Abstract
GmNAC5 is a member of NAM subfamily belonging to NAC transcription factors in soybean (Glycine max (L.) Merr.). Studies on NAC transcription factors have shown that this family functioned in the regulation of shoot apical meristem (SAM), hormone signalling, and stress responses. In this study, we examined the expression levels of GmNAC5. GmNAC5 was highly expressed in the roots and immature seeds, especially strongly in immature seeds of 40 days after flowering. In addition, we found that GmNAC5 was induced by mechanical wounding, high salinity, and cold treatments but was not induced by abscisic acid (ABA). The subcellular localization assay suggested that GmNAC5 was targeted at nucleus. Together, it was suggested that GmNAC5 might be involved in seed development and abiotic stress responses in soybean.
1. Introduction
Environmental stresses such as drought, salinity, and cold are major factors that significantly limit agricultural productivity. NAC transcription factors play essential roles in response to various abiotic stresses [1]. The N-terminal region of NAC proteins contains a highly conserved NAC domain, which can be divided into five subdomains based on sequence similarities and may function as DNA-binding region. The C-terminal regions of NAC proteins, which exhibit the transactivation activity, are highly divergent in both sequence and length [2–4]. This family of transcription factors is involved in a lot of plant developmental processes, including shoot apical meristem formation [5], hormone signaling [2, 6], regulation of cell division and cell expansion [7], control of secondary wall formation [8–10], and responses to various stresses [11–14].
The NAC family consists of several subfamilies [15]. The NAM subfamily is the best studied NAC subfamily. CUC1 and CUC2, encoding NAM subfamily proteins, are a pair of functionally redundant genes, expressed in Arabidopsis meristem and organ primordia boundary [1, 16]. The cotyledons of the transgenic seedlings overexpressing CUC1 (35S::CUC1) regularly had two basal lobes, small and round epidermal cells between the sinuses, and adventitious SAMs on the adaxial surface of this region [17]. It has been reported that CUC2 is essential for dissecting the leaves of a wide range of lobed/serrated Arabidopsis lines. Inactivation of CUC3 leads to a partial suppression of the serrations, indicating a role for this gene in leaf shaping. Morphometric analysis of leaf development and genetic analysis provide evidences for different temporal contributions of CUC2 and CUC3 [18]. The CUP played an important role in the lateral organ boundary forming snapdragon. Cupuliformis mutants are defective in shoot apical meristem formation, but cup plants overcome this early barrier to development to reach maturity. CUP encodes a NAM protein, homologous to the petunia NAM and Arabidopsis CUC proteins. The phenotype of cup mutants differs from the phenotype of NAM and CUC1 CUC2 in that dramatic organ fusion is observed throughout development [19]. Phloem transport of CmNACP mRNA was proved directly by heterograft studies between pumpkin and cucumber plants, in which CmNACP transcripts were shown to accumulate in cucumber scion phloem and apical tissues [20]. Petunia NAM proteins were mainly expressed in the meristem and primordia boundaries, which might be required by embryo and flower pattern formation [5]. For abiotic stress, it was observed that Arabidopsis AtNAC2 expression was induced by salt stress and this induction was reduced in magnitude in the transgenic Arabidopsis plants overexpressing tobacco ethylene receptor gene NTHK1. AtNAC2 was localized in the nucleus and had transcriptional activation activity. It can form a homodimer in yeast. AtNAC2 was highly expressed in roots and flowers but less expressed in other organs examined. In addition to the salt induction, AtNAC2 can be induced by abscisic acid (ABA), ACC, and NAA [21]. These showed that the NAM subfamily members not only play a regulatory role in plant development but also participate in stress responses. GmNAC5, which is a member of NAM subfamily belonging to NAC transcription factor in soybean, was cloned and analysed [22]. In order to further study the physiological and biochemical processes that GmNAC5 gene may be involved in, the soybean organ expression patterns of the gene and the relationship between GmNAC5 gene and abiotic stress were examined.
2. Materials and Methods
2.1. Plant Materials
Soybean cv. Ludou 10th was used in this study. Plants were field-grown under normal conditions in Nanjing Agricultural University. Vegetable tissues such as roots, stems, and leaves were collected from 4-week-old seedlings, while floral buds at R1 stage [23], young pods at R3 stage, and developing seeds from 15 to 50 days after flowering (DAF) were collected and frozen immediately in liquid nitrogen and stored at −80°C until use.
