Abstract
Transfer RNA (tRNA) genes and other RNA polymerase III transcription units are dispersed in high copy throughout nuclear genomes, and can antagonize RNA polymerase II transcription in their immediate chromosomal locus. Previous work in Saccharomyces cerevisiae found that this local silencing required subnuclear clustering of the tRNA genes near the nucleolus. Here we show that the silencing also requires nucleosome participation, though the nature of the nucleosome interaction appears distinct from other forms of transcriptional silencing. Analysis of an extensive library of histone amino acid substitutions finds a large number of residues that affect the silencing, both in the histone N-terminal tails and on the nucleosome disk surface. The residues on the disk surfaces involved are largely distinct from those affecting other regulatory phenomena. Consistent with the large number of histone residues affecting tgm silencing, survey of chromatin modification mutations shows that several enzymes known to affect nucleosome modification and positioning are also required. The enzymes include an Rpd3 deacetylase complex, Hos1 deacetylase, Glc7 phosphatase, and the RSC nucleosome remodeling activity, but not multiple other activities required for other silencing forms or boundary element function at tRNA gene loci. Models for communication between the tRNA gene transcription complexes and local chromatin are discussed.
Keywords: tgm silencing, Remodeling, Deacetylase, Phosphatase, RNA polymerase III, Histone
1. Introduction
Transfer RNA gene-mediated (tgm) silencing of nearby transcription by RNA polymerase II (pol II) was initially observed in yeast through experimental transplantation of pol II promoters near active tRNA genes (Hull et al., 1994). This antagonism of pol II transcription by nearby RNA polymerase III (pol III) transcription is consistent with the paucity of tRNA genes within 500 base pairs of pol II promoters in the yeast genome, though retrotransposons preferentially integrate in this environment (Bolton and Boeke, 2003) and the degree of silencing varies among pol II promoters (Hull et al., 1994). The phenomenon conforms to the formal definition of silencing, since it influences pol II transcription out of steric range and is independent of tRNA gene orientation, yet tgm silencing was not previously thought to be chromatin-mediated. It does not require many of the chromatin modifying and remodeling enzymes that were known to affect other silencing forms (Wang et al., 2005), including the SIR genes despite apparent preferential association of Sir3 and Sir4 at tRNA gene loci (Dubarry et al., 2011). In some respects tgm silencing may be incompatible with other forms of silencing, since tRNA genes are able to block propagation of silenced chromatin states caused by other silencing mechanisms (Donze and Kamakaka, 2001). It is not clear what the relationship is between tgm silencing and this “insulator” or “boundary element” function (Donze and Kamakaka, 2001; Dubey and Gartenberg, 2007; Haldar and Kamakaka, 2006; Noma and Kamakaka, 2010; Oki et al., 2004).
The investigation of this behavior led to the observation that tRNA genes are clustered at the nucleolus in Saccharomyces cerevisiae, and that mutations that disrupt nucleolar organization and release the tRNA genes from the nucleolus also release tgm silencing (Haeusler et al., 2008; Kendall et al., 2000; Rodley* et al., 2011; Thompson et al., 2003; Wang et al., 2005). This clustering is dependent on condensin complexes bound to the tRNA gene nucleoprotein complexes, and mutations in condensin subunits release both nucleolar clustering and tgm silencing (D’Ambrosio et al., 2008; Haeusler et al., 2008).
In addition to the contribution of nucleolar clustering, there were reasons to suspect that tRNA genes were affecting local pol II transcription through chromatin nucleoprotein structure. Previous studies suggested that nucleosomes interact with the pol III complexes, since tRNA genes affect nucleosome positioning in at least some contexts (Mahapatra et al., 2011; Morse et al., 1992) and chromatin structure affects the ability to transcribe tRNA genes (Dhillon et al., 2009; Ng et al., 2002; Oler et al., 2010; Parnell et al., 2008; Soutourina et al., 2006). Although deletions and mutations of several genes that affect tRNA gene transcription, including SIR2 and HTZ1, do not compromise tgm silencing (Wang et al., 2005) it is reasonable that other factors influencing nucleosome behavior surrounding tRNA genes might play a role. To test the possible involvement of nucleosomes in an unbiased manner, we screened a comprehensive library of alanine substitution mutations in the four core histones (Nakanishi et al., 2008) for the ability to release tgm silencing. Over 10% of the substitutions strongly release the silencing, with many residing in the histone H3 and H4 N-terminal tails. In addition, a large group of alleviating mutations defines a surface on the nucleosome disk that has not been implicated in other silencing forms. In light of these observations, we also tested additional mutations in a broad group of chromatin modification and remodeling enzymes, and find that the Rpd3 and Hos1 histone deacetylases, the Glc7 phosphatase, and the RSC nucleosome remodeling activity are also required for silencing.
2. Results and discussion
2.1. Histone mutations alleviate tgm silencing
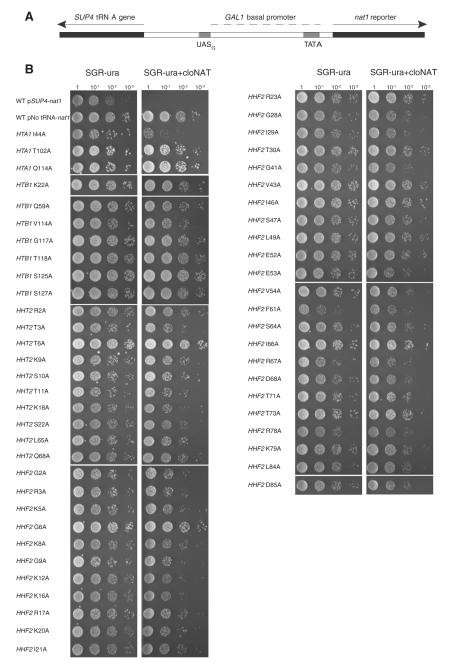
A comprehensive alanine-scanning mutation library of all four core histones (Nakanishi et al., 2008) was screened to test whether nucleosomes are involved in tgm silencing. Briefly the chromosomal copies of the relevant histone genes are deleted in a haploid strain, with the cell survival dependent on a wild type or mutant histone gene carried on a plasmid. Each strain contains a unique histone gene with one amino acid changed to alanine. Only a small number of the ala substitutions were absent because they lead to cell death or substantially slowed growth. Into each of the viable strains we placed a plasmid reporter similar to those used previously to test tgm transcriptional silencing (D’Ambrosio et al., 2008; Haeusler et al., 2008; Hull et al., 1994) (Fig. 1a).
