Abstract
Nuclear receptors (NRs) encompass a family of regulatory proteins that directly couple small-molecule signalling to transcriptional regulation. Initial studies of specific NR targets led to a model in which NRs bind highly specific DNA motifs in proximal promoter regions and strongly induce gene transcription in response to ligand binding. More recently, genome-wide studies have added to the complexity of this classic model of NR function. In particular, binding of NRs at weaker or alternate motifs is common in the context of DNA assembled into chromatin, and ligand responsiveness varies at different NR target genes. Such findings have led to proposed modifications to the classic view of NR regulation, including the “assisted loading” model in which NRs assist in opening chromatin rather than compete for binding sites, and context-specific models in which genomic and epigenomic features influence the NR function locally at each binding site. Further elucidati on of these mechanisms will be particularly important for understanding cell-specific and ligand-specific functions of each NR. Emerging genomic technologies such as ChIP-seq and GRO-seq provide insights on a larger scale leading to deeper understanding of the complexities of transcriptional regulation by NRs.
Nuclear receptors (NRs) encompass a superfamily of transcription factors (TFs), numbering 48 in humans, that bind and respond directly to hormonal, metabolic, developmental, and environmental signals1,2. NRs have proven to be tractable targets in the treatment of a variety of human diseases, ranging from hormone deficiency to diabetes and cancer1,2. Moreover, the NR superfamily provides an excellent paradigm for linking extracellular signals to changes in gene expression1,3. Basic NR structure is conserved across all metazoans, and includes both a DNA-binding domain (DBD) for direct interaction with the genome and a ligand-binding domain (LBD) that allows modulation of activity by small molecules1,2. The mechanisms of interaction with DNA, ligand molecules, and other proteins have all been studied at the atomic level for many NRs2, thereby guiding our understanding of this broad and critical component of the transcriptional regulatory network. Decades of research linking NRs to their endogenous and pharmacological ligands have produced a powerful toolkit for studying these regulators in model systems, often in combination with genetic manipulations.
The “classic model” of NR function describes NRs as simple gene regulatory switches that go from “off” in the absence of ligand to “on” in the presence of ligand due to an allosteric change in the NR protein1,3. In the case of a few steroid receptors, most notably the glucocorticoid receptor (GR), ligand binding allows the NR to enter the nucleus and bind DNA as a homodimer3, although most other NRs are constitutively nuclear and bind to DNA as heterodimers with the retinoid X receptor (RXR) subfamily of NRs1,2. Under the classic model, NRs bind specific sequences proximal to the promoters of target genes and enhance transcription in response to ligand3. However, NRs have also been found to bind distally from target gene promoters with less sequence specificity than originally proposed, often facilitated by indirect recruitment through other TFs4. Genome-wide studies, reviewed below, have demonstrated that these trends are widespread among the NR binding patterns studied to date.
The canonical model of NR function postulates that ligand-bound NRs recruit co-activator proteins to induce transcription, while ligand-free NRs either recruit co-repressor proteins or exit the nucleus5,6 (Figure 1). This has proven to be only one of many regulatory paradigms followed by NRs, and it is clear that even a single NR subtype can enact many complex regulatory programs. Co-repressors and co-activators, collectively called co-regulators, are associated with chromatin-modifying enzyme activities by which the ligand-dependent co-regulator switch remodels surrounding chromatin to regulate transcription by ligand- and target-specific mechanisms6,7. Thus, predicting the effect of a particular ligand treatment on the transcriptome has proven to be quite complicated. This complexity currently presents a major barrier to developing more precise drugs with fewer side effects2. Increasingly powerful genomic methods promise to drive new discoveries in the NR field, identifying novel mechanisms of NR action, and ultimately creating resources for teasing apart the network of NR regulation in a systems biology framework.
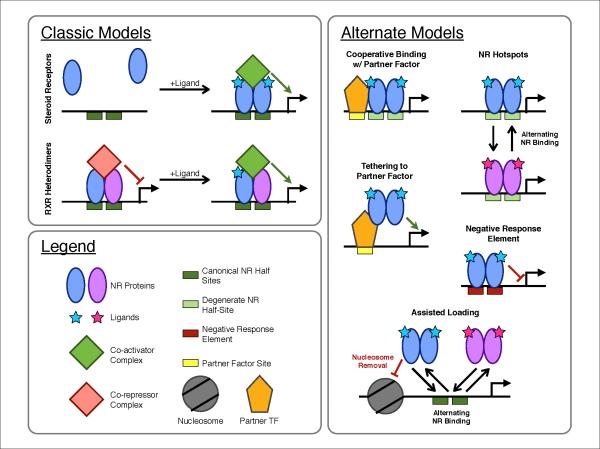
Figure 1. Models of genomic regulation by NRs.
Canonical models are based primarily on targeted studies of specific NRs and target genes. Alternate models explain genomic evidence contradicting the canonical models, and may apply only in specific cells or at specific target genes, depending on the NR.
STUDIES OF NR BINDING: NEW INSIGHTS FROM GENOMIC METHODS
Chromatin immunoprecipitation (ChIP) coupled to either genomic microarrays (ChIP-chip)8,9 or high-throughput sequencing (ChIP-seq)10,11 have made it possible to probe the genomic locations of endogenous NRs in an unbiased genome-wide manner. These studies have foremost revealed that a substantial portion of NR binding sites are distal from known gene promoters, and most likely function by long-range regulatory mechanisms4,12. Additionally, a large proportion of NR binding sites do not follow the strict sequence specificity derived from early known target genes and in vitro binding studies. The question of how an NR recognizes its genomic binding sites has proven to be a complex one, but several recurring themes have emerged from genome-wide studies of NR binding.
