Abstract
Signaling via MET receptor tyrosine kinase (MET) has been implicated in a number of neurodevelopmental events, including cell migration, dendritic and axonal development and synaptogenesis. Related to its role in the development of forebrain circuitry, we recently identified a functional promoter variant of the MET gene that is associated with autism spectrum disorder (ASD). The association of the MET promoter variant rs1858830 C allele is significantly enriched in families with a child who has ASD and co-occurring gastrointestinal conditions. The expression of MET in forebrain had been mapped in detail in developing mouse and rhesus macaque. However, in mammal, its expression in the developing brainstem has not been studied extensively throughout developmental stages. Brainstem and autonomic circuitry are implicated in ASD pathophysiology and in gastrointestinal dysfunction. To advance our understanding of the neurodevelopmental influences of MET signaling in brainstem circuitry development, we employed in situ hybridization and immunohistochemistry to map the expression of Met and its ligand, Hgf, through prenatal development of the mouse midbrain and hindbrain. Our results reveal a highly selective expression pattern of Met in the brainstem: including a subpopulation of neurons in cranial motor nuclei (nVII, nA, and nXII), B6 subgroup of the dorsal raphe, Barrington’s nucleus, and a small subset of neurons in the nucleus of solitary tract. In contrast to Met, neither full length nor known splice variants of Hgf were localized in the prenatal brainstem. RT-PCR revealed Hgf expression in target tissues of Met-expressing brainstem neurons, suggesting that MET in these neurons may be activated by HGF from peripheral sources. Together, these data suggest that MET signaling may influence the development of neurons that are involved in central regulation of gastrointestinal function, tongue movement, swallowing, speech, stress, and mood.
Keywords: autism, motor neurons, vagal complex, raphe neurons, synaptogenesis
Introduction
Met receptor tyrosine kinase (MET) and its ligand, hepatocyte growth factor (HGF) were first cloned approximately 30 years ago [1-5]. Initially, MET was characterized as a receptor tyrosine kinase with proto-oncogenic function and HGF (also named scatter factor) was found as a serum- and platelet-derived protein capable of inducing cell proliferation and migration in vitro [2,5,6]. Expression of MET and HGF has been observed in most solid tumors and MET/HGF signaling is well-characterized in human malignancies and metastasis [7,8]. Nonetheless, MET and HGF are also involved in the normal development and regeneration of several different organs [9-14].
More recently, MET signaling has been implicated in a number of neurodevelopmental events, including cell migration, dendritic and axonal development and synaptogenesis (see [15] for review). Relevant to a role for MET signaling in the development of specific forebrain circuits [16,17], a functional promoter variant of the MET gene is associated with autism spectrum disorder (ASD) [18-20]. The association of the MET promoter variant rs1858830 C allele is further enriched in families in which a child has ASD with co-occurring gastrointestinal conditions [21]. The MET promoter C variant reduces gene transcription in vitro [18], and in postmortem brains of individuals with ASD, MET transcript and protein are significantly decreased [22,23]. MET currently is categorized as a strong risk candidate for ASD ([24]; also see SFARI Gene https://gene.sfari.org/autdb/GS_Home.do).
The expression patterns of MET and its transcript (Met) in mouse forebrain was mapped in detail recently [25]. We showed that the Met transcript is located in specific populations of excitatory projection neurons within the developing mouse neocortex and subcortically in a very limited number of limbic system regions. Temporally, transcript and protein expression in the forebrain is transient, first detected late in gestation, and peaking between postnatal day 7 (P7) to 14. This is a period of active neurite outgrowth and synaptogenesis in the brain. There is strong temporal conservation of MET expression between the mouse and rhesus macaque forebrain [15,26], with conserved subcortical but different regional expression in the neocortex.
Brainstem and autonomic circuitry has been implicated in ASD pathophysiology [27-31] and in central origins of gastrointestinal dysfunction [32-35]. Since the association of the MET promoter variant is further enriched in ASD with co-occurring gastrointestinal conditions [21], MET may be a point of functional convergence of these conditions. A detailed mapping of MET gene expression in the developing brainstem may reveal how the receptor may be involved in peripheral autonomic and homeostatic functions. In mice, there has been limited Met expression mapping in several motor nuclei, i.e. trochlear motor (nIV), trigeminal (nV), superior salivatory (part of nVII), glossopharyngeal (nIX), vagus (nX), cranial accessory (nXI), and hypoglossal nuclei (nXII), from E11-E13 [36], at a time just after neuronal birth (>E9-10; [37]). While not examined beyond E13, these data are similar to its expression pattern in developing forebrain, in which Met expression rises during the period of extensive neurite growth. Supporting a functional role for MET signaling in early (prior to E13) brainstem development, HGF has been shown to be a potent axon chemoattractant in mouse mid/hindbrain explants [36]. Furthermore, limited analysis of Met and Hgf knockout mice revealed abnormal projections of hypoglossal nerve (XII) [36] and spinal motor nerves [38]. Knockdown of Met in developing zebrafish by morpholinos leads to altered facial motor neuron migration and differentiation [39]. Nonetheless, the expression pattern and function of Met in mouse brainstem development after E13, is unclear.
The expression of HGF in the developing mammalian central nervous system (CNS) is largely unknown. HGF expression analysis is rather complex, because in addition to its full length mRNA, several splice variants have been described [40-42]. Some of these HGF splice variants can bind and activate MET [40,41,43]. In human, mutations affecting several of the HGF splice variants are shown to impact hearing development [42]. The spatiotemporal regulation of MET signaling during development will depend on HGF availability in the tissue; thus, determining the developmental expression pattern of Hgf and its variants may help us to further understand the roles of HGF/MET signaling in CNS development.
In this study we examined prenatal expression of Met and Hgf in the mouse, focusing on the brainstem. Transcript and protein mapping reveal highly selective Met expression patterns, including subpopulations of neurons involved in regulating gastrointestinal function, tongue movement, swallowing, speech, stress, and mood. In contrast, Hgf and several of its splice variants were not detected in the brainstem, but instead were expressed in peripheral targets of MET-expressing brainstem neurons. In rostral aspects of the neuraxis, we found Hgf expression largely confined to periventricular regions of the forebrain. The developmental implications of these unique expression patterns are discussed.
Materials and Methods
Animals
CD1 (Crl:CD1[ICR]), Isl1tm1(cre)Sev ([44], also known as Isl1cre; generously provided by Dr. Sylvia Evan at University of California, San Diego), B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J ([45], also known as Rosa-tdTomatofx, purchased from The Jackson Laboratory, Bar Harbor, ME), and Fevtm1Esd ([46], also known as Pet-1, generously provided by Dr. Evan Deneris at Case Western Reserve University School of Medicine) mice were used in this study. Time pregnant CD-1 mice were purchased from Charles River, Wilmington, MA. Time pregnant transgenic mice were bred in-house. Mice were maintained on a 12-hour light/12-hour dark cycle with free access to food and water. The day following a time-delimited overnight pairing was considered E1. Pregnant females were deeply anesthetized with isofluorane vapors followed by rapid decapitation in order to harvest embryonic tissues. All experimental procedures using animals were approved by the Institutional Animal Care and Use Committee at the University of Southern California and conformed to US National Institutes of Health guidelines.