Fig. 1.
Phenotypes for release of tgm silencing. (A) Schematic of pSUP4-nat1 test construct. Expression of cloNAT resistance from a galactose-inducible promoter is silenced by a neighboring SUP4 tRNATyr gene. (B) Phenotypic assay of SHIMA library strains shown to alleviate tgm silencing. Cells were initially grown in SGR-ura and plated on SGR-ura + CloNAT selective media as 10 fold serial dilutions. Phenotypes were recorded after three days of growth. The wild type (WT) strain containing pSUP4-nat does not grow on media containing CloNAT, due to tgm silencing of nat1 expression. Other alanine substitutions that are not shown (Fig. 2) did not relieve silencing and had the WT phenotype, or were not present in the alanine scanning library (Nakanishi et al., 2008). “No tRNA” is a positive control lacking the SUP4 tRNA gene (Hull et al., 1994) allowing expression of nat1.
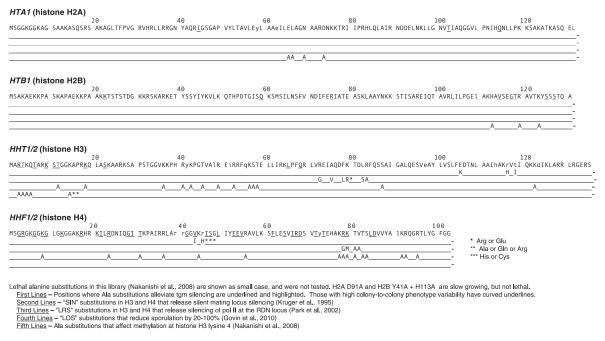
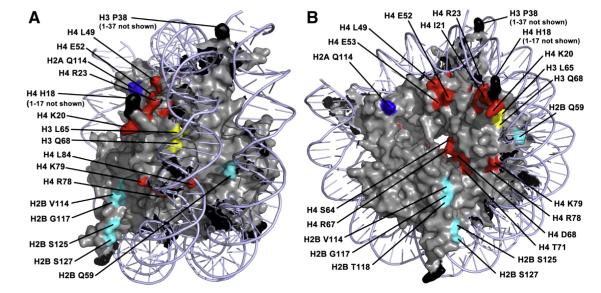
Surprisingly, over 10% of the viable alanine substitutions relieved silencing of the nat reporter gene by a neighboring tRNA gene (Fig. 1), strongly suggesting that nucleosome structure is involved. Although some of the residues coincide with regions that affect other regulatory functions of nucleosomes, many have not been found in other screens of histone mutants that affect transcriptional regulation (Fig. 2). This comparison is not comprehensive because most previous screens have not used systematic substitutions or have only examined histones H3 and H4, but the results where mutagenized regions are parallel suggest that substantially different surfaces on the nucleosome disk are involved in tgm silencing. The identified residues map to three general areas of the nucleosome structure (Figs. 2 and 3): the N-terminal tails of H3 and H4 (Campos and Reinberg, 2009; Millar and Grunstein, 2006), other residues likely in near contact with the DNA near the nucleosome dyad, and a broad surface on the nucleosome disk face. The H3 and H4 tails are particularly intensive regions of post-translational modifications that affect transcription and have the densest clusters of mutations that alleviate silencing. They emerge from the nucleosome core nearly opposite each other across the DNA helix and can interact with both the DNA and external proteins (Fig. 3). Several activities thought to modify these residues do not affect tgm silencing (Wang et al., 2005), (Tables 1 and 2), although not all activities currently known to affect nucleosome behavior were previously tested (see below). H4 R78 and K79 are the other residues that also affect multiple other nucleosome modulations (Fig. 2). They are likely in contact with the DNA and near a surface at the C terminus of H2B (S125, S127) where ala substitutions relieve tgm silencing (Fig. 3). Several additional identified residues are near the DNA in this general region: from H2B (Q59), H3 (L65 and Q68), and H4 (G28, I29, T30, G41, V43, I46, and S47). Some residues on the nucleosome disk surface or buried within the nucleosome structure are close to residues in contact with DNA, including H2A (I44, H2B S125 and S127), and H4 (K20, I21, L49, L84, and D85). The residues located on the nucleosome disk surface form a possible broad contact surface (Fig. 3) that is of particular interest because it does not closely conform to previously identified contacts (Fig. 2). In addition to those noted above, these include residues from H2B (V114, G117, and T118) and H4 (E52, E53, V54, S64, I66, R67, D68, T71, and T73). It is not known whether these surfaces are used for nucleosome packing, or for interactions with external factors such as nucleosome modification or remodeling complexes (but see below). Only two residues where alanine substitutions disrupt tgm silencing appear buried in the nucleosome crystal structure away from the DNA, H2A T102 and H4 F61.
Fig. 2.
Core histone mutations relieving tgm silencing are aligned with mutations affecting other chromatin regulation, defined below the figure. Alanine substitutions relieving tgm silencing are underlined in the top line. Wavy underline indicates high colony-to-colony variation in silencing after growth in non-selective media. Amino acid substitutions affecting other processes are indicated on the lines below, drawn from indicated publications. Viable substitutions other than alanine are indicated in the lines below the sequence. Positions at which alanine substitutions are not viable are indicated by small case letters in the sequence.
Fig. 3.