NR binding at distal regulatory regions
In the first application of genome-wide location analysis to NR binding, binding sites for estrogen receptor (ER) were mapped in the breast cancer cell line MCF-7 using a genomic tiling array covering human chromosomes 21 and 224. This study revealed that most ER binding occurs outside the proximal promoters of target genes, often with distances of 10-100kb from the regulated transcription start site (TSS). Prior to this analysis most sequence and binding analysis had been limited to proximal promoter regions within several kb of each TSS13. This surprising finding provided a major impetus to expand NR DNA-binding studies across the entire genome, which became possible with the development of whole genome tiling arrays14 and high-throughput sequencing technology10. Indeed, such studies demonstrated that distal binding by ER is a general phenomenon throughout the genome, with the vast majority (>90%) of sites binding more than 10kb away from the nearest transcription start site14–16.
Remarkably similar findings were made for other NRs, including GR17, peroxisome proliferator-activated receptor gamma (PPARγ18,19 and vitamin D receptor (VDR)20 (summarized in Table 1). While the exact proportion of distal binding events varies by NR, cell-line, and analytic methods, there is a clear trend that only a small minority of NR sites occur within 10kb of a TSS. A meta-analysis of NR cistromes showed that proximal and distal sites have similar association with ligand-responsive genes and comparable levels of evolutionary conservation21. The primary implication of this general finding is that the search space for NR response elements is many orders of magnitude larger than originally proposed13 even when focusing on ligand-responsive genes. Thus, direct measures of genome-wide binding are critical for identifying true NR binding sites, but such data are currently lacking for many NRs and relevant cell types.
Table 1. Survey of distal and non-canonical NR binding rates in genome-wide studies.
Studies are organized by NR (1st column) and cell type (2nd column). The species of the cell type is noted in parentheses (h = human, m = mouse). The published percentage of NR binding sites within ~10kb of the TSS is shown in the 3rd column (the exact distance threshold is shown in parentheses). The published percentage of NR sites containing a canonical motif is shown in the 4th column (in some studies, only the percentage of less stringent half-site motifs are reported, designated by “1/2”). Unreported values are indicated by “-”.
| NR | Cell Type | % Promoter | % Canonical Motif |
|---|---|---|---|
| ERα4 | MCF-7 (h) | - | 49% |
| ERα14 | MCF-7 (h) | 4% (1kb) | 15% |
| ERα16 | MCF-7 (h) | 7% (5kb) | 68% |
| ERα15 | MCF-7 (h) | 5% (5kb) | 71% |
| ERα23 | MDA-MB-231 (h) | 10% (1kb) | 47% |
| AR44 | LNCaP (h) | - | 10% |
| GR57 | 3T3-L1 (m) | - | 77% |
| GR17 | 3134 (m) | 7% (2.5kb) | 50% |
| GR48 | A549 (h) | 12% (10kb) | 62% |
| GR39 | Macrophages (m) | 6% | 60% |
| VDR58 | Lymphoblastoid (h) | <25% (5kb) | 67% |
| VDR59 | THP-1 (h) | 15-20% (10kb) | 32% |
| VDR20 | LS180 (h) | 2% (1kb) | 54% |
| PPARγ19 | 3T3-L1 (m) | 10% (5kb) | 44% |
| PPARγ18 | 3T3-L1 (m) | 13% (10kb) | 75% |
| PPARγ60 | Macrophage (m) | - | 70-77% |
| LXR:RXR24 | Liver (m) | 11% (5kb) | - |
| LXR61 | Macrophages (m) | 15% (500bp) | 6.3% |
| FXR62 | Liver (m) | 15% (10kb) | - |
| FXR62 | Intestine (m) | 17% (10kb) | - |
| Rev-erbα35,63 | Liver (m) | 21% (10kb) | 65% (1/2) |
| ERRα64 | Heart (m) | - | 69% (1/2) |
| ERRα65 | MCF-7, SKBr3 (h) | - | ~75% |
| ERRγ64 | Heart (m) | - | 74% (1/2) |
| RARα/γ66 | MEF (m) | - | 50% |
| RARα/γ66 | ESC (m) | - | 41% |
| RAR67 | Motor Neurons (m) | - | ~50% |
| RARα/γ26 | MCF-7 (h) | <35% (10kb) | - |
| RARα25 | MCF-7 (h) | 5% | - |
| HNF4α68 | Liver (h) | <25% (3kb) | 85% |
| HNF4α68 | Liver (m) | <25% (3kb) | 96% |
NR binding in the absence of known motifs
Initial surveys of individual NR response elements and in vitro studies of NR sequence specificity revealed a general pattern of NR binding motifs consisting of paired half-sites with specific orientation and spacing, reflecting the fact that most NRs bind DNA as dimers1. The sequence motif of each half-site varies slightly between NRs, but generally reflects the high conservation of the DBD within the NR superfamily. Binding motifs are thus classified primarily by orientation, indicated as either Direct Repeat (DR), Inverted Repeat (IR), or Everted Repeat (ER), followed by a number indicating the spacer length between the two half-sites21. For example, GR homodimers bind to a pair of half sites with inverted orientation separated by three nucleotides, denoted as “IR3”22.
Surprisingly, the initial ChIP-chip study of ER binding revealed that roughly half of the identified sites did not contain a canonical ER binding motif4, and subsequent studies have reported widely varying percentages of canonical motifs at ER binding sites (Table 1). Notably, the study reporting the highest percentage of canonical motifs at ER sites15 also reported the smallest total number of sites compared to other genome-wide studies16, suggesting that canonical motifs facilitate the strongest and most detectable binding events, but may not encompass all NR binding sites. In each study, many of the remaining sites contained an ER half-site, but the half-site motif alone is potentially recognized by many other NRs1. A comparative genome-wide study of wild-type ER versus ER containing a mutated DBD revealed that the majority of ER binding sites are dependent on a functional DBD, including many sites without a canonical motif23. Thus, genome-wide studies of ER, along with many other NRs studied to date, have challenged the notion that NRs bind only to stringent paired half-sites with highly specific orientations. The recent meta-analysis of NR cistromes confirmed a general prevalence of binding events at monomeric half-sites and other non-canonical motifs21, although many NRs remain to be analysed by genome-wide methods. A broad survey of canonical motif rates in published NR cistromes is shown in Table 1.