In situ hybridization
For in situ hybridization (ISH), embryos were dissected in cold PBS, fixed in 4% paraformaldehyde in PBS (pH 7.4) overnight at 4°C and transferred to 30% sucrose/PBS for cryoprotection. Brains were embedded in TFM tissue freezing medium (Triangle Biomedical Sciences, Inc., Durham, N.C.) over liquid nitrogen vapors and then stored at −80°C until sectioned at 20 μm with a cryostat. Digoxigenin-labeled cRNA probes were: Met probe ([25]; 2665-4051bp of GenBank #NM_008591), mouse Hgf (176-966bp of GenBank #AK042121), and mouse Isl1 (704-1307 of GenBank # BC132263). ISH on sections was performed as described [47]. Briefly, slides were fixed in 4% PFA/PBS (20 min) at room temperature (RT), washed in PBS, and followed by proteinase K treatment (1-1.25 μg/ml, RT) for 15 min. Acetylation was performed by incubating slides under agitation for 10 min in triethanolamine (TEA-HCl, pH8.0) after drop by drop addition of acetic anhydrate (0.25% of TEA volume). Slides were pre-hybridized for at least 2 hr at 60°C in hybridization solution (50% deionized formamide, 5× SSC, pH 7.0, 1× Denhardt’s, 0.1% Tween-20, 0.1% CHAPS, 5 mM EDTA, pH8.0, 100 μg/ml heparin, 300 μg/ml yeast t-RNA in DepC-H2O). The hybridization step was carried out for 16-18 hr at 60°C in hybridization solution. Slides were then washed 3 times, 45 min (3 × 45 min) at 65°C in washing solution (2×SSC, pH4.0, 50% formamide, 1% SDS in distilled water), then 3 × 15 min in TBST (25 mM Tris-HCL, pH 7.5, 136 mM NaCl, 2.68 mM KCl, 1% Tween-20 in distilled water) with light agitation at RT. Slides were blocked for 1 hr at RT with blocking reagent (100 mM Tris-HCl, pH 7.5, 150 mM NaCl containing 1.5% blocking reagent from Roche, Indianapolis, IN), and followed by incubation at 4°C overnight with alkaline phosphatase-conjugated anti-DIG Fab fragments (1:2000, Roche, Indianapolis, IN). Color development was carried out at RT in NTMT solution (100 mM NaCl, 100 mM Tris-HCl, pH 9.5, 25 mM MgCl2, 1% Tween-20, 2 mM Levamisole) with 0.2 mM 5-bromo-4-choloro-3-indolyl phosphate (BCIP) and 0.2 mM nitroblue tetrazolium (NBT) (Roche).
RT-PCR
Total RNA from the E13 and E16 CD-1 mouse whole brain, the remaining head tissue, gut, whole spinal cord, or heart was extracted with an RNeasy kit (Qiagen, Valencia, CA) per manufacturer’s recommendation and reverse-transcribed using oligoT primers and Superscript III reverse transcriptase (Life Technology, Grand Island, NY) after DNase treatment (Turbo DNA-free kit, Life Technology, grand Island, NY). Resulting cDNAs were analyzed by PCR, using the following primers:
panHgf: forward primer, 5′- GCATGTCCTCCTGCACCTCCTCCTGCT -3′;
reverse primer, 5′- TCCCCCTTCTTCCCCTCGAGGATTTCG -3′.
NK1: forward primer, 5′- TGGGGGACCAAACTTCTGCCGGTCCT -3′;
reverse primer, 5′- CGTCGGGGTAGCACCCCATGTGA -3′;
Intron4: forward primer, 5′-CCCTCGGGGTTTGCATACTG-3′;
reverse primer, 5′-CTGGGTGTTCCCCCTCCTTT-3′.
Immunostaining
Embryos, processed as for in situ analysis, were cryosectioned at 12 or 20 μm; sections were permeabilized and blocked with 0.3% Triton X-100 in 5% normal donkey serum (Jackson ImmunoResearch Laboratories) and incubated overnight at 4°C with primary antibodies: goat anti-MET (Final concentration: 2 μg/ml; R&D systems, Minneapolis, MN; AF527), 1:1000 rabbit anti-serotonin (5-HT) (Final concentration: 58 μg/ml; Sigma, St. Louis, MO; S5545), 1:1000 rabbit anti-tyrosine hydroxylase (TH) (Millipore, Billerica, MA; AB152), 1:1000 rabbit anti-corticotropin releasing hormone (CRH; kind gift of Dr. Ann Silverman at Columbia University College of Physicians and Surgeons), 1:200 rabbit anti-peripherin (Millipore, Billerica, MA; AB 1530), 1:50 rabbit anti-Pax2 (Invitrogen, CA; 18-0483). After extensive washing in PBS with 0.2% Tween-20, slides were incubated at room temperature for 1 hr in secondary antibodies: Alexa 488-conjugated donkey anti-rabbit (1:200; Life Technology), biotin-conjugated donkey anti-goat (1:500, Jackson ImunoResearch Laboratories), and Dylight488- or Dylight549-conjugated streptavidin (1:500; Jackson ImmunoResearch Laboratories).
Anatomical Mapping and Light Microscopy
The anatomical labeling of brainstem structures were determined by comparing the expression patterns on serial sections from several experiments with Chemoarchitectionic Atlas of the Developing Mouse Brain (Jacobowitz and Abbott 1997) and with Isl1 expression on adjacent sections at E13 (supplemental fig. 1). Terminology conformed to that used in the referenced atlas as well as Caton et al, 2000 [36]). Images were acquired with a Zeiss AxioCam MRm camera (Carl Zeiss, USA), using Zeiss Axiovision 4.1 software (Zeiss). Figures were prepared digitally in Adobe Photoshop CS5.1 (Adobe Systems Incorporated, San Jose, CA).
Results
Met expression in the cranial motor nuclei at late gestational stage
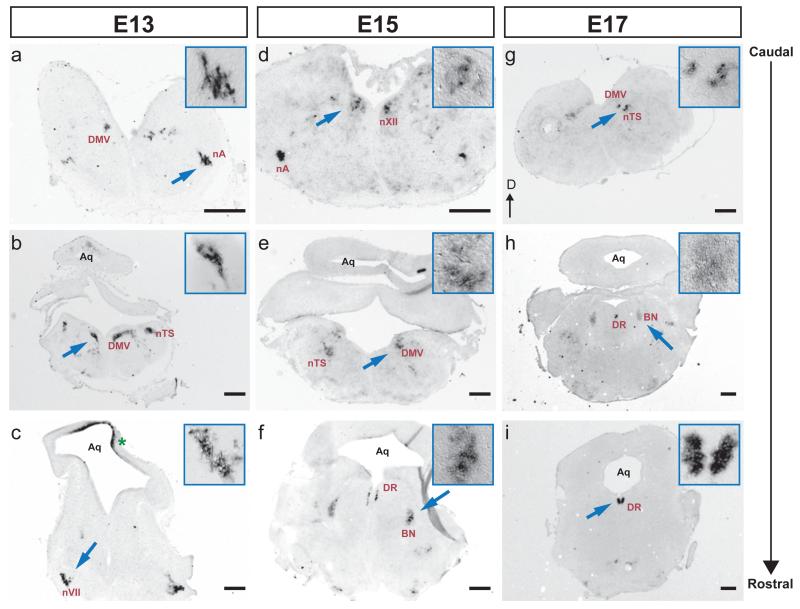
Previously, it was shown that Met is expressed by subsets of cranial motor nuclei in the early developing brainstem (mouse E11-E13, [36]). To determine whether expression of Met mRNA in these motor nuclei persists after E13, we performed in situ hybridization on serial coronal sections at E13, 15, 17, and P0. Met+ regions were defined by the expression patterns on serial sections from several experiments and the locations confirmed with the Chemoarchitectionic Atlas of the Developing Mouse Brain (Jacobowitz and Abbott 1997). At all ages examined, Met transcripts were detected in regions overlapping with subsets of motor nuclei (fig. 1a-e, and g). In all nuclei that were labeled, it appeared that not all the neurons in a specific nucleus were Met+. To confirm this, we compared the location of Met-expressing neurons with that of Islet-1 (Isl1) detected on adjacent sections (supplemental fig. 1a-f). Isl1 is expressed by all cranial motor neurons at early prenatal ages, and has been widely used as a marker of developing cranial motor neurons in several species [36], [48]. By comparing labeling patterns of Met with Isl1 expression regions, we found that at E13, Met transcripts were somatically localized to brainstem regions containing the dorsal motor nucleus of the tenth nerve (DMV or dmnX), nucleus ambiguous (nA; the motor nucleus of the spinal accessory, XI, glossopharyngeal, IX, and vagus nerves, X), nucleus of the hypoglossal nerve (nXII) as well as the nucleus of facial nerve (nVII) (fig. 1a-c, and Table 1). This is in agreement with the previous report [36] showing that Met transcripts are present in subset of cranial motor nuclei. Furthermore, similar to what was observed previously by Caton et al., Met expression in nVII did not label the neurons of branchial motor division. Rather, stained neurons occupied a position lateral to this region. The position of these Met+ neurons appears to overlap with the developing superior salivatory nucleus (visceral motor of nVII) (fig. 1c, [36]). Interestingly, Met staining was either very weak or absent in neurons residing in the oculomotor (nIII), trochlear (nIV) and trigeminal nuclei (nV) (supplemental fig. 1g and data not shown). This finding was surprising, given that expression of the Met transcript was reported in limited number of neurons present within the boundaries of nVI and nV from E11 to E13 [36]. It is possible that Met expression in these nuclei is below levels of detection on 20-micron sections compared with wholemount tissues used in previous studies.