Map of tgm silencing mutations to nucleosome structure. The positions of alanine substitutions that relieve tgm silencing were mapped to the nucleosome structure using PyMol (http://www.pymol.org/). (A) View from the edge of the disk to optimize visualization of residues in contact with DNA. (B) View optimizing visualization of residues on disk surface. Light blue = DNA. Histones are gray, except residues affecting tgm silencing. Blue = H2A, Cyan = H2B, Yellow = H3, Red = H4. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Genes tested for effects on tgm silencing.
| Methylation and demethylation | |||||||||||
| bre2 Δ a | − | jhd1 Δ b | − | sdc1 Δ a | − | set4 Δ a | − | set7 Δ a | − | swd1 Δ a | − |
| dot1 Δ b | − | jhd2 Δ b | − | set1 Δ a | − | set5 Δ a | − | shg1 Δ a | − | swd3 Δ a | − |
| fpr4 Δ b | − | rph1 Δ b | − | set2 Δ b | − | set6 Δ a | − | spp1 Δ a | − | ||
| Acetylation and deacetylation | |||||||||||
| ard1 Δ a | − | hat1 Δ a | − | hpa2 Δ a | − | * pho23 Δ b | − | sas4 Δ b | − | taf1 Δ b | − |
| * ash1 Δ | − | hat2 Δ a | − | hst1 Δ a | − | * rco1 Δ b | +++ | * set3 Δ b | − | * tod6 Δ b | − |
| * cti6 Δ | − | hda1 Δ a | − | hst2 Δ a | − | * rpd3 Δ b | +++ | * sds3 Δ | − | * tos4 Δ | − |
| * dep1 Δ | − | hda2 Δ a | − | hst3 Δ a | − | rtt109 Δ b | − | * sif2 Δ a | − | * ume1 Δ b | − |
| * dot6 Δ b | +++ | hda3 Δ b | − | hst4 Δ a | − | * rxt2 Δ b | +++ | * sin3 Δ b | + | * ume6 Δ | − |
| * eaf3 Δ b | − | hos1 Δ b | + | kat6/sas3 Δ b | − | * rxt3 Δ a | − | sir2 Δ a | − | ||
| elp3 Δ b | − | * hos2 Δ a | − | kat8/sas2 Δ b | − | * sap30 Δ a | − | * snt1 Δ a | − | ||
| esa1ts a | − | hos3 Δ a | − | nat1 Δ a | − | sas2 Δ b | − | spt7 Δ a | − | ||
| gcn5 Δ b | − | hos4 Δ b | − | nat4 Δ b | − | sas3 Δ b | − | swi8 Δ b | − | ||
| Chromatin remodeling | |||||||||||
| arp5 Δ a | − | chd1 Δ b | − | isw2 Δ b | − | ‡rsc4-Δ4aab | − | snf2 Δ b | − | swi3 Δ b | − |
| arp6 Δ b | + | esc8 Δ b | − | itc1 Δ b | − | ‡ rsc8ts b | − | snf5 Δ a | − | swp73 Δ b | − |
| ‡ arp7ts b | − | fun30 Δ b | − | rdh54 Δ a | − | ‡ rsc9ts b | + | snf6 Δ a | − | ||
| arp8 Δ b | − | ies5 Δ b | − | ‡ rsc1 Δ b | − | ‡ rsc58ts b | − | sth1ts b | +++ | ||
| ‡ arp9 Δ b | − | isw1 Δ a | − | ‡ rsc2 Δ b | − | sin4 Δ b | − | swi1 Δ b | − | ||
| Ubiquitinylation | Sumoylation | Phosphorylation | |||||||||
| cul3 Δ b | − | sas4 Δ b | − | smt3ts b | − | ubc9ts b | − | chk1 Δ b | − | snf1 Δ b | − |
| rcy1 Δ b | − | skp2 Δ b | − | uba1ts b | − | ulp1ts b | − | glc7ts b | +++ | ||
| rtt101 Δ b | − | ynl311 Δ b | − | uba2ts b | − | ulp2 Δ b | − | reg1 Δ b | +++ | ||
| Other | |||||||||||
| asf1 Δ a | − | esc2 Δ a | − | lsm1 Δ a | − | rpa12 Δ a | ± | spc1 Δ b | − | ure2 Δ b | − |
| avt4 Δ b | − | fob1 Δ a | − | maf1 Δ | +++ | rpa34 Δ a | − | srp40 Δ a | − | yap7 Δ b | − |
| bdf1 Δ a | − | gcn2 Δ b | − | mft1 Δ b | − | rpa49 Δ a | + | ssh1Δ1 | − | ycg1-2c | − |
| bdf2 Δ a | − | gcn4 Δ b | − | mig1 Δ b | − | rrn10 Δ a | + | tdp1 Δ a | − | ycs4-1c | − |
| bre1 Δ a | − | gre2 Δ a | − | nap1 Δ a | − | san1 Δ a | − | tho2 Δ | − | yhc3 Δ a | − |
| cbf5-1 d | +++ | hpr1 Δ b | − | nhp6a Δ a | − | sir1 Δ a | − | thp1 Δ a | − | yku80 Δ a | − |
| ccr4 Δ b | − | htz1 Δ a | − | paf1 Δ b | − | sir3 Δ a | − | tup1 Δ a | − | ypk3 Δ b | − |
| cdc73 Δ b | − | hul5 Δ b | − | pch2 Δ a | − | sir4 Δ a | − | uaf30 Δ a | ± | ||
| chd1 Δ a | − | isw1 Δ a | − | ptk2 Δ a | − | smc2–8c | ± | ufd2 Δ b | − | ||
| cmr1 Δ b | − | lhp1 Δ b | − | rif1 Δ a | − | smc4-1c | ± | ufd4 Δ b | − | ||
Rsc complex,
Rpd complex.
Wang et al., 2005 (13).
This study.
Haeusler et al., 2008 (14).
Kendall et al., 2000 (10).
