulation Study
To examine the empirical performance of the posterior estimates and DIC and LPML, we carry out a simulation study. Five hundred simulated data sets with n = 500 as well as n = 1, 000 were generated. In the simulation study, the baseline treatment A was generated from a Bernoulli(0.5), corresponding to a randomized trial with a 1:1 sample size allocation; two baseline covariates X1 and X2 were independently generated from a U(−1, 1) and a Bernoulli(0.6), respectively. Given A and (X1, X2), E was generated from model (2.1) with the coefficients (including an intercept) being 1.6, −1.8, 1, and 0.1, respectively. When E = 0, we simulated TD from model (2.2) with H0(t) = t, β0 = −1 and (γ01, γ02) = (1, 0.2). For E = 1, we first generated ω from a Gamma(1/τ, 1/τ ) with τ = 1. Then, TE was generated from model (2.3) with H1(t) = 5t, β1 = −0.5 and (γ11, γ12) = (1, 0) and an additional prognostic factor Z at disease progression was generated from a U(0, 10) while the selection into treatment switching (V ) for a subject in the control arm (A = 0) was from a Bernoulli(p), where p = exp(−0.5 + 0.3TE + 0.2X1 + 0.5Z)/[1+exp(−0.5+0.3TE +0.2X1 +0.5Z)]. Moreover, TG was generated from the model in (2.3) with H2(t) = t, β21 = −0.3, β22 = −0.5, and γ21 = −0.5, γ22 = 0.5, γ23 = −0.4. Finally, the censoring time was generated from a U(1, 7) and the study duration was τ* = 3. The latter yielded the average proportions of Cases 1 to 4 as 23%, 39%, 19%, and 18%.
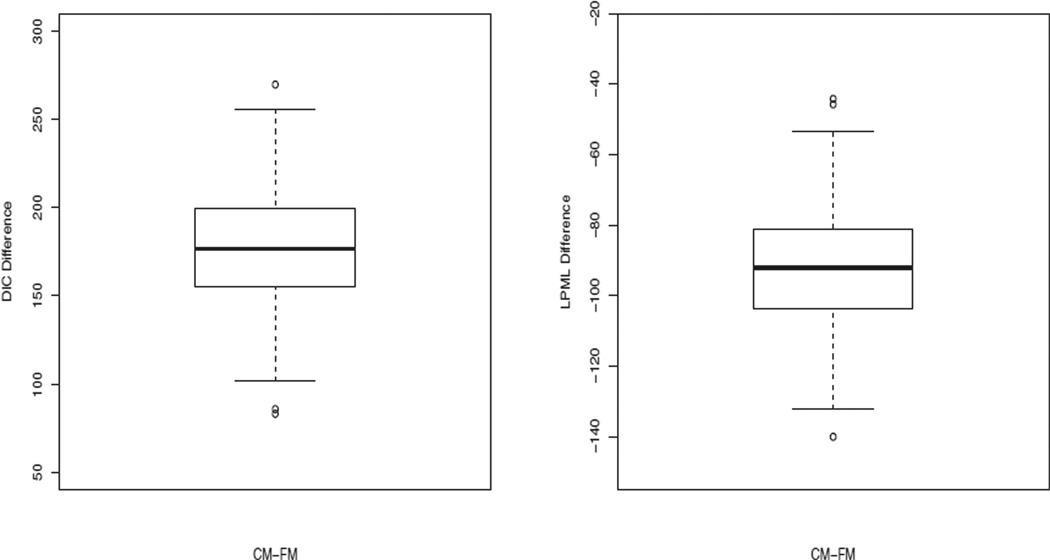
For each simulated dataset, we fit the proposed FM with various values of (J0, J1, J2) and computed DIC and LPML. The mean values of the DICs and LPMLs over the 500 simulated datasets were 2986.22 and −1493.24 for (J0, J1, J2) = (1, 1, 1); 2998.05 and −1499.39 for (J0, J1, J2) = (5, 5, 5); and 3013.21 and −1507.22 for (J0, J1, J2) = (10, 10, 10). We note that the true value of (J0, J1, J2) is (1, 1, 1). Thus, both DIC and LPML correctly identified the true model. Under the best combination of (J0, J1, J2), namely, (1, 1, 1), the average of the posterior means (EST), and the average of the posterior standard deviations (SD), the simulation standard error (SE), the root of the mean squared error (RMSE), and the coverage probability (CP) of the 95% highest posterior density (HPD) intervals for each parameter as well as Sa(t|θ) were computed. The results are given in Table 1. Table 1 shows excellent empirical performance of the posterior estimates for all the parameters as well as the survival probabilities for both n = 500 and n = 1000. In particular, the ESTs are nearly identical to the true values, the SDs are very close to the SEs, and the CPs are very close to 95%. For each simulated dataset, we also fit CM as discussed in Section 2.1 and computed the corresponding DIC and LPML for (J0, J1, J2) = (1, 1, 1). The box plots of the DIC and LPML differences between CM and FM are shown in Fig. 2. From this figure, we see that all of the DIC differences are above 0 and all LPML differences are below 0, indicating that the frailty model fits the data better than the conditional model for all 500 simulated data sets, which is expected since the data were generated from the frailty model. These results further empirically confirm that FM is indeed quite different from CM, and DIC and LPML are two effective Bayesian model comparison measures for identifying the true models.