Genome-wide studies of NR binding have produced compelling evidence that degenerate binding sites play critical roles in NR function. In two independent genome-wide studies of mouse 3T3-L1 adipocytes, the primary motif bound by PPARγ was generally more stringent in the half-site contacted by the obligate dimerization partner RXR18,19. RXR occupancy at these sites was observed early in adipogenesis, prior to PPARγ expression, and likely represents RXR binding as a homodimer or heterodimer with alternate partner factors19. Thus, the weaker sequence specificity at the PPARγ half-sites may facilitate binding by other RXR dimers, allowing them to prime sites for subsequent binding by PPARγ, which functions as the master regulator of adipogenesis.
A comprehensive study of liver X receptor (LXR) and PPARα binding in murine liver showed an unexpectedly high overlap between these two NRs, facilitated by binding to non-canonical motif sites24. LXR and PPARα both bind to DNA as heterodimers with RXR, but have sequence specificity for DR1 and DR4 elements, respectively. However, the enrichment for either such motif was particularly low at regions bound by both LXR:RXR and PPARα:RXR heterodimers, with <20% of sites containing either motif. Target gene expression patterns and physiological experiments confirmed the potential for cross-talk between these two NR pathways24. Thus, LXR and PPARα appear to synergistically co-regulate downstream pathways by binding to a high proportion of common binding sites in vivo, facilitated by lower sequence specificity. Comparison to additional NR ChIP-seq data sets in murine liver revealed that many of these sites may bind an even wider repertoire of NR proteins24. Similarly, genomic studies of ER and retinoic acid receptor (RAR) in MCF-7 cells found over 1,000 shared binding sites despite the difference in canonical binding motifs, although current results conflict on whether these co-localizations result in cooperative or antagonistic regulatory effects25,26. In 3T3-L1 pre-adipocytes, ChIP-seq profiling of multiple factors revealed ~1,000 “hotspots” bound by GR, RXR, and additional TFs27. In this study, the authors found canonical motifs for both GR and RXR enriched among the shared binding regions and proposed that each NR binds distinct sequences in close proximity. A complete investigation of degenerate binding elements has not yet been pursued using this data set.
Evidence from genome-wide studies also suggests that chromatin accessibility facilitates NR binding at degenerate recognition motifs. ChIP-seq analysis of GR showed a substantial interplay between sequence motifs, chromatin accessibility prior to ligand treatment, and GR binding17. Although GR is capable of binding nucleosomal DNA and initiating chromatin remodelling, the vast majority of sites identified were accessible to DNase I prior to ligand treatment. However, the small minority of GR-remodelled sites showed increased stringency for the canonical GR binding motif, suggesting that stringent binding motifs may be more critical for recognizing response elements in nucleosomal DNA. Interestingly, the common PPARα/LXR sites identified in liver also corresponded to regions of higher chromatin accessibility compared to sites specific to each NR, supporting the general model that chromatin accessibility facilitates binding to degenerate recognition sequences24.
The potential for binding at weaker motif sites, along with the high proportion of distal binding events, reveal the challenge of predicting individual NR binding sites and target genes from sequence alone. Thus, direct experimental evidence of genome-wide binding patterns is critical for studying NR signalling, and is currently lacking for many NRs. Furthermore, the high proportion of non-canonical motif sites is most likely a reflection of the various mechanisms facilitating NR binding, not all of which rely on strong sequence specificity. In particular, shared binding of multiple NRs to highly accessible regions with degenerate recognition motifs may facilitate cross-talk between different NR signalling pathways, although further analysis of the complete NR superfamily is needed.
Additional transcription factors assist NR binding and chromatin access
NR binding at some response elements is known to be facilitated by partner factors that make chromatin more accessible28, stabilize NR-DNA interactions, or indirectly recruit the NR protein via tethering29. Genome-wide studies have revealed that for some NRs, partner TFs have a nearly global role in determining the NR binding pattern in a specific cell type. One prominent example is the dependency of ER on the pioneer factor FoxA1 in breast cancer. Several genome-wide studies have shown that FoxA1 promotes the opening of chromatin at most ER binding sites in several breast cancer cell lines and FoxA1-mediated chromatin access is a major determinant of ER binding strength at these sites28,30. Genome-wide studies have suggested a similar role for C/EBPβ in priming PPARγ binding regions during adipogenesis27.
Genome-wide studies of GR and the partner factor AP-1 revealed an extensive overlap in binding sites and suggested that AP-1 plays a major role in maintaining the open chromatin state at these sites, in addition to recruiting GR indirectly in the absence of a canonical GR motif29. A survey of motifs at GR sites in two highly divergent cell lines showed different partner factor motifs enriched at GR sites in each cell type, with particular enrichment at pre-accessible sites lacking the canonical GR motif17. Thus, multiple partner factors likely facilitate cell-specific GR binding regions, even in the absence of strong primary binding motifs, by some combination of chromatin remodeling, stabilization of GR-DNA interactions, or indirect recruitment of GR.