2.2. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
Total RNA was extracted using a Total RNA Plant Extraction Kit (Tiangen, Beijing, China), according to the manufacturer's protocol. First-strand cDNA was synthesized using the TaKaRa PrimeScript 1st strand cDNA Synthesis Kit (TaKaRa, Dalian, China), according to the manufacturer's instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted using the SYBR Green Real-Time PCR Master Mix (TOYOBO, Osaka, Japan) on an ABI7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Gene expression was quantified using the comparative method Ct: 2−ΔΔCt method as previously described [24].
2.3. Stress Treatments
The soybean seedlings cultured with sand were moved to Hoagland nutrient solution, when growing to two true leaves. After the first cluster of fronds grew, the plants were applied with stress treatments with three replicates. For hormone treatments, the seedlings were treated with 100 μM JA and 100 μM of ABA, respectively. For salt stress, the seedlings were treated with 200 mM NaCl. For dehydration stress, the seedlings were placed on filter paper, respectively. For cold stress, the seedlings were placed in 4°C light incubator. For mechanical wounding, the seedling leaves were cut into pieces with a sharp and clean scissor. After each treatment, the leaves were harvested and frozen in liquid nitrogen immediately.
2.4. Subcellular Localization of NAC Proteins
The full-length cDNA of GmNAC5 was cloned in pBI121-GFP vector, in frame fusing with GFP reporter gene and producing the plasmid pBI-GmNAC5-GFP. After transient expression of the fusion plasmid in onion epidermal cells, the cells were observed under florescence microscope.
3. Results
3.1. Genomic Structure of GmNAC5
NAC transcription factors have been considered one of the largest families of transcription factors so far discovered in the plant genomes. GmNAC5 encodes a NAC transcription factor belonging to the NAM subfamily. It was found that the exon-intron structures were conserved among GmNAC5 homologous genes in three common species, including Arabidopsis thaliana, Zea mays, and Linum usitatissimum (Figure 1).
Figure 1.
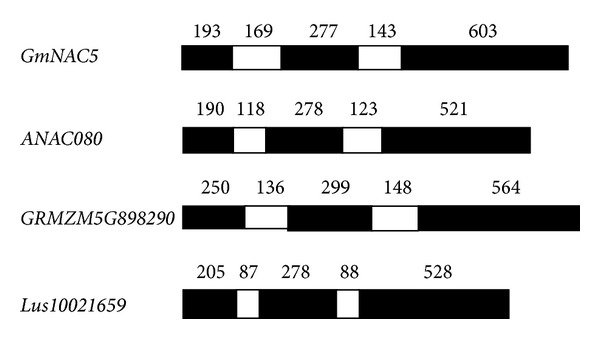
Schematic diagram of gene structures of GmNAC5 and its homologous genes in Arabidopsis thaliana (ANAC08), Zea mays (GRMZM5G898290), and Linum usitatissimum (Lus10021659). The black and white boxes indicate exons and introns, respectively.
3.2. Subcellular Localization
GmNAC5 encoding product is presumed to act as a transcription factor. If transcription factors achieve the precise adjustment of the target genes, this specific transcription factor should be located in the nucleus. Interestingly, GmNAC5 lacks the traditional nuclear localization signal (NLS); even some researchers have found that some NAC domain proteins have the nuclear localization signals [16, 25, 26]. To clarify whether soybean NAC protein GmNAC5 is located in the nucleus, the subcellular localization assay was performed (Figure 2). Despite the transient expression in the onion epidermal cells, it was observed that the GmNAC5-GFP fusion protein was located predominantly in the nucleus whereas GFP alone was localized throughout the cells (Figure 2(b)).
Figure 2.