Table 1.
Posterior estimates under FM in the simulation study
| n = 500 | n = 1,000 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | True | EST | SD | SE | RMSE | CP% | EST | SD | SE | RMSE | CP% |
|
TD model |
|||||||||||
| β0 | −1.0 | −1.00 | 0.25 | 0.24 | 0.24 | 0.95 | −1.01 | 0.18 | 0.18 | 0.18 | 0.95 |
| γ01 | 1.0 | 0.99 | 0.20 | 0.19 | 0.19 | 0.95 | 1.00 | 0.14 | 0.14 | 0.14 | 0.96 |
| γ02 | 0.2 | 0.21 | 0.23 | 0.22 | 0.22 | 0.96 | 0.21 | 0.16 | 0.15 | 0.15 | 0.96 |
|
TE model |
|||||||||||
| β1 | −0.5 | −0.53 | 0.22 | 0.22 | 0.22 | 0.94 | −0.53 | 0.16 | 0.15 | 0.15 | 0.95 |
| γ11 | 1.0 | 1.02 | 0.19 | 0.20 | 0.20 | 0.93 | 1.01 | 0.13 | 0.14 | 0.14 | 0.95 |
| γ12 | 0.0 | 0.01 | 0.21 | 0.20 | 0.20 | 0.95 | 0.00 | 0.15 | 0.15 | 0.15 | 0.95 |
|
TG model |
|||||||||||
| β21 | −0.3 | −0.31 | 0.24 | 0.23 | 0.23 | 0.96 | −0.31 | 0.17 | 0.17 | 0.17 | 0.95 |
| β22 | −0.5 | −0.49 | 0.23 | 0.23 | 0.23 | 0.95 | −0.50 | 0.16 | 0.16 | 0.16 | 0.93 |
| τ | 1.0 | 1.05 | 0.15 | 0.14 | 0.14 | 0.95 | 1.03 | 0.10 | 0.10 | 0.10 | 0.95 |
| γ22 | −0.5 | −0.49 | 0.18 | 0.19 | 0.19 | 0.95 | −0.49 | 0.13 | 0.13 | 0.13 | 0.94 |
| γ23 | 0.5 | 0.51 | 0.21 | 0.21 | 0.21 | 0.95 | 0.50 | 0.15 | 0.16 | 0.16 | 0.92 |
| γ24 | −0.4 | −0.40 | 0.32 | 0.32 | 0.32 | 0.94 | −0.40 | 0.22 | 0.23 | 0.23 | 0.95 |
|
E
model |
|||||||||||
| α0 | 1.6 | 1.63 | 0.24 | 0.24 | 0.24 | 0.95 | 1.61 | 0.17 | 0.16 | 0.16 | 0.97 |
| α1 | −1.8 | −1.81 | 0.25 | 0.26 | 0.26 | 0.94 | −1.80 | 0.18 | 0.18 | 0.18 | 0.95 |
| α21 | 1.0 | 1.01 | 0.22 | 0.24 | 0.24 | 0.92 | 1.01 | 0.15 | 0.16 | 0.16 | 0.95 |
| α22 | 0.1 | 0.11 | 0.24 | 0.25 | 0.25 | 0.94 | 0.12 | 0.17 | 0.17 | 0.17 | 0.94 |
| Estimated Survival function
of control arm |
|||||||||||
| S0(τ*/2) | 0.44 | 0.44 | 0.03 | 0.03 | 0.03 | 0.95 | 0.44 | 0.02 | 0.02 | 0.02 | 0.96 |
| S0(τ*) | 0.26 | 0.28 | 0.03 | 0.03 | 0.03 | 0.93 | 0.27 | 0.02 | 0.02 | 0.02 | 0.95 |
| Estimated Survival function
of treatment arm |
|||||||||||
| S1(τ*/2) | 0.57 | 0.57 | 0.03 | 0.03 | 0.03 | 0.95 | 0.57 | 0.02 | 0.02 | 0.02 | 0.93 |
| S1(τ*) | 0.36 | 0.37 | 0.03 | 0.03 | 0.03 | 0.93 | 0.37 | 0.02 | 0.02 | 0.02 | 0.91 |
Fig. 2.