An integrative study of ER and androgen receptor (AR) cistromes and the corresponding genome-wide changes in DNase I sensitivity after treatment with each ligand demonstrated that the impact of open chromatin on NR binding and conversely the impact of NR binding on chromatin structure both vary depending on the NR31. In particular, ER was shown to preferentially bind to regions that are accessible to DNase I digestion prior to ligand treatment, while AR bound primarily to inaccessible regions and had a bigger impact on remodelling these regions into an open state. Moreover, the partner factor FoxA1 was associated with different changes in chromatin structure at binding sites for these NRs. In the case of ER, co-localization with FoxA1 was associated with larger increases in chromatin accessibility after ligand treatment, while for AR, co-localization with FoxA1 was associated with weaker increases in chromatin accessibility. Interestingly, the behaviour of AR and its relationship to FoxA1 also appear to be distinct in different cell types and chromatin contexts. FoxA1 has also been shown to assist AR in mimicking the binding profile and regulatory targets of ER in a subclass of ER-negative breast cancer32, as well as mask AR binding sites in prostate cancer cells33. Collectively, these studies demonstrate that NRs with inherent ability to access binding sites in nucleosomal DNA are still dependent on chromatin state overall, and other epigenomic features may be able to block access to their binding sites in specific cell types. These results also demonstrate that the relationships between NRs and their partner factors can be quite complex, showing both cooperative or pioneer relationships at some sites, and antagonistic roles at others.
Recent work from the Hager lab has led to an “assisted loading” model in which one NR can initiate chromatin remodelling to a more open state and rapidly dissociate from the DNA, leaving the binding region open to occupancy by additional NRs34. This was demonstrated in an artificial system in which the ER gene was mutated to mimic the DBD of GR (called “ER-pBox”) and thus bind preferentially to glucocorticoid response elements (GREs) in response to estrogen. ER-pBox and GR protein constructs, each tagged with a different fluorescent label, were co-expressed in cell lines and their occupancy of various exogenous and endogenous GREs was measured in response to ligands. Each NR construct bound GREs in response to their corresponding ligand, but the ligand-induced binding of ER-pBox was further enhanced by simultaneous treatment with the GR ligand at GREs where GR is known to mediate chromatin remodelling to increase accessibility. At GREs where the chromatin is already accessible in the absence of either NR there was still no evidence of competitive binding, i.e., ligand-induced occupancy of GR did not disrupt the ligand-induced binding of ER-pBox. The fact that both NRs bind the same sequence element in this system indicates that the NR proteins must rapidly dissociate from DNA, even in cases where the NR protein mediates chromatin remodelling34. Whether these results can be generalized to other NRs remains an open question, but the NR hotspots containing degenerate motifs in liver discussed above24 support a role for cooperative, dynamic binding of multiple NRs at shared response elements. Additionally, the adipocyte “hotspots” bound by GR, RXR, and other TFs showed evidence of cooperative binding, wherein reduction of one TF disrupted the binding of other TFs at nearby elements27, consistent with the assisted loading model. To further explore this model in an endogenous context, the binding dynamics of multiple wild-type NRs with different sequence specificities will need to be examined at shared or proximal NR binding sites under various combinations of ligand treatments.
Genome-wide studies of NR co-regulator recruitment
The primary mechanism by which NRs influence transcriptional regulation is through the recruitment of co-regulators, many of which coordinate alterations of post-translational marks on histone tails5–7. Genome-wide location analysis can also be used to measure the recruitment of co-regulators and the NR-mediated histone modifications at specific NR binding sites11.
In a genome-wide study of Rev-erbα-mediated gene repression in mouse liver, the co-regulator NCoR and the histone deactylase HDAC3 were shown to co-localize almost exclusively with Rev-erbα, driving circadian rhythms in histone acetylation at thousands of genomic sites35. This finding was particularly surprising given that NCoR/HDAC3 complexes are believed to be general co-repressors for the entire superfamily of NRs6. Quantitative analysis of these ChIP-seq profiles has demonstrated a strong global correlation between the binding signal of Rev-erbα, NCoR, and HDAC3, suggesting that Rev-erbα tends to recruit NCoR/HDAC3-containing complexes at all binding sites in liver, dependent only on the strength of the primary Rev-erbα-DNA interaction36.
Several studies of ER co-activators in breast cancer cells have revealed the potential for selective co-regulator recruitment at a subset of binding sites, raising the possibility that chromatin context affects the ability of a bound NR protein to recruit co-regulators at a given site. A ChIP-chip study of ER and steroid receptor co-activators (SRCs) showed a strong correlation of ER and SRC binding signal at the promoters of estrogen-induced genes, while ER-bound genes repressed by estrogen treatment did not show concomitant recruitment of SRCs37. However, this study relied on a promoter tiling array rather than a genome-wide tiling array, and therefore does not cover the large proportion of ER binding events at distal sites.
In a subsequent study, Lupien and colleagues focused on co-activator associated arginine methyltransferase 1 (CARM1), which methylates arginine residues on the H3 tail as well as on the SRC3 protein38. Although genomic localization of CARM1 could not be measured directly, CARM1-specific changes in histone methylation were observed after E2-treatment and used to identify a subset of “active” ER sites, accounting for 70% of the total ER cistrome in MCF-7 cells. These sites were more strongly associated with gene activation, recruitment of additional co-activators, and changes to chromatin structure after ligand treatment, suggesting that the remaining 30% of ER sites lacking CARM1 activity were inactive in response to ligand. Interestingly, a subset of CARM1-activated ER sites did not co-localize with the pioneer factor FoxA1, discussed above, and these sites were associated with both ER-mediated activation and repression of target genes. Surprisingly, CARM1 was also required to mediate repression of some target genes, despite its canonical role as a co-activator. Similarly, a ChIP-seq study in macrophages demonstrated that only half of GR sites are bound by SRC2 in response to GR ligand, and co-activator-bound sites were associated with both ligand-activated and ligand-repressed genes39.
Overall, these studies demonstrate an additional layer of complexity resulting from the interactions between NRs and co-regulators. In some cases, NRs show genome-wide correlation with specific co-regulators, and a single NR may play a dominant role in utilizing a co-regulator in a particular cell type, as for Rev-erbα, NCoR, and HDAC3 in liver35,36. In other cases, co-regulators may only be recruited to a subset of NR binding sites, as observed for ER38 and GR39. Selective co-regulator recruitment could be due to conformational differences in the bound NR proteins40 or due to recognition of additional chromatin features. Furthermore, co-regulator complexes alter surrounding histone modifications, which may alter the accessibility of the surrounding chromatin, thereby facilitating or stabilizing NR binding5,7.