Fig. 1.
Distribution of Met mRNA in the developing mouse brainstem. Series of coronal sections through the caudo-rostral extent of E13 (a, b, c), E15 (d, e, f), and E17 (g, h, i) brainstem hybridized with DIG-labeled riboprobe to the MET receptor tyrosine kinase. Arrows point to areas shown at higher magnification (boxed). Dorsal (D) is to the top. DMV, dorsal nucleus of the tenth nerve (motor, vagus); nA, ambiguous nucleus; nTS, nucleus of the solitary tract; DR, dorsal raphe nuclei; BN, Barrington’s nucleus; Aq, Aqueduct; nVII, nucleus of the seventh nerve; nXII, nucleus of the twelfth nerve (hypoglossal). Met expression was also detected in the superior colliculus (asterisk). The anatomical labeling of brainstem structures were determined by comparing the expression patterns on serial sections from several experiments with Chemoarchitectionic Atlas of the Developing Mouse Brain (Jacobowitz and Abbott 1997) and with Isl1 expression on adjacent sections at E13 (supplemental fig. 1). Scale bars=30 μm. Insert: a-d and g, magnified 3 times; e, f, h, and i, magnified 4 times.
Table 1.
Summary of Met expression in the mid/hindbrain nuclei at different developmental periods
| Nuclei | Function | E13 | E14-E17 | E18-P0 |
|---|---|---|---|---|
| nVII | GVE, SVA, SVE/BE | + | +/− | − |
| nA | SVE/BE | + | + | +/− |
| DMV | GVA, GVE, SVA | + | + | +/− |
| nXII | GSE | + | + | +/− |
| nTS | GVA | + | + | +/− |
| BN | Visceral related, stress related | − | + | +/− |
| DR | Stress related, mood, autonomic regulation | − | + | + |
GVE, general visceral efferent: regulates autonomic innervation of smooth muscles, cardiac muscles, and glands
SVA, special visceral afferent: mediate sensations of taste from tongue
SVE/BE, control gill-related muscles of face, pharynx, larynx, and neck
GVA, general visceral afferent: mediates sensations of pain and temperature from visceral organs
GSE, general somatic efferent: controls muscles derived from glossal muscles
+, expression detected
−, no expression detected
+/−, weak expression detected
We were most interested to know whether Met expression in these motor nuclei was maintained at E15 and beyond. We observed that with the exception of nVII, in which Met mRNA levels greatly diminished after E13, expression of the transcript was evident at E15 and E17 (fig. 1d, e and g). However, by P0, detectable Met transcript labeling in neurons located within regions of nA, DMV, and nXII had largely disappeared (Table 1).
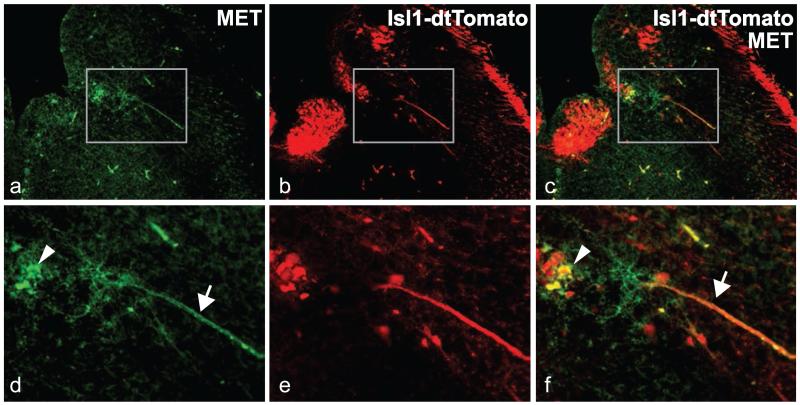
In a second set of developmental mapping studies, we used an antibody to MET combined with an endogenous fluorescent reporter strategy, in order to more precisely determine the cellular localization of the MET protein. Immunofluorescence staining was performed on sections through the mid- and hindbrain from Isl1cre/Rosa-tdTomatofx/+ embryos obtained by crossing Isl1cre/+ mice [44] with Rosa-tdTomatofx/fx mice [45]. In Isl1cre/Rosa-tdTomatofx/+ embryos, all neurons in cranial motor nuclei express tdTomato, which we confirmed by double staining using an antibody directed against peripherin, a peripheral and motor neuron specific type III intermediate filament that is widely used as a marker for postmitotic cranial motor neurons ([49-51]; supplemental fig. 2 and data not shown). Cellular immunolocalization of MET was compared with neuronal labeling of cranial motor nuclei by tdTomato. Similar to in situ hybridization observations, at E16 only a subpopulation of neurons in the DMV, nXII, and nA was both tdTomato+ and MET+ (fig. 2). In DMV, double-immuno-positive neurons tended to be located in ventral regions (fig. 2j, k; arrowheads). No MET immunoreactivity was found colocalized with tdTomato+ in nIII, nIV, nV, and nVI (supplemental fig. 3). As noted previously for forebrain neurons [25], MET protein was located at a cellular level in both neuronal somata (fig. 3, arrowheads) and their axons, which were evident coursing through the brainstem toward the periphery (white arrows).
Fig. 2.
MET protein expression in neurons residing in DMV, nXII, and nTS. Micrographs show MET immunofluorescence in a series of coronal sections harvested from Isl1cre/Rosa-tdTomatofx/+ mice at E16. a-c, endogenous tdTomato fluorescence, expressed under the control of Isl1cre at 3 successive levels of the medulla; d-f, MET immunostaining on the same sections; g-i overlay of tdTomato and MET immunofluorescence. Regions in white boxes are presented at higher magnification in j-l. VN, vestibular nucleus. Arrowheads indicate subsets of MET+/tdTomato+ neurons in nX (j and k) and in nA (l). Arrows point to a subset of MET+/tdTomato− neurons located lateral to DMV. An almost complete overlap of tdTomato and MET immunofluorescence is observed in the nA (l). No MET-immunoreactivity was found in the main VN (g) and in rostral part of DMV (i), which is indicated by an asterisk. Scale bar, a-i: 50 μm, j-l: 10 μm.
Fig. 3.
MET protein is localized to somata and axons in a subpopulation of ventrolaterally situated neurons within the boundary of DMV. Higher magnifications of boxed areas in a-c are shown in c-f. Immunostaining for MET (a, d) was performed on hindbrain section harvested from Isl-1cre/Rosa-tdTomatofx/+ E16 embryos. a and d, MET immunoreactivity; b and e, tdTomato fluorescence; c and f, overlay. Arrowheads point to MET+/tdTomato+ soma and arrows to axons that are double-labeled by anti-MET and tdTomato.