Box Plots of the DIC and LPML Differences between CM and FM.
5 Analysis of the Panitumumab Study
We carry out here a detailed analysis of a subset of the data from the panitumumab study (PMAB408) conducted by Amgen Inc. (van Cutsem et al., 2007 and Amado et al., 2008). PMAB408 was an open label, randomized, phase III multicenter study designed to compare the efficacy and safety of panitumumab plus best supportive care (P+BSC) versus BSC alone in subjects with EGFr-expressing metastatic colorectal cancer who had documented disease progression during or after prior standard treatment with fluoropyrimidine, irinotecan, and oxaliplatin chemotherapy. Subjects were randomly assigned to receive P+BSC (treatment) or BSC (control). The baseline covariates include initial treatment (P+BSC versus BSC), age in years at screening, baseline Eastern Cooperative Oncology Group (ECOG) score (score 0 or 1 versus ≥ 2 (bECOG01)), primary tumor diagnosis type (rectal versus colon (Rectal)), gender, and region (western Europe (WesternEU), eastern and central Europe (CenEstEU), and rest of the world). In the subset of the data, there were 223 and 231 patients in the control and treatment arms, respectively. There were 424 subjects who died (208 and 207 in the control and treatment arms, respectively), 387 subjects (201 and 186 in the control and treatment arms, respectively) who developed disease progression, and 59 subjects (18 and 41 in the control and treatment arms, respectively) who died without disease progression. The median age was 62.5 years with interquartile range (55, 69) years. There were 388 patients with ECOG score 0 or 1, 287 were males, 151 had rectal cancer, 352 were from Western Europe, 39 were from Eastern and Central Europe, and 63 were from the rest of the world. The median follow-up time was 189.5 days and the interquartile range of the follow-up time was (93, 334) days. Among those 387 patients who developed disease progression, the median disease progression time is 53 days and the interquartile range is (45, 84) days. Of these 201 patients who developed disease progression in the control arm, 167 patients were switched to the treatment arm at the time of disease progression.
The model for the time in months to disease progression includes all the baseline covariates. Among the 387 patients who developed disease progression, the median age at the time of disease progression was 62.1 years with interquartile range (55.0, 69.1), the numbers of patients who had partial response, stable disease, and progressive disease were 19, 86, and 282, respectively. There were 348 patients with baseline ECOG score 0 or 1, 286 patients had a last ECOG score on or prior to disease progression 0 or 1, and 180 patients had grade 2 or above adverse events. The covariates for the time in months from disease progression to death include treatment, bECOG01, age at disease progression, best tumor response with partial response (BTR PR) or stable disease (BTR SD) versus progressive disease according to investigator assessment, last ECOG score on or prior to disease progression (score 0 or 1 versus ≥ 2 (LECOG01)), and adverse events (AE).
We fit both FM and CM with different values of J0, J1 and J2 to the panitumumab data. The DIC and LPML values are given in Table 2. We see from Table 2 that (J0, J1, J2) = (1, 30, 5) achieves the smallest DIC value and the largest LPML value among the 7 combinations of (J0, J1, J2) considered here under both FM and CM and the best DIC and LPML values were 3475.27 and −1741.32 under FM and 3482.62 and −1746.76 under CM, respectively. We also observe that for each of these seven combinations of (J0, J1, J2), FM consistently has a smaller DIC value and a larger LPML value than CM, implying that FM fits the panitumumab data better than CM.
Table 2.