Changes in NR binding correlate with disease progression
Recent genome-wide studies have found that NR binding profiles and regulatory programs undergo dramatic changes during disease progression, with profound implications for disease treatment. In one landmark study, Wang and colleagues showed that AR binding sites differed substantially between cell culture models of androgen-dependent and androgen-independent prostate cancers41. These differences in binding were correlated with differences in AR-dependent gene expression and different mechanisms of binding site recognition, with the partner factor FoxA1 playing a more prominent role in androgen-independent cells. These findings have direct clinical relevance, because androgen-independent prostate cancer is resistant to hormone-based treatments that are effective in treating androgen-dependent prostate cancer, even though both cancer types require AR expression.
In a similar study, ChIP-chip revealed substantially different binding profiles when ER was activated by classic agonist treatment as compared to activation through the epidermal growth factor (EGF) pathway 42. The EGF pathway is an alternate activation signal for ER that allows breast cancer to escape standard drug treatments, as indicated by drug-resistance associated with ER+ERBB2+ tumors. Lupien and colleagues observed thousands of ER sites in MCF-7 cells bound only after activation through the EGF pathway, adjacent to genes associated with ER/EGF-dependent induction and expression patterns seen in poor prognosis cases. Interestingly, the EGF-specific ER binding sites were primarily enriched for the partner factors AP-1 and FoxA1, rather than the canonical ER motif, suggesting that ER bound to these sites primarily through tethering mechanisms.
The application of ChIP-seq to clinical samples has the potential to directly explore the correlation between NR binding patterns and disease progression. Most notably, a survey of ER binding in various primary breast cancers and related metastases showed distinct patterns of genomic localization in samples with different outcomes and prognoses43. Importantly, ER+ tumors that are resistant to the ER partial antagonist tamoxifen still showed ER binding to the genome, indicating that the drug-resistance was not due to a loss of ER activity. However, ER took on a substantially different binding pattern in the drug-resistant tumors and the newly acquired binding sites occurred preferentially around target genes associated with poor prognosis. Motif enrichments and follow-up studies in cell culture models suggested that FoxA1 mediates the acquisition of new ER binding sites in drug-resistant and metastatic cancer cells, echoing the association with AR binding patterns seen in androgen-independent prostate cancer cells.
Thus, NRs can shift their binding profiles in a given cell type, thereby shifting regulatory behaviour and altering physiology without changing the intrinsic DNA binding affinity of the NR protein. These altered cistromes can drive progression and in some cases allow tumors to escape standard NR antagonist treatments. In particular, partner factors such as FoxA1 and AP-1 may play a critical role in the acquisition of “disease-specific” binding sites, and therefore present intriguing therapeutic targets in conjunction with traditional hormone treatments.
LINKING NR BINDING TO TRANSCRIPTIONAL REGULATION ON A GENOME-WIDE SCALE
The combined application of genome-wide location and expression analyses within the same experimental system has revealed that NR binding does not guarantee ligand-induced activation of the nearest target gene. In many studies, a substantial portion of NR binding sites cannot be linked to a ligand-responsive gene, even when considering potential target genes up to 50kb from each binding site12,16,39,44. Additionally, there is often NR binding enrichment around both ligand-activated and -repressed genes within the same experiment (Table 2). These studies, reviewed below, have largely revealed that the canonical model of NRs as genome-wide on/off switches is oversimplified. Instead, these studies support a more complex system in which transcriptional regulation by NR signalling is highly dependent on cell type, ligand, and the epigenomic context of a particular binding site—although specific rules relating these contextual factors to regulatory output remain elusive.
Table 2. Survey of NR binding enrichment near ligand-responsive genes.
Summary of studies in which NR binding and ligand-dependent transcriptional changes were both measured in the same experimental system. Many of these studies demonstrate that NRs can be associated with both gene activation and repression in response to the same ligand treatment. The table is organized by NR and cell type, as in Table 1. The 3rd column briefly explains the criteria for assigning NR sites to target genes. The proportions of ligand-activated and ligand-repressed genes bound by the NR are shown in columns 4 and 5, respectively.
| % of Regulated Genes w/ Assoc. Binding Site | ||||
|---|---|---|---|---|
| NR | Cell Type | Criteria for linking NR binding sites to target genes | % Activ. | % Repr |
| ERα16 | MCF-7 (h) | ERα site within 50kb of genes showing E2-dependent RNAPII enrichment | 89% | 47% |
| ERα14 | MCF-7 (h) | ERα site within 50kb of E2-responsive genes (multiple time points) | 18-33% | 17-24% |
| ERα52 | MCF-7 (h) | ERα site within 10kb of E2-responsive TSS in GRO-seq (multiple time points) | 33%-50% | 8% |
| ERα23 | MDA-MB-231 (h) | ERα site within 50kb of E2-responsive gene | 53% | 27% |
| ERα69 | MCF-7 (h) | ERα+Cohesin site within 20kb of E2-responsive gene | ~30% | ~40% |
| GR48 | A549 (h) | GR site within 10kb of dex-responsive gene | 47% | 8% |
| VDR59 | THP-1 (h) | VDR site within 400kb of ligand-responsive gene | 72% | 43% |
| PPARγ18 | 3T3-L1 (m) | PPARγ site assigned to nearest gene within 50kb | ~60% | ~20% |
| PPARγ60 | Macrophages (m) | PPARγ site assigned to nearest gene within 100kb | 39% | 15% |
| LXR24 | Liver (m) | LXR:RXR sites assigned to nearest TSS | 63% | 70% |
| RARα/γ66 | MEF (m) | RAR bound to ChIP-chip probe associated with regulated gene | 12% | 5% |
Linking distal binding sites to target genes
The discovery that the vast majority of NR binding occurs many thousands or even millions of nucleotides away from target genes has raised the question of how such regulatory elements ultimately influence transcription, and presents a barrier to the accurate prediction of true NR targets based on their genome-wide binding profiles. In limited cases, targeted mutations in endogenous response elements have been used to directly confirm the loss of ligand-responsiveness at the target TSS, however this method is not tractable for large-scale confirmation of distal response elements45. One indirect indicator that a distal NR binding site regulates a particular target gene is the formation of a chromatin loop structure that brings the element and target TSS into close spatial proximity within the nucleus46. Initially, the “chromosome conformation capture” (3C) assay was used to detect loop formation between specific response elements and target genes, e.g. in the confirmation of ER target genes predicted from ChIP-chip data4. Additionally, enrichment for multiple NR binding sites around ligand-responsive genes has also been shown, e.g. for PPARγ47 and GR48, and thus the number of adjacent sites may also be a powerful criteria for identifying true NR target genes based on genome-wide binding profiles. However, the observation of multiple functionally redundant response elements around some target genes also presents a barrier to testing the individual contribution of each binding site.