Met expression in other mid/hindbrain nuclei
In addition to cranial motor nuclei, Met transcripts were detected in several other mid/hindbrain nuclei that had not been described previously. Specific Met neuronal expression was observed in regions that coincided with the nucleus of solitary tract (nTS), the caudal part of the dorsal raphe (DR), and in a group of neurons near the locus coeruleus (LC), later identified as Barrington’s nuclei (BN; see below). The spatiotemporal patterns of Met expression in these various nuclei are detailed below:
nTS
In situ hybridization consistently showed Met transcripts were present from E13 to E17 in a group of neurons located lateral to the DMV (fig. 1 b, e, g). An E16, MET immunostaining in Isl1cre/Rosa-tdTomatofx/+ mice revealed the presence of MET+/dtTomato-neurons lateral to the MET+/dtTomato+ DMV neurons (fig. 2k, arrow). Based on their position in the developing brainstem, these may constitute a subpopulation of neurons within the nTS. The nTS is a brainstem sensory nucleus that sends and input from the facial nerve (VII) as well as receiving afferents from glossopharyngeal (IX) and vagus (X) nerves [52]. The nTS emerges rostrally from the dorsomedial edge of the spinal trigeminal nucleus and extends caudally to the border of spinal cord [53]. It has been shown that, during development, nTS neurons can be subdivided to two main groups based on the combination of transcription factors expressed [54]. At E13, the majority of nTS neurons express Phox2b, but a small subset of nTS neurons arising in rhombomere 7 (where DMV nuclei is located during early developmental stage) does not express Phox2b [54,55]. Instead these neurons express Pax2 [54]. We found that, at E13, the MET+ neurons adjacent to the DMV expressed Pax2 (supplemental fig. 4), suggesting that they situated within the developing nTS. As in the case of other nuclei, MET labeling was present only in a small subpopulation of nTS neurons.
Dorsal Raphe Nuclei (DR)
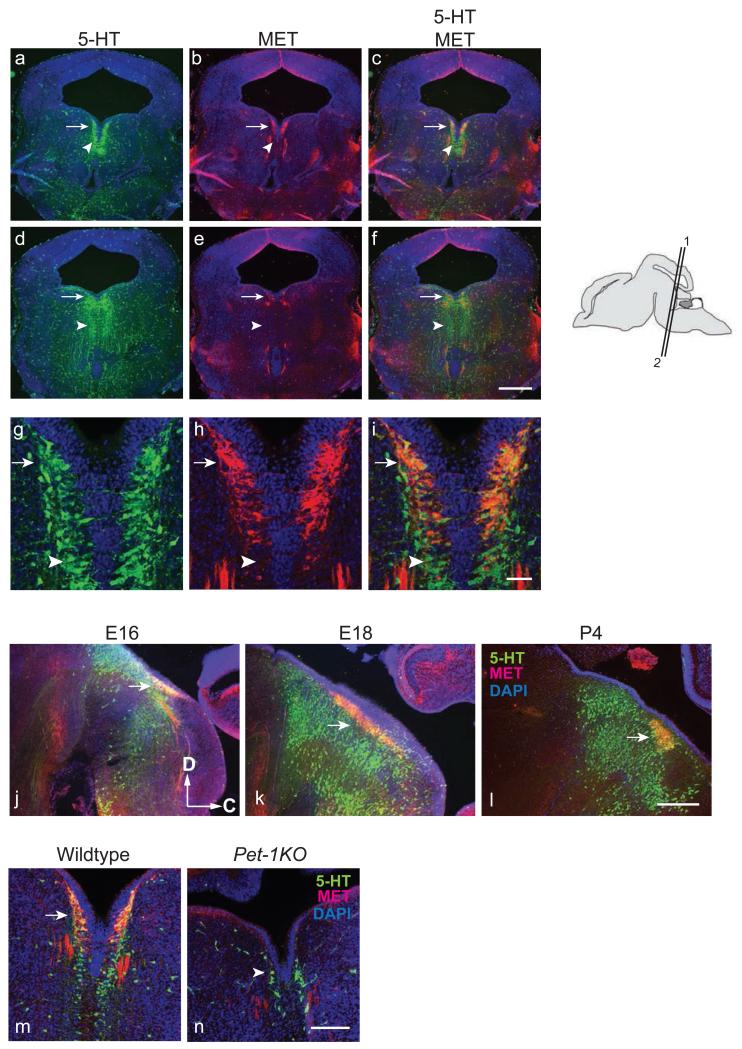
Specific expression of Met in the DR region first emerged by E15 (fig. 1f). Intense Met mRNA expression was maintained in DR neurons up to P0 (fig. 1 h and i and Table 1). Double-label immunostaining with antibodies against MET and 5-HT showed that MET immunoreactivity was present in a subset of DR 5-HT neurons (fig. 4 a-f). When examined under higher magnification, all of the MET+ neurons in DR were 5HT+ (fig. 4 g-i). Staining of serial coronal sections at E16 and sagittal sections from E16 to P4 showed that MET was expressed mainly in the caudal part of DR as paired nuclei situated just below the aqueduct (fig. 4 a-c and j-l). Based on their rostro-caudal and dorso-ventral locations, 5-HT neurons within the midline raphe nuclei are divided into 9 subgroups, B1-9, in the adult rat [56]. The MET+ neurons were located in a region of the DR that includes the B6 and B7 groups. B6, also designated as nucleus raphe dorsalis-caudalis [57], lies caudal to B7. Unlike B7, whose nuclei are partially fused in the midline at E17-E18 in rat (approximately E15-E16 for mouse) and completely fused at P1, B6 nuclei remain paired in adulthood [58]. Judged by the position (caudal aspect of DR) and the paired nuclei where they were observed (fig. 1f, h, i; 4a, b, and j-l), the MET+/5-HT+ neurons most likely belong to the B6 serotonergic subgroup. To further delineate which subset of 5-HT neurons express MET, double immunostaining on sections from Pet-1 knockout mice (Pet-1KO) was performed. The ETS oncogene family protein, Pet-1 (also known as FEV), is expressed in all 5-HT neurons in DR and it is required for the differentiation of approximately 70% of 5-HT neurons in the raphe (fig. 4n, arrowhead; [46]). Remarkably, there was no detectable MET expression in the DR of homozygous Pet-1 null mice (fig. 4d, arrow), even though residual 5-HT+ neurons were observed (fig. 4n, arrowhead). These data suggest that MET expression in B6 subgroup of DR 5-HT neurons is Pet-1 dependent.
Fig. 4.
MET expression in the caudal DR serotonergic neurons is Pet-1 dependent. a to i, Immunostaining for MET and 5-HT on E16 coronal sections through the midbrain in CD-1 mice. a, d, and g, immunostaining with 5-HT antibody; b, e, and h, immunostaining with MET antibody; c, f, and i, merged images of 5-HT and MET immunostaining; a-c are taken from the same section; d-f are taken from the same section. The corresponding plane of section is indicated on the cartoon showing a sagittal view on the right. a-c correspond to “1”; d-f correspond to “2”. g-i are higher magnification images of the of DR region pictured in a-c. Arrows denote MET+/5-HT+ neurons. Arrowheads denote 5-HT+, MET− neurons. j – l, MET expressing 5-HT neurons are located at the caudal region of the DR. Immunostaining for MET and 5-HT on sagittal sections from E16, E18, and P4 brain. Dorsal (D) to the top; caudal (C) to the right. m and n, MET expression in 5-HT neurons requires Pet-1. In Pet-1KO mice, no MET expression was observed in the DR. Arrow indicates the MET-expressing serotonergic neurons in DR in E15 wild type mice. Arrowhead denotes the residual serotonergic neurons in the Pet-1KO. No MET protein was localized in these Pet-1-independent 5-HT neurons. Scale bar, a-f and j-l = 50 μm; g-i = 5 μm; m-n = 30 μm.