DIC and LPML Values for the Panitumumab Data
| Parameter | FM | CM | ||||||
|---|---|---|---|---|---|---|---|---|
| J0 | J1 | J2 | DIC | pD | LPML | DIC | pD | LPML |
| 1 | 30 | 5 | 3475.27 | 67.13 | −1741.32 | 3482.64 | 67.32 | −1746.76 |
| 3 | 30 | 5 | 3479.02 | 69.20 | −1742.90 | 3486.14 | 69.48 | −1748.76 |
| 5 | 30 | 5 | 3480.09 | 71.24 | −1743.92 | 3486.92 | 71.44 | −1748.94 |
| 1 | 25 | 5 | 3493.38 | 61.83 | −1749.55 | 3500.00 | 62.20 | −1755.11 |
| 1 | 35 | 5 | 3477.89 | 72.31 | −1743.16 | 3484.61 | 72.54 | −1748.62 |
| 1 | 30 | 3 | 3484.99 | 65.15 | −1746.02 | 3488.20 | 65.37 | −1749.05 |
| 1 | 30 | 10 | 3481.34 | 72.23 | −1744.74 | 3490.95 | 72.46 | −1751.73 |
Table 3 shows the posterior estimates of the model parameters under FM with (J0, J1, J2) = (1, 30, 5). The 95% HPD intervals for treatment were (−1.753, −0.484) under the E model, (−1.145, 0.175) under the TD model, (−1.733, −1.148) under the TE model, and (−1.479, −0.441) under the TG model.
Table 3.
Posterior Estimates for the Panitumumab Data under FM with (J0, J1, J2) = (1, 30, 5)
| Parameter | EST | SD | 95% HPD | Parameter | EST | SD | 95% HPD |
|---|---|---|---|---|---|---|---|
|
E
Model |
TD Model |
||||||
| Intercept | 1.512 | 0.989 | (−0.438, 3.463) | ||||
| Treatment | −1.115 | 0.324 | (−1.753, −0.484) | Treatment | −0.481 | 0.337 | (−1.145, 0.175) |
| Age | −0.010 | 0.014 | (−0.039, 0.017) | Age | 0.023 | 0.015 | (−0.008, 0.052) |
| bECOG01 | 1.987 | 0.346 | (1.336, 2.699) | bECOG01 | −0.617 | 0.296 | (−1.202, −0.034) |
| Rectal | 0.303 | 0.337 | (−0.342, 0.967) | Rectal | −0.060 | 0.319 | (−0.696, 0.546) |
| Male | −0.317 | 0.330 | (−0.944, 0.342) | Male | −0.326 | 0.306 | (−0.923, 0.279) |
| CenEastEU | 0.001 | 0.628 | (−1.171, 1.301) | CenEastEU | −0.308 | 0.638 | (−1.545, 0.951) |
| WesternEU | 0.334 | 0.422 | (−0.498, 1.149) | WesternEU | 0.231 | 0.395 | (−0.546, 0.991) |
|
TE Model |
TG Model |
||||||
| Treatment | −1.443 | 0.150 | (−1.733, −1.148) | Treatment | −0.975 | 0.265 | (−1.479, −0.441) |
| Age | −0.019 | 0.006 | (−0.031, −0.007) | V*(1-Treatment) | −1.475 | 0.256 | (−1.968, −0.962) |
| bECOG01 | −0.869 | 0.220 | (−1.297, −0.431) | PR Age | −0.007 | 0.007 | (−0.020, 0.006) |
| Rectal | −0.112 | 0.133 | (−0.379, 0.138) | BTR PR | −0.254 | 0.347 | (−0.942, 0.413) |
| Male | −0.129 | 0.132 | (−0.386, 0.131) | BTR SD | −0.088 | 0.192 | (−0.460, 0.296) |
| CenEastEU | 0.190 | 0.274 | (−0.351, 0.727) | bECOG01 | −0.445 | 0.242 | (−0.908, 0.034) |
| WesternEU | −0.074 | 0.188 | (−0.446, 0.286) | LECOG01 | −1.186 | 0.177 | (−1.522, −0.829) |