Table 2.
Comparison of histone tail residues and chromatin modification enzymes affecting tgm silencing.
| H3 residue | PTMs | Enzymes | H4 residue | PTMs | Enzymes |
|---|---|---|---|---|---|
| A1 | – | – | S1 | P | CK2 (not tested) |
| R2 a | Me | ? | G2 a | – | – |
| T3 a | – | – | R3 a | Me | Hmt1 |
| K4 | Ac, Me | Gcn5–Jhd2-Rtt109-Set1 | G4 | – | – |
| Q5 | – | – | K5 a | Ac | Esa1-Hos2-Rpd3b |
| T6 a | – | – | G6 a | – | – |
| A7 | – | – | G7 | – | – |
| R8 | – | – | K8 a | Ac | Esa1-Hos2-Rpd3b |
| K9 a | Ac, Me | Gcn5-Hda1-Hos2-Rpd3b | G9 a | – | – |
| S10 a | P | Snf1-Glc7b | L10 | – | – |
| T11 a | – | – | G11 | – | – |
| G12 | – | – | K12 a | Ac | Esa1-Hos1b-Hos2-Rpd3b |
| G13 | – | – | G13 | – | – |
| K14 | Ac | Gcn5-Hda1-Hos2-Rpd3b | G14 | – | – |
| A15 | – | – | A15 | – | – |
| P16 | – | –- | K16 a | Ac | Esa1-Hos2-Hst1-Sas2-Sir2 |
| R17 | – | – | R17 a | – | – |
| K18 a | Ac | Gcn5-Hda1-Hos2-Rpd3b | H18 | – | – |
| Q19 | – | – | R19 | – | – |
| L20 | – | –- | K20 a | Ac | Esa1-Hos2-Hst1-Sas2-Sir2 |
| A21 | – | – | I21 a | – | – |
| S22 a | – | – | L22 | – | – |
| K23 | Ac | Gcn5-Hda1-Hos2-Rpd3b | R23 a | – | – |
PTMs — Post-translational modifications Ac = acetylation, Me = methylation, P = phosphorylation.
(Bold) — residues in the H3 and H4 N-terminal tails that affect tgm silencing.
(Bold) — enzymes that are required for tgm silencing.
This difference between residues affecting tgm and other silencing forms (Kruger et al., 1995; Park et al., 2002) is consistent with our previous observation that tgm silencing is indifferent to deletion of many other genes affecting silencing at silent mating loci, telomeres, and the ribosomal RNA gene (RDN) locus. Also, the relationship of tgm silencing to the boundary element function of tRNA genes in blocking propagation of other silencing is not yet clear. The mechanisms giving rise to these two phenomena in chromosomes are likely not identical, since tgm silencing and boundary function have distinct requirements for tRNA gene transcription and other protein complexes. It remains possible that there is some overlap in mechanisms.
2.2. Chromatin modification and remodeling activities affecting tgm silencing
To further explore activities that might alter local nucleosome structure to cause silencing, we surveyed a more extensive collection of gene deletions and conditional mutations that were linked to tRNA gene behavior, as well as additional chromatin modification activities. We previously tested a number of gene deletions and mutations that affected silencing near telomeres, silent mating loci and the RDN locus (Wang et al., 2005), concluding that the same type of chromatin modifications was not essential for tgm silencing. With the clear involvement of histones, we canvased a wider group of mutations, including defects in several genes that were not known to affect chromatin structure at the time of our earlier work.
The results are summarized in Table 1 for both current tests of chromatin-modifying enzymes and relevant previous tests. The gene products already known to affect tgm silencing, other than the known pol III transcription components, were primarily nucleolar proteins (Cbf5, Rpa49, Rrn10, Uaf30) and subunits of the condensin complex (Brn1, Smc2, Smc4, Ycg1, Ycs4), all of which release the clustering of tRNA genes at the nucleolus (D’Ambrosio et al., 2008; Kendall et al., 2000; Wang et al., 2005). This disruption of nuclear organization coincided with poor growth of the yeast and possible widespread effects on gene expression. Also included in previously described alleviators is a deletion of the gene for Maf1, a known negative regulator of pol III transcription that is responsive to TOR signaling. Maf1 does not appear to be required for tRNA gene localization, but releases tgm silencing (Moir et al., 2006) and might create a signal that affects local chromatin in addition to its known interactions with pol III (Vannini et al., 2010). Deletion of several additional gene products that might have been expected to contribute to tgm silencing had no effect. Several of these (Fkh1, Hda1, Yap6, Nhp6a, Reb1) were tested because they were recently found to be associated upstream of tRNA genes in a genome-wide survey (Venters et al., 2011), but none of them were required for tgm silencing. Others were candidates because they affected either tRNA gene transcription or boundary function, including Htz1 (H2A.Z), Gcn5, Sas2, Dot1, and Sir2 (Kirkland and Kamakaka, 2010; Wang et al., 2005), but these also did not appear to be required.