Barrington’s nucleus
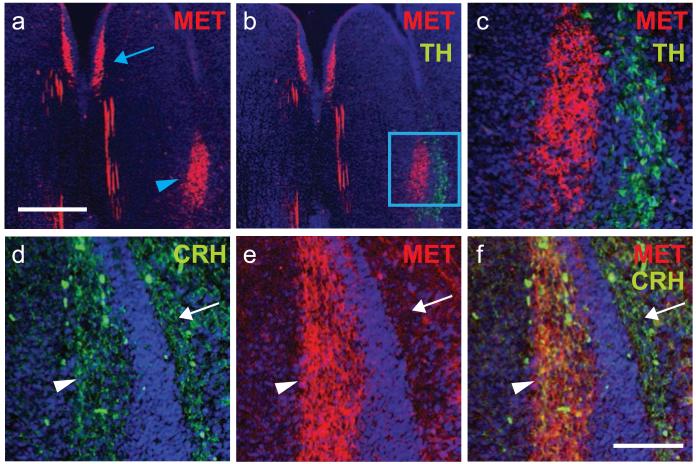
In situ hybridization staining revealed that Met mRNA expression were evident for a brief prenatal time period at E15 and E17 in the lateral pons at the level of the locus coeruleus (LC); labeling was not detectable by P0 (fig. 1f and h). Based on this localization, we initially suspected that Met was expressed in the tyrosine hydroxylase (TH)-containing LC neurons. Surprisingly, double-immunostaining revealed that MET+ and TH+ neurons were completely exclusive of each other (fig. 5b and c). Instead, MET+ neurons were located medial to the LC, overlapping with neurons in the Barrington’s Nucleus (BN). BN regulates several visceral organs and is a central regulator of micturition [59-62]. Neurons in the BN express corticotropin-releasing hormone (CRH) [63-65]. In order to verify that MET is expressed in BN neurons, we performed double immunostaining with anti-MET and anti-CRH antibodies. Co-localization confirmed that nearly all CRH+ neurons of BN expressed MET (fig. 5c-f).
Fig. 5.
MET is expressed by neurons in Barrington’s nucleus. Immunostaining for MET, tyrosine hydroxylase (TH), and corticotropin releasing hormone (CRH) at E16. (a-c) Arrowhead denotes MET+ neurons situated medial to the TH+ locus coeruleus. Note the absence of double-labeled neurons. The arrow in (A) indicates MET+ neurons in the dorsal raphe. (d-f) In contrast, immunostaining for CRH and MET revealed co-localization in neurons residing within Barrington’s nucleus (arrowhead), but not in the more laterally situated locus coeruleus (arrow). Scale Bar: top = 30 μm; bottom = 10 μm.
Hepatocyte growth factor (HGF) is expressed in the developing pineal gland and cells lining the lateral ventricles
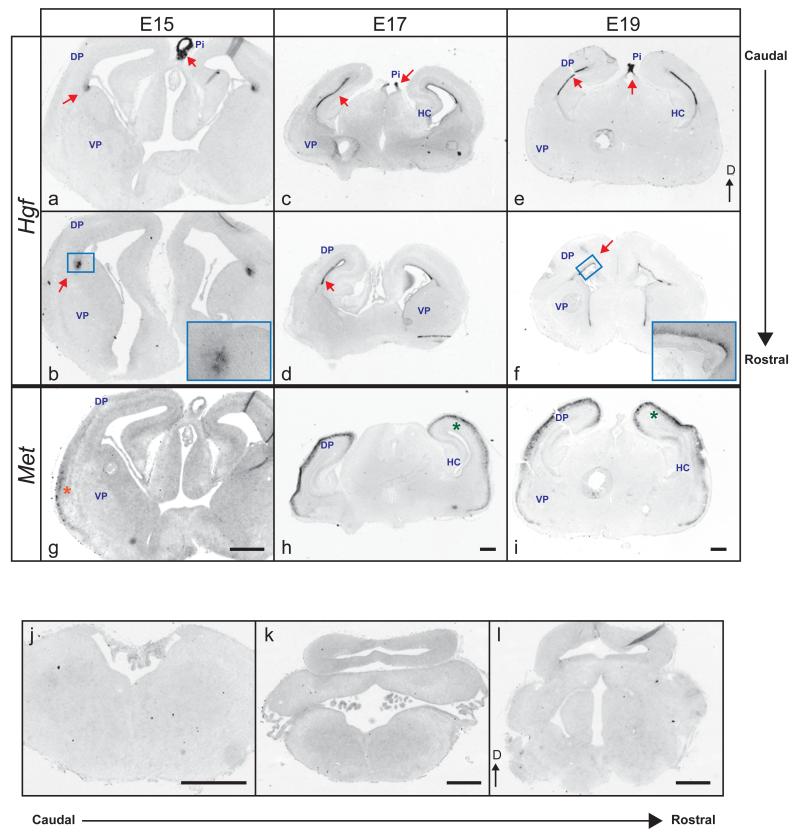
HGF is the only known ligand for MET [66,67]. To detect Hgf transcripts, two DIG-labeled riboprobes encompassing non-overlapping regions of mouse Hgf cDNA were used (supplemental fig. 5a). Confirming specificity, both Hgf riboprobes readily detected Hgf expression in limb buds, as described previously (supplemental fig. 5b and [38]). Furthermore, both probes used separately on adjacent sections yielded identical expression patterns (data not shown). Therefore, only ISH probe #1 was used in subsequent experiments. In the forebrain, we detected Hgf expression in the dorsal pallium at E15 and present through P0, the last age examined. More specifically, Hgf transcripts were confined to cells residing along the ventricular surface at the pallio-subpallial boundary at E15 (fig. 6a and b), where radial glial fibers originate [68]. After E15, expression of Hgf extended through the entire ventricular/subventricular zone (fig. 6c to f). Hgf expression was also detected weakly in the developing epithalamus that contains the emerging pineal gland at E13. Staining became very intense by E15 and remained at high levels through the end of gestation (fig. 6a, c, and e).
Fig. 6.
Distribution of Hgf mRNA in the prenatal mouse forebrain. Coronal sections from E15 (a, b), E17 (c, d), and E19 (e, f) brains hybridized with a DIG-labeled riboprobe to full-length Hgf transcripts. g-i, Met mRNA expression on adjacent sections of a, c, and e respectively. DP, dorsal pallium; VP, ventral pallium; HC, hippocampus; Pi, developing pineal gland. Higher magnification images of boxed areas are shown in the right bottom corner of each micrograph. Arrows denote areas of intense Hgf labeling. Stars indicate Met transcript localization. j-l, Similarly stained caudal sections at E16 show that Hgf is not expressed in the midbrain and hindbrain. Absence of labeling was also evident at earlier and later prenatal ages. Dorsal (D) is at the top. Scale bar = 50 μm.
HGF is expressed in target tissues of MET+ brainstem neurons
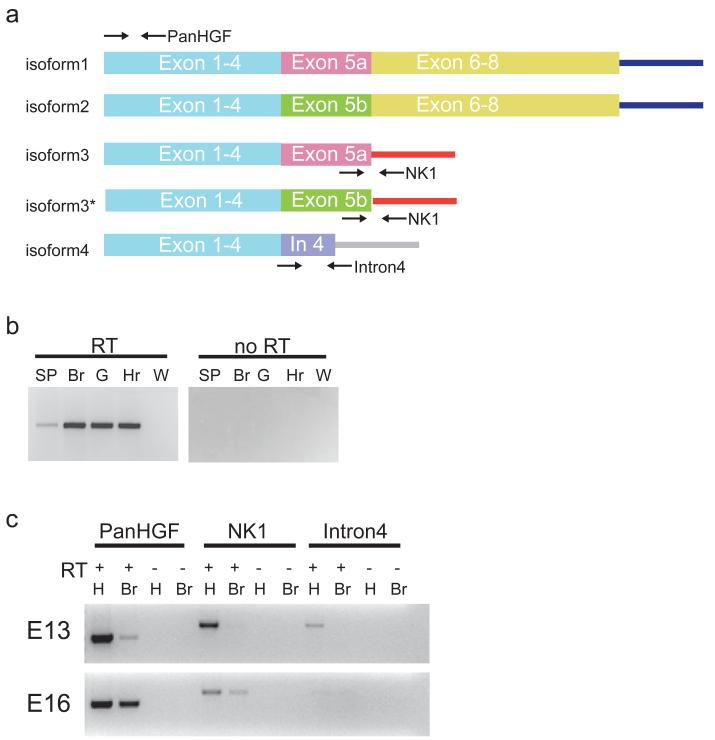
The expression pattern of Hgf in the hindbrain/midbrain was also investigated by in situ hybridization at E13, 15, 17, and 19. Unexpectedly, we did not detect any Hgf transcript in the developing hindbrain and midbrain (fig. 6j - l and data not shown) at any age tested. We reasoned that like spinal motor neurons that utilize somite-derived Hgf [38], MET+ motor neurons in the brainstem might utilize peripheral sources of HGF. Using RT-PCR, Hgf transcripts were detected at E16 in the developing gut (innervation target for DMV and some nA neurons), heart (nA neurons), head mesenchymal/muscle tissues (neurons residing in nVII, nXII, and nA), as well as the developing spinal cord (neurons in BN) (fig. 7b and c). Thus Hgf transcripts were present in peripheral tissues specifically targeted by MET-expressing brainstem neurons at E16.