Another genome-wide approach, called “chromatin-interaction analysis by paired-end tag sequencing” (ChIA-PET), couples ChIP, 3C-like proximity ligation, and high-throughput sequencing to simultaneously examine both primary DNA interactions and chromosome looping structures involving a specific factor. In particular, this method has been applied to study ER in MCF-7 cells, demonstrating that many previously identified distal sites were structurally linked to a target gene TSS through chromosomal looping12. Most of the observed interactions involved regions on the same chromosome, often within 100kb. Importantly, genes directly linked to distal ER sites by this method showed significant enrichment for estrogen-induced RNA Polymerase II (RNAPII) occupancy and expression, broadly supporting the model that distal ER binding sites can regulate target genes many kb away through the spatial re-arrangement of chromatin. Furthermore, siRNA-mediated knock-down of ER resulted in loss of spatial interactions around target genes and corresponding loss of estrogen-inducible expression, confirming that ER plays an integral role in forming some chromosomal loop structures. A subsequent ChIA-PET study of RNAPII in the same cell line demonstrated that additional loops exist between ER-linked promoters and nearby gene promoters, forming multi-gene complexes that can propagate the estrogen-induction signal across multiple genes49. Thus, genome-wide maps of chromosomal looping structures are a powerful tool in linking NR binding elements to their true target genes.
Direct genome-wide measurements of transcription rate in response to NR signalling
One major limitation in studying gene regulation by NRs is the use of genome-wide measures of mRNA concentration, e.g. microarrays and RNA-seq, as proxies for changes in transcription rate. While this is generally a reasonable assumption given sufficient time for mRNA levels to reach a new steady state, this also leads to a convolution of primary and secondary regulatory effects14. In other words, the observed changes in mRNA concentration after ligand treatment represent both direct transcriptional regulation by a particular NR as well as the secondary effects of downstream regulators. Several new genomic technologies that overcome this limitation are being used to more accurately define the primary effects of NR signalling on transcriptional regulation.
Transcription of most protein-coding genes is carried out by RNAPII, which is regulated at several steps including recruitment, initiation, and elongation50. In addition to studying NR localization, ChIP-chip and ChIP-seq have also been useful for measuring changes in RNAPII occupancy genome-wide, particularly in the core promoters and bodies of known genes. This technique provides an alternate measure of transcription rate that can detect changes at earlier time points compared to measures of mature mRNA14,16. A more direct method to sequence and map nascent transcripts originating from all polymerase classes genome-wide is called “Global Run-On Sequencing” (GRO-seq)51. This technique carries the same advantages of measuring RNAPII occupancy and also identifies changes mediated by RNA polymerases I and III. GRO-seq applied to ER signalling in MCF-7 cells revealed widespread and rapid changes after ligand treatment, encompassing ~25% of the transcriptome52. Importantly, comparison of these changes to ER ChIP-seq data in the same cell type16 revealed substantial enrichment of ER binding adjacent to transcripts up-regulated after 10-40 minutes of estrogen treatment, but not adjacent to transcripts up-regulated after 160 minutes. The 10-40 minute changes in nascent transcript levels observed by GRO-seq typically returned to basal level by 160 minutes of estrogen treatment, and correlated most strongly with mature transcript expression levels measured after 3 hours of estrogen treatment, indicating that the primary transcriptional response regulated by genomic ER is transient and takes additional time to affect mature transcript levels. Importantly, the GRO-seq method also identified widespread and rapid up-regulation of rRNAs and tRNAs, highlighting that estrogen signalling plays a direct role in controlling RNA Polymerases I and III in addition to the more widely studied RNAPII.
An additional advantage of the GRO-seq method, utilized by Wang and colleagues to study AR signalling, is the ability to detect changes in eRNA levels53. eRNAs are a recently discovered class of short RNAs transcribed symmetrically from active enhancer sites54. In the AR study, the ligand-induced changes in eRNA levels around AR sites were a better indicator of changes in adjacent gene expression compared to other chromatin and structural markers such as H3K4me1, p300, and Mediator. By analyzing eRNA sequences, the authors were able to show AR and FoxA1-mediated changes in enhancer activities, some of which were independent of major restructuring events. These results indicate that enhancer formation and activation can be regulated separately in the context of NR signalling.
Overall, these studies highlight the additional insight gained from measuring RNAPII occupancy and nascent transcripts compared to mature mRNA levels. In particular, the short time periods before observable changes allow strong separation of primary and secondary effects of ligand signalling52. GRO-seq can also be used to differentiate changes in transcription initiation versus elongation as well as changes in enhancer activity53, which can not be distinguished by traditional gene expression microarrays or sequencing of mature mRNAs.