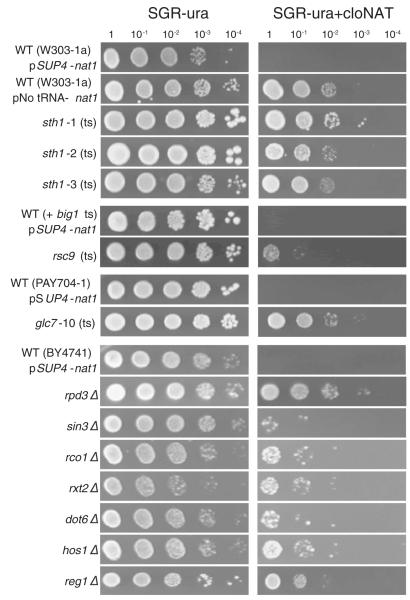
In contrast we found clear evidence that four activities affecting nucleosomes were required for tgm silencing: the RSC nucleosome remodeling activity, the Rpd3 deacetylase, the Hos1 deacetylase, and the Glc7 phosphatase. Phenotypic results for mutants that alleviated silencing in this new screen are shown in Fig. 4. RSC was a suspected activity because it is needed for the boundary element/insulator function of tRNA genes in blocking propagation of other silencing signals, is preferentially associated with pol III transcription units, and can affect local nucleosome behavior (Dhillon et al., 2009; Mahapatra et al., 2011; Ng et al., 2002; Parnell et al., 2008; Soutourina et al., 2006). Despite this, we initially did not believe RSC was involved because we tested several viable RSC subunit mutations and deletions (rsc8ts, rsc1Δ, rsc2Δ, rsc58ts, arp7ts, arp9Δ) that did not relieve tgm silencing ((Ben-Aroya et al., 2008; Wang et al., 2005), this study). The physical and functional association of RSC activity with tRNA genes nevertheless caused us to pursue the question further and test multiple conditional mutations in the essential catalytic subunit. All three distinct ts mutations in STH1 (sth1-1, sth1-2, sth1-3 (Du et al., 1998)) released tgm silencing even at permissive growth temperatures (Fig. 4). We also found that a conditional mutation in one other RSC subunit, Rsc9, weakly alleviated tgm silencing at permissive growth temperatures (rsc9ts in Fig. 4 and Table 1). Interaction of the Sth1 subunit of RSC with the nucleosome residues identified in our screen would be consistent with the low-resolution structure of the complex derived from cryo-electron microscopy (Chaban et al., 2008). In particular the Sth1 ATPase subunit is proposed to contact an extensive surface near the H3/H4 dimers where they meet the DNA at the nucleosome dyad axis, consistent with the concentration of histone residues that alleviate tgm silencing in the disk surface and in contact with the DNA. The H3 and H4 N-terminal tails and several residues shown in Figs. 2 and 3 also exist at this approximate surface. It is not clear why mutations in the catalytic Sth1 subunit of RSC release tgm silencing while mutation or deletion of the other tested subunits does not. Possible explanations range from other individual subunits of RSC not being required for the recognition events in tgm silencing, to Sth1 and Rsc9 being part of another complex that has not yet been described. In addition, conditional mutations in essential RSC subunits can only be tested under permissive or semi-permissive conditions so lack of silencing phenotype is not conclusive.
Fig. 4.
Release of tgm silencing by mutations in chromatin modification and remodeling activities. Deletions and conditional mutations in genes encoding chromatin modification and remodeling complex members were tested for their ability to alleviate tgm silencing as in Fig. 1. Results are shown from mutations that relieve silencing, along with the relevant parental wild type strains for the deletions (BY4741) and conditional mutants (W3031a for sth1 (Du et al., 1998), an unrelated ts mutation (big1) from the same library (Ben-Aroya et al., 2008) for rsc9, PAY704-1 for glc7 (Andrews and Stark, 2000)). For the parental strains, the positive control showing cloNAT resistance with a plasmid construct lacking the tRNA gene is shown. The mutant strains shown demonstrate cloNAT resistance with the tRNA gene present (pSUP4-nat1), and are also cloNAT resistant with the tRNA gene absent (not shown).
Two deacetylase activities were required for tgm silencing (Table 1). One or more of the Rpd3 deacetylase complexes are involved, though the pattern of required subunits did not fit conveniently into previously defined Rpd3 complexes. The Rpd3 deacetylase subunit itself is needed, but this is a component of all Rpd3 complexes, by definition. Deletions of other subunits from both the Rpd3L and Rpd3S complexes (dot6Δ, rco1Δ, rxt2Δ, sin3Δ; 34) also relieve tgm silencing, but it was found that other subunits from both complexes (and the extended Rpd3L complex) are not required (ash1Δ, cti6Δ, dep1Δ, eaf3Δ, hos2Δ, pho23Δ, rxt3Δ, sap30Δ, sds3Δ, set3Δ, snt1Δ, sif2Δ, tod6Δ, ume1Δ, ume6Δ). It is possible that a previously undefined, hybrid subcomplex is at work at the tRNA genes, and that additional subunits are not essential to this function even if they are physically present. It is not particularly surprising to detect interaction between the RSC remodeler and some form of Rpd3 deacetylase complex, since an interaction has been previously documented in a different context (Chen et al., 2012).
The Hos1 histone deacetylase also appeared to be required, possibly through deacetylation of H4 K12 or other residues (Robyr et al., 2002). Since Hos1 is also thought capable of interacting with Sin3 (Grigat et al., 2012), it is possible that both Rpd3 and Hos1 are recruited through Sin3. There is also a possibility that Hos1 functions through another target — for example the Smc3 subunit of cohesin is a known interacting partner and might indirectly affect tgm silencing through affecting chromosome architecture. We consider this particular hypothesis unlikely, however, since we find that conditional mutations in cohesin do not release tgm silencing (unpublished).
Both the Glc7 phosphatase and its regulatory subunit, Reg1, appear to be required for tgm silencing. This could be due to a requirement for histone dephosphorylation, notably at H3 S10 (Hsu et al., 2000), but the full range of Glc7 substrates is not known (reviewed in Santangelo (2006)) and other mechanisms are possible. It is unlikely that the Glc7 effect is exerted through the Gal promoter elements in the reporter construct, since Glc7-dependent activation of Gal-responsive promoters occurs through action on the Mig1 repressor and MIG1 deletion does not release tgm silencing.