Fig. 7.
Expression of full length and splice variants of Hgf in the developing brain and visceral organs. a, Diagram of the various forms of Hgf; relative positions of primer sets used for RT-PCR are indicated by arrows. Isoform 3* was newly discovered by sequencing the PCR product and it is not reported in UCSC mouse genome browser. b, RT-PCR analysis of E16 CNS and visceral organ tissues for Hgf transcript expression using a pan Hgf primer set. SP, whole spinal cord; Br, whole brain; G, whole gut; Hr, whole heart. c, RT-PCR expression analysis of Hgf splice variants in E13 and E16 whole brain and head muscle/mesenchymal tissues. RT, reverse transcriptase; H, head muscle/mesenchymal tissues; Br, whole brain; Pan Hgf, NK1, and Intron 4 primer sets are shown in a.
Dynamic expression of Hgf alternative spliced-variants in the embryonic brain
Alternative spliced variants of Hgf are capable of binding and activating the MET receptor [40,41,43]. These variants however, are not detectable using our Hgf riboprobe (fig. 7a and supplemental fig. 5); thus their expression in the developing brain remains to be investigated. There are ten known mouse Hgf transcripts encoding four different protein isoforms reported in UCSC mouse genome browser (fig. 7a). Isoforms 1 and 2 (detectable with the probes used in this study) encode the full-length proHGF that can be activated via membrane-bound proteases. Isoform 3 encodes a truncated version of HGF (also called HGF/NK1; [69]) and was shown to bind and transduce downstream signals through MET in cultured cells. Isoform 4 encodes an even shorter form of HGF (compared to NK1) and its ability to interact with MET has never been determined. Two primer sets were designed to detect the presence of isoform 3 and 4 (fig. 7a) via RT-PCR. As shown in fig. 7c, Hgf/NK1 transcripts were expressed in both brain and head tissues at E16 and in head tissue only at E13; Hgf isoform 4 cDNA was detected in E13 head tissues only. Interestingly, direct sequencing of the PCR products revealed two Hgf/NK1 splice variants, both using the alternative exon 5 splice site (one includes exon 5a and the other exon 5b; see fig. 7a, isoform 3 and isoform 3*). The data thus reveal a complex combination of alternatively spliced forms of Hgf that are expressed in the developing embryonic brain and head tissue, which could serve as ligands for activating MET.
Discussion
The present study reveals specific spatiotemporal expression patterns of Met transcript, protein and multiple splice variants of Hgf transcripts in the developing fetal mouse brainstem. The data reveal several new potential MET ligand sources, including full length HGF in peripheral regions targeted by MET+ brainstem motor neurons, and locally expressed alternative splice forms of the ligand. The data demonstrate a relatively selective and transient pattern of Met expression in developing brainstem sensory and motor neurons that are part of complex circuits that participate in visceral organ (eg. DMV, nA, BN, and nTS) and oral functions (nVII, nA, and nXII). While challenging to do precisely in fetal development, neuronal position, axonal trajectories and synaptic targets are used to categorize cranial motor neurons as somatic motor, branchiomotor or visceral motor [36]. Our results indicate that Met/MET expression in the brainstem is located mainly in subpopulations of neurons that reside in branchiomotor and visceral motor nuclei. Most interestingly, Met expression was also observed in neurons situated in two brainstem nuclei involved in homeostatic, mood and stress regulation, the B6 subgroup of the DR and BN. We found consistently that within each labeled nucleus, Met was only expressed in specific subsets of neurons, verified by using double-labeling strategies with specific markers of neuronal subtypes (see below). Surprisingly, we did not detect any Met transcript or protein in the trigeminal nucleus (nV) and observed only very faint in situ signals in the trochlear motor nucleus (nIV) (supplemental fig. 1g and data not shown). Using wholemount tissue, Canton et al. reported colocalization of Met and Isl1 expression in these two nuclei between E11-E13 [36]. However, detailed illustration of the staining was not provided in that report, making it difficult to determine the extent of Met expression in these nuclei was difficult to assess. Using our in situ and immunolocalization methods on tissue sections, the results suggest that nV and nIV cranial nerve nuclei do not express detectable levels of MET after E13, though we cannot rule out that some cells with very weak Met expression could be present in these nuclei.
Importantly, our study reveals the existence of several new potential sources of HGF ligand capable of activating MET receptor expressed prenatally by brainstem neurons. Whereas MET-expressing forebrain neurons may utilize full-length Hgf expressed locally, peripheral and local alternatively spliced forms of Hgf may serve the signaling requirements of MET in the developing brainstem. Differences in distribution patterns of alternatively spliced forms of HGF suggest that MET signaling may perform different functions in the forebrain and hindbrain during development.
MET, HGF and the visceral sensory/motor connection
Our results reveal the expression of Met in neurons located in developing DMV, nA, nTS, and BN. These nuclei are involved in sensory and motor connections to visceral organs and the spinal cord. This has important functional implications, particularly in the context of neurodevelopmental disorders in which MET dysfunction has been implicated as a risk factor, such as ASD [18-20]. Expression of Met in these nuclei was detected first at E11 for DMV and nA [36], E13 for nTS, and E15 for BN, at the time when the corresponding neurons are postmitotic, have initiated axonal extensions, and are beginning to target peripheral structures [37]. Previous studies have shown that MET activation regulates axonal outgrowth by spinal and cranial facial motor motor neurons in vitro [36,38]. In vivo, axonal targeting by neurons in nXII and spinal motor neurons innervating cutaneous maximus is disrupted by the absence of Met [36] [70]. Nonetheless, widespread axon pathfinding defects have not been described in either Hgf or Met knockout mice. Because constitutive deletion of Hgf or Met results in early lethality [36] [70], conditional deletion of Met will be required to examine its role in mediating maturation of brainstem visceral sensory/motor neurons during pre- and postnatal development.
The role of HGF-MET signaling during development in peripheral organs is not fully understood, though an early study first reported Hgf expression in fetal kidney, intestine, lung, liver, pancreas, and stomach [67]. This expression was confirmed in the present study using RT-PCR (fig. 7). Given the localization of MET in motor neurons that innervate these structures, one may speculate a potentially important developmental impact of MET signaling on the extent of, or innervation patterns of motor axons. For example, the vagal nerve controls GI peristalsis, while nTS receives the sensory information from the GI tract, relaying input to DMV and nA. In this context, it is noteworthy that there is an increased association of MET rs1858830 C allele in children with ASD who exhibit co-occurring gastrointestinal conditions [21]. These children exhibit severe constipation as their primary GI condition [32] [33] [71], consistent with some contribution from the atypical development of MET-expressing circuits that are involved in mediating GI motility and visceral sensory functions. Moreover, in the context of expression in additional brainstem neuron groups, other autonomic problems are widely reported in children with ASD [29] [30] [31].