| AE | 0.348 | 0.141 | (0.082, 0.631) | ||||
| τ | 0.322 | 0.083 | (0.163, 0.490) | ||||
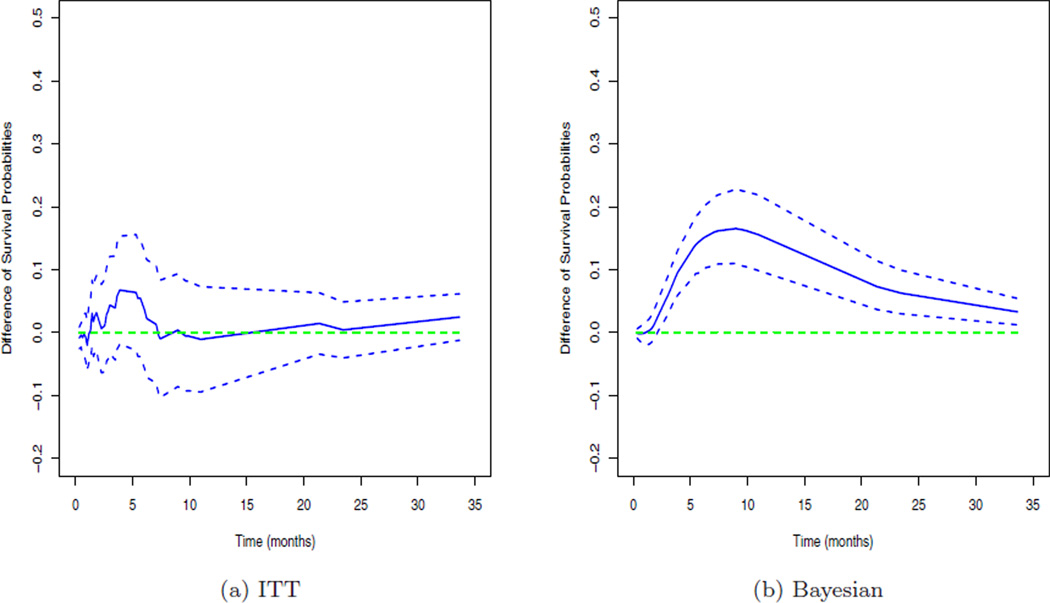
These results imply that treatment is associated with E, TE, TG but not with TD. The other important prognostic factors include bCOG01 under the E, TD, and TE model, LECOG01 under the TG model, age under the TE model, and AE under the TG model as their corresponding 95% HPD intervals do not contain 0. The treatment switching variable, V, is also associated with TG. The posterior mean and 95% HPD interval for τ were 0.322 and (0.163, 0.490), which implies that there is a moderate dependence between TE and TG. We also fit the best CM with (J0, J1, J2) = (1, 30, 5) to the panitumumab data and the posterior mean and 95% HPD interval for γ21 in (2.4) were −0.083 and (−0.161, −0.008), which implies that there is a positive association between TE and TG. Panel (a) in Figure 3 shows the estimated differences of the survival probabilities and their pointwise 95% confidence intervals (CIs) between the two treatment groups of P+BSC and BSC using the intent-to-treat (ITT) Kaplan-Meier approach and Panel (b) plots the posterior estimates, E[S1(t|θ)−S0(t|θ)|Dobs], where S0(t|θ) and S1(t|θ) are given in (3.2), and the corresponding pointwise 95% HPD intervals of S1(t|θ) − S0(t|θ) between these two treatment groups. From Panel (a) of Figure 3, we see that the ITT approach yields no difference between two treatment groups as all 95% CIs contain 0. In contrast, as shown in Panel (b) of Figure 3, all the posterior estimates of S1(t|θ) − S0(t|θ) are above 0 and the corresponding 95% HPD intervals are above 0 after 2.25 months. We note that the maximum estimated difference E[S1(t|θ) − S0(t|θ)|Dobs] was attained at approximately 9 months and the corresponding posterior mean and 95% HPD interval were 0.165 and (0.110, 0.227). These posterior estimates indicate that P+BSC does yield a higher survival probability than BSC.
Fig. 3.
The estimated differences with 95% intervals of the survival curves between the treatment and control arms.
In all of the Bayesian computations, we used 20,000 Gibbs samples after a burn-in of 1000 for each model to compute all the posterior estimates, including posterior means, posterior standard deviations, and 95% HPD intervals. Codes were written for the FORTRAN 95 compiler using IMSL subroutines with double precision accuracy. The convergence of the Gibbs sampler was checked using several diagnostic procedures discussed in Chen et al. (2000). The autocorrelations for all model parameters disappeared before lag 10.