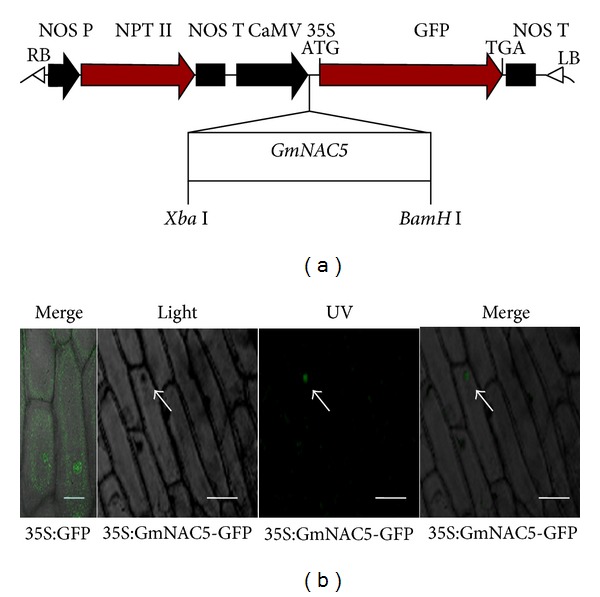
Subcellular localization of GmNAC5. (a) The structure of 35S:GmNAC5-GFP vector. (b) Subcellular localization of GmNAC5-GFP fusion protein. The arrow indicates the location of the nucleus. Bars: 40 μm in 35S:GFP; 80 μm in 35S:GmNAC5-GFP.
3.3. Tissue-Specific Expression of GmNAC5
In order to analyze the physiological and biochemical processes that GmNAC5 gene may involve, qRT-PCR approach was used to analyze GmNAC5 gene expression in soybean in different tissues and organs. GmNAC5 was mainly expressed in the roots and seeds in soybean development and weakly expressed in the other organs (Figure 3). GmNAC5 has the lowest expression level in the stems, but the highest expression level in soybean seeds of 40 days after flowering. The difference in GmNAC5 expression level of each period in soybean seed development is obvious. The highest expression level was found 40 days after flowering (DAF), but only weak expression in the seeds of 15 days and 50 days after flowering, which indicates that the GmNAC5 may participate in the middle stage of soybean seed development. We found that GmNAC5 has strong expression in roots, but expression levels in stems, leaves, and pods are weak.
Figure 3.
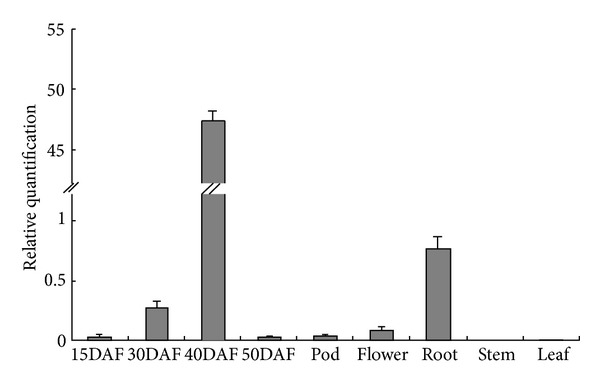
Real-time RT-PCR analysis of GmNAC5 expression in various soybean tissues. DAF: days after flowering.
3.4. Expression of GmNAC5 in Soybean under Various Stresses
GmNAC5 was weakly expressed in leaves in soybean under normal growth condition. The real-time qRT-PCR was performed to detect the expression of GmNAC5 in soybean under various stresses (Figure 4). For jasmonic acid treatment, GmNAC5 was significantly induced after 3 h of JA treatment (Figure 4(a)). For mechanical wounding, expression of GmNAC5 was sharply induced after 1 h of treatment (Figure 4(b)). For NaCl treatment, GmNAC5 expression was markedly upregulated by 8-fold after 3 h of treatment and then decreased (Figure 4(c)). Under drought treatment, expression of GmNAC5 showed a weak increase and then declined (Figure 4(d)). For cold stress, it was found that GmNAC5 expression was gradually increased and reached the maximum after 12 h of treatment (Figure 4(e)). In order to reveal whether stress responsive expression of GmNAC5 was involved in ABA pathway, we studied expression of GmNAC5 under ABA treatment (Figure 4(f)). The qRT-PCR assay suggested that expression of GmNAC5 was not markedly affected by ABA, suggesting that GmNAC5 may participate in ABA-independent signaling pathway in soybean under abiotic stresses.
Figure 4.