MET, HGF and the oral sensory/motor circuitry
Neurons located in nVII, nA, and nXII innervate muscles in the oral tract such as tongue, pharynx, and larynx that control taste, eating, swallowing, and speech. The specific expression of Met in subsets of neurons in these nuclei thus warrants further investigation in animals carrying conditionally deleted Met alleles. The function of MET may be even more selective and complex, however, because only subpopulations of neurons in these motor nuclei express Met. What may be the functional significance of such differential Met expression? It is possible that MET signaling acts only on certain subtypes of neurons in these motor nuclei. For example, in the dorsal root ganglia (DRG), MET signaling promotes peptidergic identity in a subset of nociceptors due to its ability to suppress Runx1 expression [72]. In contrast, Runx1 represses Met expression in nonpeptidergic neurons. This counter balance between Runx1 and MET signaling creates mixed populations of DRG neurons, consistent with MET signaling regulating the survival and differentiation of functionally related subsets of motor neurons in these brainstem nuclei. Cranial motor neurons develop early and have reached their target tissues by E13. Interestingly, only nXII nerves showed targeting defect at E12 in Hgf knockout mice [36]. Our results show that MET expression in these central motor nuclei extends through birth, suggesting that MET/HGF signaling may exert a role beyond the initial chemoattraction of cranial motor nerves.
Met and dorsal raphe 5-HT projection
Expression of Met transcript and protein occurs in a highly circumscribed region of the caudal dorsal raphe 5-HT that includes neurons corresponding to the B6 subgroup. 5-HT neurons are involved in homeostatic, stress and mood regulation. Tracing experiments have shown specific projections from B6 5-HT neurons to the nucleus accumbens, amygdala, hypothalamus, and medial prefrontal cortex, structures involved in emotional state and stress regulation [73], [74]. Furthermore our results show an absence of MET expression in the Pet-1 knockout mouse, suggesting that Met transcription is downstream of this factor shown to be critical for the differentiation of the majority of rostrally projecting raphe neurons. The role of MET signaling in 5-HT neurons is not known. While not yet explored, the high level of G-protein coupled 5-HT receptors in 5-HT neurons suggest a ligand-independent, alternative way to activate MET receptor as has been demonstrated in human hepatocellular and pancreatic carcinoma cells [75].
HGF as a Putative CNS Signaling Molecule in Development
Although MET activity has been linked to a variety of histogenic events [15], the developmental expression of its only known ligand, HGF, had not been examined thoroughly. In this study, we found that Hgf expression in the fetal CNS specifically localized to 2 regions: developing pineal gland (starting at E13, fig. 6a, c, and e) and cells along the ventricular/subventricular zone of the lateral ventricles. The current report did not examine early postnatal expression in the forebrain, a time when high levels of MET signaling influence later events of dendrite and synapse development [16]. Met deletion conditionally from the dorsal pallium does not result in major disruption of forebrain architecture and axon patterning [16]. Thus, the function of MET activation via HGF in this region of the developing forebrain appears to be more subtle. In addition to its abundance in serum, HGF also is present in cerebrospinal fluid (CSF) [76] [77]. This raises the possibility that HGF produced near the lateral ventricles and in the developing pineal gland, situated above the third ventricle, may be secreted into the CSF and initiate signaling in distant regions of the embryonic brain. Interestingly, HGF in human CSF can be detected, and levels were decreased in children with ASD [78].
The present data also provide the first evidence that there are multiple forms of Hgf transcript expressed during fetal brain development. In human, according to UCSC human genome browser (www.genome.ucsc.edu), seven HGF mRNA splicing isoforms encode 7 different protein isoforms. Besides the two full length forms of HGF that have been confirmed as MET ligands, the two smaller isoforms, HGF/NK1 and HGF/NK2 also can activate the receptor in vitro [79] [41] [80]. In addition, HGF/NK2 can act as an antagonist to full length HGF in vitro [81]. In mice, only 4 HGF protein isoforms generated from alternative splicing are reported in UCSC mouse genome browser (fig. 7a). Our result shows that in addition to Hgf/NK1 (fig. 7a, isoform 3), another short Hgf splice variant (isoform 4, fig. 7a) is present in the developing mouse brain. These smaller polypeptides are similar in length (less than 25 KDa), to other growth factors, including FGFs, neurotrophines and TGFβ family members. Future functional studies will determine whether short HGF isoforms exert biological actions on MET-expressing neurons.
Conclusion
Expression mapping studies such as the one presented here provide a foundation for experimental manipulations that probe function. The challenge will be to translate our findings of robust MET expression in a limited number of brainstem nuclei that participate in homeostatic, visceral and autonomic regulation, combined with the very restricted expression of HGF to relevant peripheral structures and ventricular regions of the forebrain. MET signaling is pleiotropic, and thus may influence a complex number of histogenic events in brainstem nuclei. The variety of HGF ligands in the brain adds to the challenge of understanding MET developmental function.
Supplementary Material
Supplemental fig. 1 – Comparison of the localization of Met and Isl1 in the brainstem at E13. a-c, Isl1 riboprobes were used to detect Isl1 expression on adjacent sections of those used in fig. 1. The Isl1+ structures were compared with those in Chemoarchitectonic Atlas of the Developing Mouse Brain (Jacobowitz and Abbott 1997). d-f are the same sections shown in fig. 1 a-c. Arrows denote Met-expressing neurons that are positioned comparably to the Isl1+ motor neurons. Scale bar = 30 μm. Corresponding planes of sections from an E13 brain in sagittal view are shown on the cartoon drawing on the right. Section 1 = a and d; section 2 = b and e; section 3 = c and e. g, Very weak signal for the Met transcript was detected in a limited number of neurons in the area containing the nucleus of trochlear nerve (nIV). Scale bar = 30 μm and 10 μm for the box.
Supplemental fig. 2 – Peripherin expression overlaps with tdTomato-labeled cranial motor nuclei in Isl1cre/Rosa-tdTomatofx/+. Immunostaining using an antibody against peripherin was performed on sections adjacent to those used in fig. 2. a, d, and g are tdTomato fluorescence expressed under the control of Isl1cre in 3 successive sections. The level of the section is indicated on the bottom in a cartoon showing a sagittal view of the E16 mouse brain. a is at plane 1; d at plane 2; g at plane 3. b, e, and h are immunostaining using anti-peripherin. c, f, and i are overlapping images of tdTomato fluorescence and anti-peripherine. Peripherin immunoreactivity was seen overlapping with tdTomato+ cranial motor nuclei at E16. Scale bar = 20 μm.
Supplemental fig. 3 – No detectable MET immunoreactivity in dtTomato labeled neurons residing in nIII, nVI, nV, and nVI. Micrographs show MET immunofluorescence in a series of coronal sections harvested from Isl1cre/Rosa-tdTomatofx/+ mice at E13. (a, d, and g), endogenous tdTomato fluorescence, expressed under the control of Isl1cre at 3 successive levels; (b, e, and h), MET immunostaining on the same sections; (c, f, and i), overlay of tdTomato and MET immunofluorescence. Arrowhead in “a” indicates the spinal trigeminal nerve (V) labeled with tdTomato. Arrowhead in b points to the same region depicted in a, showing no MET immunoreactivity. Arrows indicate tdTomato+ neurons that are MET− within the boundaries of nVI, nIV, and nIII. Doubled arrowheads denote a MET-expressing region in superior colliculus that was also seen by Met in situ hybridization (fig. 1c, asterisk). Scale bar=30 μm.
Supplemental fig. 4 – Pax2 expression in some MET+ neurons. a, double immunofluorescence staining with anti-MET and anti-Pax2 on E13 coronal sections. V4, 4th ventricle. The approximate location of the section shown is indicated on the cartoon on the right. b, higher magnification of boxed region in a. Arrows denote some cells that are both Pax2+ and MET+. Scale bar, A = 10 μm; B = 5 μm.
Supplemental fig. 5 – Expression of Hgf in E10 limb bud. a, schematic drawing of the relative locations of two Hgf DIG-riboprobes used for staining. Identical labeling patterns were found with both probes. For consistency, we used probe #1 in this report. b, as a positive control, the ISH probes detect Hgf mRNA in the E10 limb buds.