6 Discussion
In this paper, we have proposed a novel semi-competing risks Bayesian frailty model that accommodates treatment switching and dependence between the progression time and survival time. This type of scenario arises often in clinical trials in which, once a patient experiences an event, such as progression, they immediately switch to the experimental treatment. As a result of the switch, the model attempts to capture the treatment effect when no subjects would have switched treatment. The innovation in the model lies in the fact that the observed data likelihood is modeled and is based on four possible scenarios, and the model itself has three components. This type of model is quite different from what has been proposed in the literature. Another innovation here lies in the Bayesian approach to fit the model. Efficient MCMC methods based on the collapsed Gibbs sampler facilitate a flexible Bayesian model that is computationally feasible and identifiable. Such a model does not appear computationally feasible from a frequentist perspective. As shown in the simulation studies and real data analysis, our proposed model has several advantages over the conditional model (CM) proposed by others in the literature. It appears to have better performance under certain scenarios and produces a better model fit according to DIC and LPML. The proposed model is useful for practitioners encountering treatment switching studies in the presence of semi-competing risks where one is interested in assessing the treatment effect on overall survival.
Acknowledgements
The authors wish to thank the Editor-in-Chief, the Associate Editor, and the referee for their helpful comments and suggestions, which have led to an improved version of this article. This research was partially supported by NIH grants #GM 70335 and #CA 74015.
Appendix A: Proofs of Theorems
Proof of Theorem 1
Using (3.1) with the prior distributions assumed in the theorem, we have
Using (2.9), it is easy to show that
where ,
| (A.1) |
| (A.2) |
and
| (A.3) |
To prove propriety of the posterior, it is sufficient to show (a) ∫ La(α|Dobs)dα < ∞; (b) and (c) . dβ1dγ1dλ1dβ21dβ22dγ2dλ2dτ < ∞.
Under Conditions (i) and (ii), Theorem 2.1 of Chen and Shao (2001) directly leads to (a). Let ji be an index such that s0,ji−1 < yi ≤ s0ji. Then we have δi0j = 1 for j = ji and δi0j = 0 for j ≠ ji and
| (A.4) |
where M1 > 0 is a constant. Consider the transformation ξ0j = log(λ0j), where for j = 1, …, J0. Under condition (iii), there exist J0 + p0 distinct i1, …, iJ0+p0 ∈ 𝒩1 such that the (J0 + p0) × (J0+p0) matrix , which has rows ) for ℓ = 1, …, J0+p0, is of full rank. Using (A.4), we have
| (A.5) |
where M12 > 0 is a constant and ξ0 = (ξ01, …, ξ0J0 )′. Now, we take a one-to-one transformation . Using (A.5), we have
| (A.6) |
where M13 > 0 is a constant, which completes the proof of (b).
For (c), we first rewrite (A.2) as follows:
where
| (A.7) |
| (A.8) |
and . Let ji denote the index such that s2,ji−1 < yGi ≤ s2,ji. Then, we have
| (A.9) |
Observing that for all r > 0 and v > 0, we obtain
| (A.10) |
Under condition (iii), X2 is of full rank. Therefore, there exist J2 + p2 distinct i1, …, iJ2+p2 ∈ 𝒩2 such that the (J2 +p2)×(J2 +p2) matrix , which has rows () for ℓ = 1, …, J2 + p2, is of full rank. Let 𝒩21 = {i1, …, iJ2+p2} and 𝒩22 = 𝒩2 − 𝒩21. Using (A.10), we have
| (A.11) |
where M21 > 0 is a constant. Similar to (A.5) and (A.6), using (A.11), we can show that
| (A.12) |
where M22 and M23 are two positive constants. Now, using (A.2), (A.3), (A.7), and (A.12), we obtain
| (A.13) |
The right hand side of (A.13) is precisely the kernel of the posterior distribution of (β1, γ1, λ1, τ ) under the GORH model of Banerjee et al. (2007). Thus, under conditions (iii) and (iv), following the proof of Theorem 3.1 of Banerjee et al. (2007), we can show that the integration of the right hand side of (A.13) over (β1, γ1,λ1, τ) is finite, which completes the proof of Theorem 1.
Proof of Theorem 2
For the posterior conditional distribution, the first derivative of the log-likelihood function is given by . The second derivative is given by .