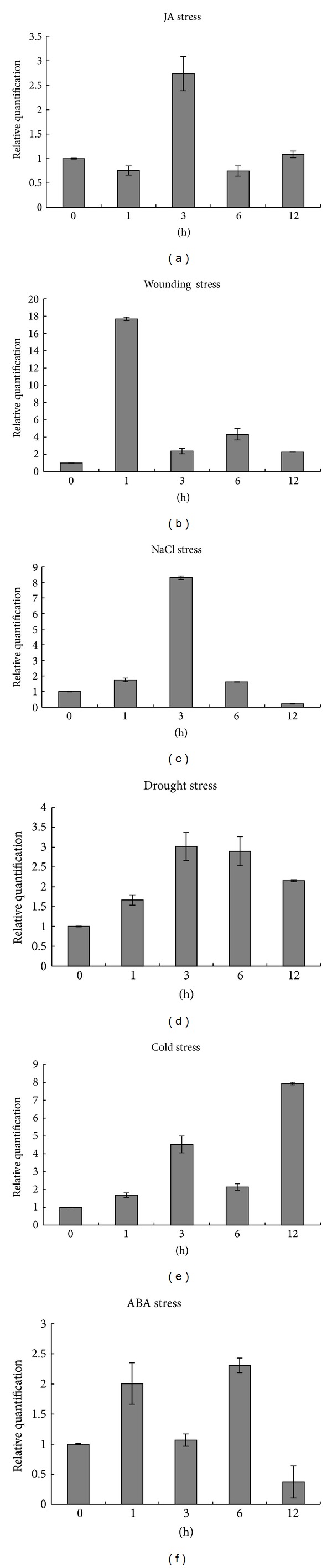
Expression of GmNAC5 in soybean seedlings under various stresses. The soybean seedlings were stressed with 100 μM JA (a), wounding (b), 200 mM NaCl (c), drought (d), 4°C (e), and 100 μM ABA (f).
4. Concluding Remarks
It has been documented that the plant-specific NAC (for NAM, ATAF1, 2, and CUC2) transcription factors play an important role in plant development and stress responses [27]. GmNAC5 belongs to the NAM subgroup and is most closely related to CUC1, CUC2, and NAM, which are involved in developmental events, maintenance of shoot meristem, and cotyledons separation [28]. In this study, we observed some new clues involved in the functions of GmNAC5. Tissue-specific expression analysis indicated that GmNAC5 was highly expressed in immature seeds at 40 DAF and in the roots, suggesting the involvements of GmNAC5 in seed development and root development. It was also found that transcripts of Arabidopsis AtNAC2 were accumulated at the late stages of seed development [29].
It is also possible that higher expression of GmNAC5 in soybean roots is associated with abiotic stress responses. Arabidopsis AtNAC2 expression was highly in roots and induced by salt stress [21]. Further studies suggested that AtNAC2 functioned downstream of ethylene and auxin signaling pathways and regulated lateral root development under salt stress. Expression of GmNAC5 was significantly induced by multiple abiotic stresses but not by ABA, suggesting that GmNAC5 may be involved in ABA-independent stress responses in soybean under abiotic stresses. It was previously reported that NAC transcription factor involves the control of plant senescence and transient expression of GmNAC5 in tobacco leaves induced senescence and necrosis, suggesting that GmNAC5 may play a role in the regulation of stress promoted senescence. Through microarray analysis, it was found that Arabidopsis AtNAC2 regulated many senescence-related genes and the majority of them are also regulated by salt stress, a major promoter of plant senescence [29]. Whether GmNAC5 plays a regulatory role in stress regulated root development or stress promoted senescence still needs to be further analyzed.
Conflict of Interests
The authors declare no conflict of interests. The authors do not have a direct financial relation with the commercial identity mentioned in the current paper.
Authors' Contribution
Hangxia Jin and Guangli Xu contributed equally to this work.
Acknowledgments
This work was supported in part by the National Basic Research Program of China (973 Program) (2010CB125906) and the National Natural Science Foundation of China (30800692, 31171573, and 31201230).