NR function varies across genomic binding sites
Although many of the initial NR target genes were activated by the presence of ligand, the rules governing NR-mediated regulation in general have proven far more complex. Genome-wide technologies have been a critical technique in revealing this complexity, and are now laying the foundation to systematically dissect the various regulatory programs for each NR.
Genome-wide studies combining NR location analysis and ligand-dependent transcription changes demonstrate that a substantial proportion of NR binding sites do not appear to mediate adjacent gene expression in the cell type or condition examined. For example, Welboren and colleagues showed that while estrogen-activated genes were the most enriched for ER binding events within 50kb, the majority of ER binding sites were adjacent to a gene that did not change in response to agonist treatment16. Similar trends have been observed for AR44 and GR39. As discussed above, most distal sites need to form chromatin loops bringing them in close spatial proximity to a target gene TSS in order to influence transcription. In MCF-7 cells, the majority of ER binding sites, particularly those showing weaker enrichment, were not part of a chromosomal interaction with a TSS12. Even among the set of ER target genes forming a loop between TSS and ER binding site, only 60% of these genes showed induced expression after estrogen treatment. These results demonstrate that many distal NR sites are not spatially engaged with target genes in a single condition or cell type, and even when response elements are engaged with a TSS, they do not necessarily affect the transcriptional machinery.
Other genome-wide studies have revealed that the transcriptional targets of NR signalling in a given cell type are dependent on the activating signal. As discussed above, this specificity can be driven in part by differences in the NR binding profile, but signal-specific binding sites do not completely explain gene regulation patterns. For example, in the comparison of estrogen-induced and EGF-induced ER signalling, the two alternate activation signals were associated with co-activator-mediated histone acetylation at distinct subsets of the ER binding sites induced by both signals42. Thus, the upstream activation signal can also modulate subsequent regulatory steps, such as co-activator recruitment, at specific subsets of NR binding sites.
In the genome-wide comparison of AR binding in androgen-dependent and -independent cell lines introduced above, transcriptomic analyses of siRNA-mediated AR silencing demonstrated that in androgen-independent cells, AR promoted a constitutive gene expression program41. These ligand-independent target genes were enriched for regulators of cell cycle, and were substantially different from the ligand-induced AR target genes expressed in androgen-dependent cells. Thus, AR not only acquires a different binding program in androgen-independent cells, but it also switches to a predominantly ligand-independent mode of regulation, potentially facilitated by partner factors such as FoxA1.
Another major finding, supported by the newer measures of transcription rate changes, is that NRs can simultaneously activate and repress distinct sets of target genes within the same cell type and ligand treatment. Targeted molecular methods have already established potential mechanisms for NR-mediated repression, including co-repressor recruitment in the absence of ligand and antagonist ligands inducing a repressive protein conformation6, but these mechanisms were initially thought to function in a genome-wide manner, affecting all NR target genes equally. However, genome-wide methods have revealed the surprising commonality of NR-dependent gene repression in response to ligands previously considered agonists, even in the presence of a classic NR response element. For example, in the genome-wide study of LXR function in murine liver introduced above, a strict set of LXR-dependent target genes were determined by examining ligand-dependent RNAPII recruitment in wild-type and LXR-null livers24. Surprisingly, LXR binding was equally enriched around LXR-dependent ligand-activated and ligand-repressed genes. The recent meta-analysis of cistromic and transcriptomic data revealed similar trends for ER and RAR21. Additional examples of NR binding associated with both gene activation and repression in response to classic agonist ligands are shown in Table 2.
One possible mechanism for NR site-specific regulatory effects is that the primary binding site sequence may influence regulatory potential. While a single NR protein can bind to a wide variety of sequences, it is possible that binding to different sequences has an allosteric effect on NR protein conformation, thus altering regulatory function40. Surjit and colleagues recently described a “negative GRE” (nGRE) sequence element that confers ligand-induced repression rather than activation55. The nGRE sequence was enriched at GR binding sites adjacent to glucocorticoid-repressed genes in a previously published genome-wide study48, and the glucocorticoid-dependent repression of these genes was shown to require the GR DBD, in contrast to the previously characterized mechanism of transrepression via tethering to NF-κB and AP-156. However, another study of GR-mediated repression opposed this hypothesis, suggesting instead that the co-localizing factors around GREs are the primary determinants of activating versus repressive functions, with a particular focus on the enrichment of IRF3 binding co-localizing with GR around glucocorticoid-repressed genes39. Both studies did agree that GR has repressive functions that require direct DNA binding and are unrelated to NF-κB/AP-1 tethering. Whether repressive GR sites are specified by the binding site sequence or determined by the presence of additional partner factors remains an open question, and indeed both models may be correct at a subset of elements or in different cell types.
Conclusion
Genome-wide studies of NR binding in chromatin have revealed much about NR biology, yet much remains to be learned. One major issue is that NR binding profiles, while enriched for canonical response element motifs, also encompass a substantial number of sites that do not follow the previously proposed sequence constraints. While some NRs have been shown to bind indirectly to DNA by tethering to other TFs, it is unclear whether such mechanisms can explain the totality of non-canonical binding sites. Another possibility is that highly accessible chromatin conformations and/or partner factors can stabilize NR binding to degenerate response elements, leading to the occurrence of NR hotspots24,27. Increased chromatin access at these sites is potentially regulated through assisted loading mechanisms, in which one active NR protein remodels chromatin into an open state, but rapidly dissociates to allow access by other NR subtypes34.