Assuming that , we will always have . Therefore, the conditional density of τ* given by (3.7) is log-concave. Letting , then we have , where B1, B2 and B3 are defined in Theorem 2. After some algebra, the solution is given by . The reasons are as follows: with bτ > 0, bkj > 0 and , it is obvious that B1 < 0; and with the previous assumption that , then B2 > 0, so we have (B1 + B2 +B3)2 −4B1B2 > 0. Therefore, the equation has two roots. Since τ* > 0, then we only keep the positive solution τ̂* since the other root is negative. Since we just showed that the conditional density of τ* given by (3.7) is log-concave, it follows that the mode of (3.7) is analytically available and given by τ̂*.
Appendix B: Computational Development
B.1. Derivation of the Potential Survival Function
After some algebra, we obtain
When J0 = J1 = J2 = 1, we obtain P(TD > t|A = a, x, E = 0, β0, γ0, λ0) = exp{−tλ0 exp(Aβ0 + x′γ0)}, ∫ω P(TE>t | A = a, x, E = 1, β1, γ1, λ1, ω) f (ω | τ)dω = ∫ω exp[−ωt λ1 exp(Aβ1 + x′γ1)]f(ω|τ) , and .
For the more general case, where the values of J0, J1 and J2 are not specified, we have
Before the next derivation, we need to align the partitions of the time axis for h1 and h2. Let 0 < s3,1 < s3,2 < … s3, J3 be the ordered distinct values of sk, Jk, where k = 1, 2. For a given time point t, there exists jt such that t ∈ (s3, jt−1, s3,jt). In order to facilitate the computation, let s3, J3+1 = s3, J3, s3, J3 = s3, J3−1, …, s3, jt = t. Then the corresponding constant hazards for each interval are as follows: λ3kj = λkl if sk,l−1 ≤ s3, j−1 < s3, j ≤ skl for j ∈ (1, J3 + 1) and l ∈ (1, Jk), where k = 1, 2. Next let
and
While u ∈ (s3,j−1, s3,j], we have (t − u) ∈ (t − s3,j, t − s3,j−1], then there exist jl and ju such that (t − s3,j) ∈ (s3,jl−1, s3,jl] and (t − s3,j−1) ∈ (s3,ju−1, s3,ju]. Let r = ju − jl. Then the range of r is from 0 to J3. Therefore, when r = 0, we have
Then
When r ≥ 1, we have
for which
B.2. Sampling from the Conditional Posterior Distributions
In the posterior computation section, a series of conditional posterior distributions are listed. Now we will show how to sample from these distributions.
-
(i)
. It is easy to see that conditional on (β0, γ0, β1, γ1, β2, γ2,w, E*, Dobs), () are independent. Therefore, the conditional distributions can be sampled separately. Let δikj = 1 if the ith subject failed or was censored in the jth interval for j = 1, 2, …Jk and 0 otherwise. It can be shown that (ia) [λ0j |β0, γ0, E*, Dobs] ~ Gamma(), where , and exp exp ; (ib) ; and (ic) ~ Gamma (), where and exp .
-
(ii)
[].
-
(iia)
[]. From the joint posterior distribution, it is obvious that conditional on [], the parameters [β0, γ0], [β1, γ1], [β2, γ2] are independent. Therefore, we can sample the following conditional posterior distributions separately.
-
(iiaa)[β0, γ0|λ0, E*, Dobs]. This density is proportional to
It is easy to show that this conditional distribution is log-concave in each component of β0 and γ0.
-
(iiab)[]. This density is proportional to
It can also be shown that this conditional distribution is log-concave in each component of β1 and γ1.
-
(iiac)[]. This density is proportional to
Similarly, this conditional distribution can be shown to be log-concave in each component of β2 and γ2.
-
(iiaa)
-
(iib)[]. This conditional posterior distribution is given by
-
(iic)
[]. This one is shown in Theorem 2.
-
(iid)
. To sample from this conditional distribution, we can sample instead. It can be shown that [] ~ Gamma(aw, bw), where
-
(iii)[α|E*, Dobs]. This conditional posterior distribution is given by
It is easy to show that this density is log-concave in each component of α. Therefore, we apply the adaptive rejection algorithm of Gilks and Wild (1992) to draw α.
Contributor Information
Yuanye Zhang, Novartis Institutes for BioMedical Research, Inc., 220 Massachusetts Avenue, Cambridge, MA 02139.
Ming-Hui Chen, Email: ming-hui.chen@uconn.edu, Department of Statistics, University of Connecticut, 215 Glenbrook Road, U-4120, Storrs, CT 06269.