References
- 1.Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. Journal of Experimental Botany. 2012;63(8):2933–2946. doi: 10.1093/jxb/err462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie Q, Frugis G, Colgan D, Chua N-H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes and Development. 2000;14(23):3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takada S, Hibara K-I, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128(7):1127–1135. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- 4.Duval M, Hsieh T-F, Kim SY, Thomas TL. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Molecular Biology. 2002;50(2):237–248. doi: 10.1023/a:1016028530943. [DOI] [PubMed] [Google Scholar]
- 5.Souer E, Van Houwelingen A, Kloos D, Mol J, Koes R. The no apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85(2):159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 6.Fujita M, Fujita Y, Maruyama K, et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant Journal. 2004;39(6):863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 7.Sablowski RWM, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA . Cell. 1998;92(1):93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 8.Iwase A, Hideno A, Watanabe K, Mitsuda N, Ohme-Takagi M. A chimeric NST repressor has the potential to improve glucose productivity from plant cell walls. Journal of Biotechnology. 2009;142(3-4):279–284. doi: 10.1016/j.jbiotec.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Mitsuda N, Iwase A, Yamamoto H, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19(1):270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant Journal. 2008;56(5):768–778. doi: 10.1111/j.1365-313X.2008.03633.x. [DOI] [PubMed] [Google Scholar]
- 11.Nogueira FTS, Schlögl PS, Camargo SR, et al. SsNAC23, a member of the NAC domain protein family, is associated with cold, herbivory and water stress in sugarcane. Plant Science. 2005;169(1):93–106. [Google Scholar]
- 12.Ohnishi T, Sugahara S, Yamada T, et al. OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes and Genetic Systems. 2005;80(2):135–139. doi: 10.1266/ggs.80.135. [DOI] [PubMed] [Google Scholar]
- 13.Hu H, Dai M, Yao J, et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(35):12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, He G, Zhao S, Guo C, Lu M. Expression analysis of two NAC transcription factors PtNAC068 and PtNAC154 from poplar. Plant Molecular Biology Reporter. 2012;30(2):370–378. [Google Scholar]
- 15.Ooka H, Satoh K, Doi K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana . DNA Research. 2003;10(6):239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 16.Taoka K-I, Yanagimoto Y, Daimon Y, Hibara K-I, Aida M, Tasaka M. The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant Journal. 2004;40(4):462–473. doi: 10.1111/j.1365-313X.2004.02238.x. [DOI] [PubMed] [Google Scholar]
- 17.Hibara K-I, Takada S, Tasaka M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant Journal. 2003;36(5):687–696. doi: 10.1046/j.1365-313x.2003.01911.x. [DOI] [PubMed] [Google Scholar]
- 18.Hasson A, Plessis A, Blein T, et al. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell. 2011;23(1):54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B. Cupuliformis establishes lateral organ boundaries in Antirrhinum . Development. 2004;131(4):915–922. doi: 10.1242/dev.00993. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development. 1999;126(20):4405–4419. doi: 10.1242/dev.126.20.4405. [DOI] [PubMed] [Google Scholar]
- 21.He X-J, Mu R-L, Cao W-H, Zhang Z-G, Zhang J-S, Chen S-Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant Journal. 2005;44(6):903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 22.Meng Q, Zhang C, Gai J, Yu D. Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.) Journal of Plant Physiology. 2007;164(8):1002–1012. doi: 10.1016/j.jplph.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 23.Fehr WR, Caviness CE, Burmood DT, Pennington JS. Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Science. 1971;11(6):929–931. [Google Scholar]
- 24.Huang F, Chi Y, Gai J, Yu D. Identification of transcription factors predominantly expressed in soybean flowers and characterization of GmSEP1 encoding a SEPALLATA1-like protein. Gene. 2009;438(1-2):40–48. doi: 10.1016/j.gene.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Ernst HA, Olsen AN, Skriver K, Larsen S, Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Reports. 2004;5(3):297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran L-SP, Nakashima K, Sakuma Y, et al. Isolation and functional analysis of arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng H, Cheng H-Y, Yu X-W, et al. Characterization of a chickpea (Cicer arietinum L.) NAC family gene, CarNAC5, which is both developmentally- and stress-regulated. Plant Physiology and Biochemistry. 2009;47(11-12):1037–1045. doi: 10.1016/j.plaphy.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science. 2005;10(2):79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Balazadeh S, Siddiqui H, Allu AD, et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant Journal. 2010;62(2):250–264. doi: 10.1111/j.1365-313X.2010.04151.x. [DOI] [PubMed] [Google Scholar]