Another major issue is the apparent disconnect between NR binding and observable changes in gene regulation. While genome-wide binding analyses have revealed thousands of NR binding sites in a single cell type, the majority of these sites cannot be linked to transcriptional changes after ligand treatment, even when considering distal regulation over distances of 50-100kb. The most likely explanation in this case is that gene regulation by NRs is modulated at multiple steps. Binding site accessibility is one major determinant of the cell-specific NR regulatory program, but NR binding alone is clearly not sufficient for ligand-dependent gene regulation. A separate regulatory step may be required to bring the bound response element into close spatial proximity with the target TSS. Additionally, binding site sequence, partner factors, and chromatin state may modulate the ability of a NR to recruit the co-regulators needed to influence transcription. Only by studying these determinants in an unbiased genome-wide way can we reliably deduce the molecular rules governing transcriptional regulation by NRs. Furthermore, the available data are heavily biased towards a small subset of frequently studied NRs, and it is currently unknown whether the observed trends are truly representative of the entire NR superfamily.
While the classic model of NR regulation has proven to be a useful tool in deciphering the mechanism of action for certain drugs, genome-wide methods have now demonstrated that the classic model explains only a small portion of gene regulation by NRs—the role of these regulators in the transcriptional network has proven to be far more complex than originally proposed. As genomic techniques become less expensive and more widely used, they will allow us to probe the multi-faceted mechanisms of NR regulation with unprecedented depth and throughput. Based on the results above, we propose several guiding principles for interpreting genome-wide studies of NR binding and gene regulation. First, any single genomic experiment captures an amalgam of different regulatory mechanisms. Genome-wide location analysis identifies NR binding sites with distinct roles in transcriptional regulation and transcriptomic studies often capture both direct and indirect regulatory targets. Second, the dynamics of chromatin state and binding information, e.g. before and after ligand treatment, are often more informative than a single condition, as they tend to allow partitioning of sites by different molecular mechanisms. Third, the integration of orthogonal data sets, e.g. NR binding, chromatin dynamics, and gene expression patterns, allow further sub-classification of binding elements and potential target genes that can separate distinct modes of NR-mediated regulation. Finally, the accurate dissection of these distinct regulatory mechanisms has direct implications for precisely targeting NRs in human disease. Ultimately, genome-wide studies must be implemented in a systems biology framework to fully tackle the complex role that NRs play in the overall cellular network.
Acknowledgements
The authors thank David J. Steger for critical reading of the manuscript. Studies of cell-specific integration of nuclear receptor function at the genome in the Lazar lab are funded by DK43806, DK45586, and DK49780. L.J.E. was supported by DK095526.
Footnotes
Further Reading/Resources
For additional reading on NR biology, we refer readers to the special issue of Pharmacological Reviews (December 2006, Volume 58:4) published in collaboration with the International Union of Pharmacology, which contains comprehensive reviews on each NR subfamily. For additional information about genome-wide location analysis techniques beyond the reviews cited above, see the special ENCODE Project issues of Nature (September 2012, Volume 489) and Genome Research (September 2012, Volume 22). Genome-scale data and other resources related to the NR superfamily are hosted by the Nuclear Receptor Signaling Atlas (NURSA: www.nursa.org). An extensive catalogue of genome-wide data sets for NRs and their related co-factors and histone marks is available at: http://cistrome.dfci.harvard.edu/NR_Cistrome/
Contributor Information
Logan J. Everett, Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA, USA Institute of Diabetes, Obesity, and Metabolism, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA, USA
Mitchell A. Lazar, Division of Endocrinology, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA, USA Institute of Diabetes, Obesity, and Metabolism, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA, USA
References
- 1.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore JT, Collins JL, Pearce KH. The nuclear receptor superfamily and drug discovery. ChemMedChem. 2006;1:504–523. doi: 10.1002/cmdc.200600006. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 4.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Lazar MA. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 8.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 9.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 10.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 11.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourdeau V, et al. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 14.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 15.Lin C-Y, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welboren W-J, et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefterova MI, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen R, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol. 2012;26:37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Q, et al. A comprehensive view of nuclear receptor cancer cistromes. Cancer Res. 2011;71:6940–6947. doi: 10.1158/0008-5472.CAN-11-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strähle U, Klock G, Schütz G. A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc Natl Acad Sci U S A. 1987;84:7871–7875. doi: 10.1073/pnas.84.22.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stender JD, et al. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boergesen M, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross-Innes CS, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siersbæk R, et al. Extensive chromatin remodelling and establishment of transcription factor “hotspots” during early adipogenesis. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biddie SC, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He HH, et al. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22:1015–1025. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson JLL, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahu B, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss TC, et al. Dynamic Exchange at Regulatory Elements during Chromatin Remodeling Underlies Assisted Loading Mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Z, Feng D, Everett LJ, Bugge A, Lazar M. a Circadian Epigenomic Remodeling and Hepatic Lipogenesis: Lessons from HDAC3. Cold Spring Harb Symp Quant Biol. 2011;76:49–55. doi: 10.1101/sqb.2011.76.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kininis M, et al. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupien M, et al. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol. 2009;29:3413–3423. doi: 10.1128/MCB.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlenhaut NH, et al. Insights into Negative Regulation by the Glucocorticoid Receptor from Genome-wide Profiling of Inflammatory Cistromes. Mol Cell. 2013;49:158–171. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupien M, et al. Growth factor stimulation induces a distinct ER(alpha) cistrome underlying breast cancer endocrine resistance. Genes Dev. 2010;24:2219–2227. doi: 10.1101/gad.1944810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010;285:15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, De Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 47.Soccio RE, et al. Species-Specific Strategies Underlying Conserved Functions of Metabolic Transcription Factors. Mol Endocrinol. 2011;25:694–706. doi: 10.1210/me.2010-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy TE, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li G, et al. Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 51.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surjit M, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 56.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 57.Steger DJ, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramagopalan SV, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heikkinen S, et al. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011;39:9181–9193. doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lefterova MI, et al. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas AM, et al. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bugge A, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dufour CR, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Deblois G, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–6157. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 66.Delacroix L, et al. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231–244. doi: 10.1128/MCB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mahony S, et al. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 2011;12:R2. doi: 10.1186/gb-2011-12-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt D, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]