Joseph G. Ibrahim, Department of Biostatistics, University of North Carolina, Chapel Hill, NC 27599
Donglin Zeng, Department of Biostatistics, University of North Carolina, Chapel Hill, NC 27599.
Qingxia Chen, Department of Biostatistics, Vanderbilt University, Nashville, TN 37232.
Zhiying Pan, Amgen Inc., One Amgen Center Drive, Thousand Oaks, CA 91320.
Xiaodong Xue, Amgen Inc., One Amgen Center Drive, Thousand Oaks, CA 91320.
References
- Aalen OO, Johansen S, Johansen S. An empirical transition matrix for non-homogeneous Markov chains based on censored observations. Scandinavian Journal of Statistics. 1978;5:141–150. [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. Journal of Clinical Oncology. 2008;28:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- Andersen PK, Borgan O, Gill RD, Keiding N. Statistical Models Based on Counting Processes. New York: Springer; 1993. [Google Scholar]
- Banerjee T, Chen MH, Dey DK, Kim S. Bayesian analysis of generalized odds-rate hazards models for survival data. Lifetime Data Analysis. 2007;13:241–260. doi: 10.1007/s10985-007-9035-3. [DOI] [PubMed] [Google Scholar]
- Chen MH, Shao QM. Propriety of posterior distribution for dichotomous quantal response models with general link functions. Proceedings of the American Mathematical Society. 2001;129:293–302. [Google Scholar]
- Chen MH, Shao QM, Ibrahim JG. Monte Carlo Methods in Bayesian Computation. New York: Springer; 2000. [Google Scholar]
- Day R, Bryant J. Adaptation of bivariate frailty models for prediction, with application to bio- logical markers as prognostic indicators. Biometrika. 1997;84:45–56. [Google Scholar]
- Fine JP, Jiang H, Chappell R. On semi-competing risks data. Biometrika. 2001;88:907–919. [Google Scholar]
- Gilks WR, Wild P. Adaptive rejection sampling for Gibbs sampling. Appl Stat. 1992;41:337–348. [Google Scholar]
- Ghosh D. Semiparametric inferences for association with semi-competing risks data. Statistics in Medicine. 2006;25:2059–2070. doi: 10.1002/sim.2327. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG, Chen MH, Sinha D. Bayesian Survival Analysis. New York: Springer; 2001. [Google Scholar]
- Liu JS. The collapsed Gibbs sampler in Bayesian computations with applications to a gene regulation problem. Journal of the American Statistical Association. 1994;89:958–966. [Google Scholar]
- Mandel M. The competing risks illnessCdeath model under cross-sectional sampling. Biostatistics. 2010;11:290–303. doi: 10.1093/biostatistics/kxp048. [DOI] [PubMed] [Google Scholar]
- Marcus SM, Gibbons RD. Estimating the efficacy of receiving treatment in randomized clinical trials with noncompliance. Health Services & Outcomes Research Methodology. 2001;2:247–258. [Google Scholar]
- Oakes D. Bivariate survival models induced by frailties. Journal of the American Statistical Association. 1989;84:487–493. [Google Scholar]
- Peng L, Fine JP. Regression Modeling of Semicompeting Risks Data. Biometrics. 2007;63:96–108. doi: 10.1111/j.1541-0420.2006.00621.x. [DOI] [PubMed] [Google Scholar]
- Shen Y, Thall PF. Parametric likelihoods for multiple non-fatal competing risks and death. Statistics in Medicine. 1998;17:999–1015. doi: 10.1002/(sici)1097-0258(19980515)17:9<999::aid-sim785>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with Discussion) Journal of the Royal Statistical Society, Series B. 2002;64:583–639. [Google Scholar]
- Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyn B, Canon JL, Van Laethem JL, Maurel J, Richardson G, Wolf M, Amado RG. Open-label Phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. Journal of Clinical Oncology. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- Wang W. Estimating the association parameter for copula models under dependent censoring. Journal of the Royal Statistical Society, Series B. 2003;65:257–273. [Google Scholar]
- Zeng D, Chen Q, Chen MH, Ibrahim JG Amgen research group. Estimating treatment effects with treatment switching via semi-competing risks models: An application to a colorectal cancer study. Biometrika. 2012;99:167–184. doi: 10.1093/biomet/asr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. Unpublished Ph.D dissertation. Simon Fraser University; 2009. Multi-state Processes with Duration-dependent Transition Intensities: Statistical Methods and Applications. [Google Scholar]