The histone residues that are known to be modified in yeast and affect tgm silencing are clustered exclusively in the N-terminal tails of H3 and H4. Table 2 shows these residues (in bold) along with the enzymes thought to be responsible for the modifications or removal of modifications. Rpd3 has multiple possible targets among the residues that affect tgm silencing, including H3K9, H3K18, H4K5, H4K8 and H4K12. In contrast Hos1 and Glc7 have only one identified target each in this region, at H4K12 and H3S10, respectively. One reasonable model for the requirements for Rpd3, Hos1, and Glc7 is that they are required to deacetylate and dephosphorylate residues in the H3/H4 tails to allow subsequent association and action of Sth1/RSC. On the surface, a requirement for deacetylation would seem at odds with data suggesting histone H3 N-terminal tail acetylation encourages RSC association through its bromodomain (Chatterjee et al., 2011), but the extensive number of potential target residues in the tails, especially for Rpd3, suggests that a more textured understanding of modification combinations might be required. Alternatively, acetylation and/or phosphorylation might affect other proteins, which in turn affect RSC activity. Rpd3, in particular, has genetic interactions with a large set of genes and affects acetylation of an overlapping set of gene products (Duffy et al., 2012). Among these are genetic interactions with RSC components Rsc1, Rsc3 and Rsc30, and acetylation of Rsc3, Rsc4, and Rsc30, suggesting that direct modification of RSC could contribute to the silencing even if individual subunits are not absolutely required. Similar alternative explanations might be possible for Hos1 and Glc7 as well, including the possibility that members of the extended pol III complex itself are targets. To address these questions convincingly, extensive analysis will be required of the modified histone and other protein residues adjacent to active vs. inactive tRNA genes under conditions that promote or alleviate silencing.
2.3. Communication between the tRNA gene and the chromatin
It is not clear at this time how the presence of an active tRNA gene transcription complex is communicating with nearby chromatin, presumably through these modification and remodeling activities. One intriguing possibility is suggested by the fact that Maf1, a direct target of the TOR signaling pathway, is required for tgm silencing. Activated Maf1 is thought to bind to pol III in the nucleus and interfere in communication between pol III and transcription factors, decreasing tRNA gene transcription (Desai et al., 2005; Moir et al., 2006; Roberts et al., 2006). When MAF1 is deleted, tRNA transcription increases, which we originally expected would intensify silencing near tRNA genes. When tgm silencing was instead abolished by maf1Δ (Park et al., 2002), it indicated Maf1 was exerting an influence beyond its physical interaction with pol III and the tRNA gene-bound transcription factors. A link between Maf1 and RSC is thought to be mediated through the Rsc9 subunit (Damelin et al., 2002), consistent with the involvement of the RSC complex in transcriptional regulation in response to TOR signaling. The fact that Rsc9 is the only non-Sth1 subunit that affects tgm silencing suggests a link between Maf1 and RSC, though no direct physical interactions have been reported.
The exclusion of nucleosomes from tRNA genes by the massive pol III transcription complexes, as well as the tendency of nucleosomes to reside fixed distances from tRNA genes has been reported in yeast (Hull et al., 1994; Venters et al., 2011). The present results, in combination with studies of tRNA genes as chromatin boundary elements, indicate that there is an interaction with chromatin modification machinery to influence the behavior of those nucleosomes. Unlike boundary element function, which requires only bound TFIIIC (Noma et al., 2006; Simms et al., 2008; Valenzuela et al., 2009), tgm silencing seems to require the presence of the full RNA polymerase III complex (pol III, TFIIIC, TFIIIB, 1), containing at least 25 polypeptides. Thus, it is not surprising that silencing near the tRNA genes involves interactions with activities not observed in boundary function. Since both tRNA genes and some tRNA-derived short interspersed elements (SINEs) have been shown to have boundary element properties in larger eukaryotic genomes (Lunyak et al., 2007; Raab et al., 2011), it will be interesting to examine whether the other interactions with local chromatin structure are retained as well in these highly reiterated elements.
3. Methods
3.1. Silencing reporter plasmid construction
The pSUP4-nat1 plasmid was created by S. cerevisiae gap repair of a CEN/URA3 plasmid previously used to test silencing of a HIS3 reporter gene (Hull et al., 1994) by gap repair, precisely replacing the HIS3 coding region with that of nat1. The nat1 gene was amplified from pAG25 (Goldstein and McCusker, 1999) with 50 bp of sequence on each end precisely corresponding to the sequence before and after the HIS3 coding region in the plasmid. The nat1 insert was transformed into BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Brachmann et al., 1998) along with pSUP4-o (Hull et al., 1994) that had been cleaved at unique SfiI and BsiI sites in the HIS3 ORF. Gap repaired plasmids were selected on SD-ura. Insertions in the resulting plasmid candidates were verified by sequencing, and as expected the nat1 expression was fully silenced in BY4741 by the presence of the tRNA gene. Generally, we find the nat1 reporter phenotype to be more sensitive to release of silencing than the HIS3 reporter.
3.2. Screening for histone mutations that result in alleviation of tRNA gene-mediated silencing
Individual yeast strains in the scanning histone mutagenesis with alanine (SHIMA) library (Nakanishi et al., 2008) were used to test tgm silencing, and have been described previously. Briefly, H2A and H2B substitutions were constructed in FY406: [MATa (hta1-htb1)::LEU2, (hta2-htb2)::TRP1, his3Δ200, leu2Δ1, ura3-52, trp1Δ63, lys2-128]. Wild type histone genes were present on plasmid pSAB6 (HTA1-HTB1-URA3). In strains bearing mutant histones, a plasmid containing the single alanine substitution in one of the histone genes (pZS145 HTA1-Flag + HTB1, CEN HIS3) was added by transformation and the original pSAB6 removed by growth on 5-fluoroorotic acid (5-FOA). H3 and H4 mutations similarly were tested in YBL574: [MATa, leu2Δ1, his3Δ200, ura3-52, trp1Δ63, lys2-128δ, (hht1-hhf1) Δ::LEU2, (hht2-hhf2) Δ::HIS3, Ty912Δ35-lacZ::his] originally containing plasmid pDM9 (HHT1-HHF1-URA3) that had been replaced with pWZ414-F12 (HHT2 + HHF2, TRP1) containing the single alanine substitution. SHIMA library strains were transformed with pSUP4-nat1 using a modified version of lithium acetate transformation (Becker and Lundblad, 1993). Transformed cells were recovered in SD-ura, plated to SD-ura, and grown on SGR-ura (+ galactose + raffinose, -uracil) synthetic media prior to phenotypic assay. Library strain transformants were assayed on SGR-ura + cloNAT (nourseothricin 150 μg/mL, Werner BioAgents) at 30 °C and compared to control strains that contained plasmids with wild type HTA1 and HTB1 or HHT2 and HHF2 along with pSUP4-nat1. To eliminate the possibility that a cis mutation in the pSUP4-nat1 plasmid caused the phenotype, the strains were cured of pSUP4-nat1 on 5-FOA, retransformed with the pSUP4-nat1 plasmid and the phenotypes retested.
To further ensure the identity of the SHIMA library mutants and eliminate the possibility of second-site mutations in the genome, the cured strains were used to prepare DNA (Kaiser et al., 1994) that was transformed into Escherichia coli, followed by selection for the plasmid. E. coli transformants were grown in liquid LB + ampicillin (100 μg/mL) and used to prepare plasmid DNA (Qiagen, QIAprep Spin Miniprep Kit). Histone genes were sequenced using the following primers: HTA1seqF, HTA1seqR, HTB1seqF, HTB1seqR, HHT2seqF, HHT2seqR, HHF2seqF, HHF2seqR (Oler et al., 2010) The sequenced mutant SHIMA histonebearing plasmids were re-assayed for their ability to alleviate silencing in the original histone-deleted strains containing pSUP4-nat1. The hta1 and htb1 mutant plasmids were transformed into FY406a that was then cured of pSAB6 containing wild type HTA1 and HTB1 genes, by successively culturing the cells twice on 5′FOA media. The cured cells were transformed with pSUP4-nat1 and assayed as previously described. The same procedure was followed for mutant hht2 and hhf2 plasmids with the exception that YBL574a was used.
Many of the histone mutants displayed a silencing alleviation phenotype that was variable in terms of the percentage of cells that could grow on SGR-ura + cloNAT. Due to this variability, all the histone mutants were tested multiple times for alleviation of tgm silencing. Alanine substitutions that alleviated tgm silencing were mapped to the nucleosome structure (White et al., 2001) using PyMol (http://www.pymol.org/), with two views shown in Fig. 3. PyMol files of the structures are available on request.
3.3. Testing additional mutations for alleviating tgm silencing
A number of mutations and gene deletions were previously tested for alleviating tgm silencing (D’Ambrosio et al., 2008; Wang et al., 2005) and are summarized in Table 1 along with newly tested mutants. Mutations in additional genes summarized in Table 1 were tested compared to parental wild type strains in all cases, using the same tRNASUP4-nat1 reporter plasmid. Strains with the indicated gene deletions were obtained from the S. cerevisiae deletion library (Open Biosystems). Conditional sth1 mutants (Ben-Aroya et al., 2008; Titus et al., 2010) were the gift of Susan Wente. The three ts mutations have been characterized previously (Ben-Aroya et al., 2008), and two additional sth1 conditional mutants (BLY491 containing sth1-L1346A and SWY4143 containing sth1-F793S, (White et al., 2001)) gave similar loss of silencing phenotypes (not shown). The parent and mutant strains having the conditional glc7-10 allele (Andrews and Stark, 2000) were the gift of Michael Stark. Conditional rsc9 and rsc58 mutants and multiple control strains (Ben-Aroya et al., 2008) were the gift of Philip Hieter. Loss of the URA3 marker in these strains (Ben-Aroya et al., 2008) was selected on 5-fluoroorotic acid media and the strains were retested for retaining the ts phenotype before transformation with the tRNASUP4-nat1 test plasmid with a URA3 marker. Conditional uba2-ts and ubc9-ts mutants and parent (JD52) strains were obtained from Erica Johnson (Johnson and Blobel, 1997). Conditional esa1-ts mutant (LPY4679) and parent (LPY5) were obtained from Lorraine Pillus (Chang and Pillus, 2009). Mutants in smt3, ulp1, and uba1 and parent strain (FY86) were obtained from David Tollervey (Lambermon et al., 2002; Russell and Tollervey, 1992). Other mutant strains that did not relieve silencing are from the sources noted in Table 1.
Acknowledgments
We thank Ray Trievel for help with nucleosome modeling and for critical reading of the manuscript, Li Wang for technical assistance with assays. We are grateful to Philip Hieter, Erica Johnson, Lorraine Pillus, Michael Stark, Laura Titus, David Tollervey, and Susan Wente for gifts of yeast strains, and Ali Shilatifard for the gift of the SHIMA library and helpful discussions. This work was supported by NIH Grant GM082875 to DRE, by the NIH University of Michigan Genetics Predoctoral Training Grant (T32GM07544) to DAP, and by support from the University of Michigan Undergraduate Research Opportunity Program for SRR and BC.
Abbreviations
- Ac
acetylation
- ATPase
enzyme hydrolyzing adenosine triphosphate
- C terminus
carboxy terminus
- CK2
casein kinase 2
- FOA
fluoroorotic acid
- H2A
histone H2A
- H2A.Z
histone variant H2A.Z
- H2B
histone H2B
- H3
histone H3
- H4
histone H4
- Me
methylation
- ORF
open reading frame
- P
phosphorylation
- pol II
RNA polymerase II
- pol III
RNA polymerase III
- Rpd3L
protein complex containing Rpd3 protein
- Rpd3S
protein complex containing Rpd3 protein
- RSC
chromatin remodeling complex
- SD
synthetic yeast media with dextrose
- SGR
synthetic yeast media with galactose and raffinose
- SHIMA
scanning histone mutagenesis with alanine
- SINE
short interspersed element
- tgm
tRNA gene-mediated
- TFIIIB
RNA polymerase III transcription factor IIIB
- TFIIIC
RNA polymerase III transcription factor IIIC
- TOR
Target of Rapamycin
- tRNA
transfer RNA
- ura
uracil
Footnotes
Competing financial interests The authors declare no competing financial interests.
Author contributions: P.D.G. designed and either directly supervised or carried out the majority of the experiments.
A.K. helped design, carry out and interpret the experiments.
J.I.-H. carried out the original screen of the library to indicate nucleosome structure was involved.
E.L.M. re-created recombinant structures and strains and retested phenotypes.
D.A.P. contributed to interpretation of the results and creation of the manuscript.
S.R. back-checked the mutant histone sequences and retransformed strain phenotypes.
B.C. re-tested the phenotypes of many of the mutants that did and did not alleviate silencing for quality control and to provide figure panels.
D.R.E. supervised design and interpretation of the experiments and participated in writing and editing the manuscript.
All authors have read and provided editing suggestions for the manuscript.
References
- Andrews PD, Stark MJR. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J. Cell Sci. 2000;113:507–520. doi: 10.1242/jcs.113.3.507. [DOI] [PubMed] [Google Scholar]
- Becker DM, Lundblad V. Introduction of DNA into yeast cells. Curr. Protoc. Mol. Biol. 1993:13.7.1–13.7.2. doi: 10.1002/0471142727.mb1307s27. [DOI] [PubMed] [Google Scholar]
- Ben-Aroya S, Coombes C, Kwok T, O’Donnell KA, Boeke JD, Hieter P. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell. 2008;30:248–258. doi: 10.1016/j.molcel.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton EC, Boeke JD. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 2003;13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Chaban Y, et al. Nat. Struct. Mol. Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Pillus L. Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae. Genetics. 2009;183:149–160. doi: 10.1534/genetics.109.103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Sinha D, Lemma-Dechassa M, Tan S, Shogren-Knaak MA, Bartholomew B. Histone H3 tail acetylation modulates ATP-dependent remodeling though multiple mechanisms. Nucleic Acids Res. 2011;39:8378–8391. doi: 10.1093/nar/gkr535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-F, et al. The Rpd3 core complex is a chromatin stabilization module. Curr. Biol. 2012;22:56–63. doi: 10.1016/j.cub.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, et al. The genome-wide localization or Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell. 2002;9:563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- Desai N, Lee J-H, Upadhya R, Chu Y, Moir RD, Willis IM. Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J. Biol. Chem. 2005;280:6455–6462. doi: 10.1074/jbc.M412375200. [DOI] [PubMed] [Google Scholar]
- Dhillon N, et al. DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J. 2009;28:2583–2600. doi: 10.1038/emboj.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Wnf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry M, Loiodice I, Chen CL, Thermes C, Taddei A. Tight protein–DNA interactions favor gene silencing. Gene Dev. 2011;25:1365–1370. doi: 10.1101/gad.611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey RN, Gartenberg MR. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 2007;21:2150–2160. doi: 10.1101/gad.1583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy SK, et al. Exploring the yeast acetylome using functional genomics. Cell. 2012;149:936–948. doi: 10.1016/j.cell.2012.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grigat M, Jaschke Y, Kliewe F, Pfeifer M, Walz S, Schuller HJ. Multiple histone deacetylases are recruited by corepressor Sin3 and contribute to gene repression mediated by Opi1 regulator of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics. 2012;287:461–472. doi: 10.1007/s00438-012-0692-x. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar D, Kamakaka RT. tRNA genes as chromatin barriers. Nat. Struct. Mol. Biol. 2006;13:192–193. doi: 10.1038/nsmb0306-192. [DOI] [PubMed] [Google Scholar]
- Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Mol. Cell. Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9 is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 1994. p. 234. [Google Scholar]
- Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13108–13113. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland JG, Kamakaka RT. tRNA insulator function: insight into inheritance of transcription states? Epigenetics. 2010;5:96–99. doi: 10.4161/epi.5.2.10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W, et al. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- Lambermon MHL, Fu Y, Kirk DAW, Dupasquier M, Filipowicz W, Lorkovic ZJ. Uba1 and Uba2, two proteins that interact with Ubp1, a multifunctional effector of pre-mRNA maturation in plants. Mol. Cell. Biol. 2002;22:4346–4357. doi: 10.1128/MCB.22.12.4346-4357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Dewari PS, Bhardwaj A, Bhargava P. Yeast H2A.Z, FACT complex and RSC regulate transcription of tRNA gene through differential dynamics of flanking nucleosomes. Nucleic Acids Res. 2011;39:4023–4034. doi: 10.1093/nar/gkq1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Moir RD, Lee JH, Haeusler RA, Desai N, Engelke DR, Willis IM. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf 1. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15044–15049. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RH, Roth S, Simpson RT. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell. Biol. 1992;12:4015–4025. doi: 10.1128/mcb.12.9.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Kamakaka RT. The human pol III transcriptome and information flow. Nat. Struct. Mol. Biol. 2010;17:539–541. doi: 10.1038/nsmb0510-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol. Cell. Biol. 2004;24:1956–1967. doi: 10.1128/MCB.24.5.1956-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Cosgrove M, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab JR, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2011;31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR. Dephosphorylation and genome-wide association of Maf with Pol III-transcribed genes during repression. Mol. Cell. 2006;22:633–644. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, et al. Microarray deacetylation maps determine genome-wide functions for histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Rodley* CDM, Pai* DA, Mills TA, Engelke DR, O’Sullivan JM. tRNA gene identity affects nuclear positioning. PLoS One. 2011;6:e29267. doi: 10.1371/journal.pone.0029267. (* = equal contribution) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell ID, Tollervey D. Nop3 is an essential yeast protein that is required for rRNA processing. J. Cell Biol. 1992;119:737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms TA, et al. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot. Cell. 2008;7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J, et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:139–141. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol. Biol. Cell. 2010;21:1072–1087. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–148. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Venters BJ, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR. Silencing near tRNA genes requires nucleolar localization. J. Biol. Chem. 2005;280:8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Sutto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]