Acknowledgements
We would like to thank Drs. Alexandre Bonnin and Kathie Eagleson (University of Southern California) and Linda Rinaman (University of Pittsburgh) for helpful discussions and comments on the manuscript. This work was supported by the Hearing Health Foundation (formally Deafness Research Foundation) Research Grant and Zumberge Individual awards (H-H. Wu) and NIMH grant MH067842 (P. Levitt).
Reference
- 1.Cooper CS, Blair DG, Oskarsson MK, Tainsky MA, Eader LA, Vande Woude GF. Characterization of human transforming genes from chemically transformed, teratocarcinoma, and pancreatic carcinoma cell lines. Cancer Res. 1984;44:1–10. [PubMed] [Google Scholar]
- 2.Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, Vande Woude GF. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 3.Stoker M, Perryman M. An epithelial scatter factor released by embryo fibroblasts. J Cell Sci. 1985;77:209–223. doi: 10.1242/jcs.77.1.209. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura T, Nawa K, Ichihara A, Kaise N, Nishino T. Purification and subunit structure of hepatocyte growth factor from rat platelets. FEBS Lett. 1987;224:311–316. doi: 10.1016/0014-5793(87)80475-1. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura T, Nishizawa T, Hagiya M, Seki T, Shimonishi M, Sugimura A, Tashiro K, Shimizu S. Molecular cloning and expression of human hepatocyte growth factor. Nature. 1989;342:440–443. doi: 10.1038/342440a0. [DOI] [PubMed] [Google Scholar]
- 6.Stoker M, Gherardi E, Perryman M, Gray J. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature. 1987;327:239–242. doi: 10.1038/327239a0. [DOI] [PubMed] [Google Scholar]
- 7.Abounader R, Laterra J. Scatter factor/hepatocyte growth factor in brain tumor growth and angiogenesis. Neuro Oncol. 2005;7:436–451. doi: 10.1215/S1152851705000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 10.Birchmeier C, Gherardi E. Developmental roles of hgf/sf and its receptor, the c-met tyrosine kinase. Trends Cell Biol. 1998;8:404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- 11.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 12.Gentile A, Trusolino L, Comoglio PM. The met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 13.Trusolino L, Bertotti A, Comoglio PM. Met signalling: Principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 14.Benvenuti S, Comoglio PM. The met receptor tyrosine kinase in invasion and metastasis. J Cell Physiol. 2007;213:316–325. doi: 10.1002/jcp.21183. [DOI] [PubMed] [Google Scholar]
- 15.Judson MC, Eagleson KL, Levitt P. A new synaptic player leading to autism risk: Met receptor tyrosine kinase. J Neurodev Disord. 2011;3:282–292. doi: 10.1007/s11689-011-9081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Judson MC, Eagleson KL, Wang L, Levitt P. Evidence of cell-nonautonomous changes in dendrite and dendritic spine morphology in the met-signaling-deficient mouse forebrain. J Comp Neurol. 2010;518:4463–4478. doi: 10.1002/cne.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu S, Anderson CT, Levitt P, Shepherd GM. Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated met receptor tyrosine kinase. J Neurosci. 2011;31:5855–5864. doi: 10.1523/JNEUROSCI.6569-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, Persico AM, Levitt P. A genetic variant that disrupts met transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell DB, Li C, Sutcliffe JS, Persico AM, Levitt P. Genetic evidence implicating multiple genes in the met receptor tyrosine kinase pathway in autism spectrum disorder. Autism Res. 2008;1:159–168. doi: 10.1002/aur.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson PB, Boccuto L, Skinner C, Collins JS, Neri G, Gurrieri F, Schwartz CE. Further evidence that the rs1858830 c variant in the promoter region of the met gene is associated with autistic disorder. Autism Res. 2009;2:232–236. doi: 10.1002/aur.87. [DOI] [PubMed] [Google Scholar]
- 21.Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM, Levitt P. Distinct genetic risk based on association of met in families with co-occurring autism and gastrointestinal conditions. Pediatrics. 2009;123:1018–1024. doi: 10.1542/peds.2008-0819. [DOI] [PubMed] [Google Scholar]
- 22.Campbell DB, D’Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, Persico AM. Disruption of cerebral cortex met signaling in autism spectrum disorder. Ann Neurol. 2007;62:243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- 23.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu LM, Li JR, Huang Y, Zhao M, Tang X, Wei L. Autismkb: An evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 2012;40:D1016–1022. doi: 10.1093/nar/gkr1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judson MC, Bergman MY, Campbell DB, Eagleson KL, Levitt P. Dynamic gene and protein expression patterns of the autism-associated met receptor tyrosine kinase in the developing mouse forebrain. J Comp Neurol. 2009;513:511–531. doi: 10.1002/cne.21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Judson MC, Amaral DG, Levitt P. Conserved subcortical and divergent cortical expression of proteins encoded by orthologs of the autism risk gene met. Cereb Cortex. 2011;21:1613–1626. doi: 10.1093/cercor/bhq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth DA, Muchnik C, Shabtai E, Hildesheimer M, Henkin Y. Evidence for atypical auditory brainstem responses in young children with suspected autism spectrum disorders. Dev Med Child Neurol. 2012;54:23–29. doi: 10.1111/j.1469-8749.2011.04149.x. [DOI] [PubMed] [Google Scholar]
- 28.Ploeger A, Raijmakers ME, van der Maas HL, Galis F. The association between autism and errors in early embryogenesis: What is the causal mechanism? Biol Psychiatry. 2010;67:602–607. doi: 10.1016/j.biopsych.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Geschwind DH. Advances in autism. Annu Rev Med. 2009;60:367–380. doi: 10.1146/annurev.med.60.053107.121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porges SW, Furman SA. The early development of the autonomic nervous system provides a neural platform for social behavior: A polyvagal perspective. Infant Child Dev. 2011;20:106–118. doi: 10.1002/icd.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bal E, Harden E, Lamb D, Van Hecke AV, Denver JW, Porges SW. Emotion recognition in children with autism spectrum disorders: Relations to eye gaze and autonomic state. J Autism Dev Disord. 2010;40:358–370. doi: 10.1007/s10803-009-0884-3. [DOI] [PubMed] [Google Scholar]
- 32.Pang KH, Croaker GD. Constipation in children with autism and autistic spectrum disorder. Pediatr Surg Int. 2011;27:353–358. doi: 10.1007/s00383-010-2680-8. [DOI] [PubMed] [Google Scholar]
- 33.Wang LW, Tancredi DJ, Thomas DW. The prevalence of gastrointestinal problems in children across the united states with autism spectrum disorders from families with multiple affected members. J Dev Behav Pediatr. 2011;32:351–360. doi: 10.1097/DBP.0b013e31821bd06a. [DOI] [PubMed] [Google Scholar]
- 34.Mayer EA. Gut feelings: The emerging biology of gut-brain communication. Nat Rev Neurosci. 2011;12:453–466. doi: 10.1038/nrn3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayer EA, Tillisch K. The brain-gut axis in abdominal pain syndromes. Annu Rev Med. 2011;62:381–396. doi: 10.1146/annurev-med-012309-103958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caton A, Hacker A, Naeem A, Livet J, Maina F, Bladt F, Klein R, Birchmeier C, Guthrie S. The branchial arches and hgf are growth-promoting and chemoattractant for cranial motor axons. Development. 2000;127:1751–1766. doi: 10.1242/dev.127.8.1751. [DOI] [PubMed] [Google Scholar]
- 37.Pierce ET. Time of origin of neurons in the brain stem of the mouse. Prog Brain Res. 1973;40:53–65. doi: 10.1016/S0079-6123(08)60679-2. [DOI] [PubMed] [Google Scholar]
- 38.Ebens A, Brose K, Leonardo ED, Hanson MG, Jr., Bladt F, Birchmeier C, Barres BA, Tessier-Lavigne M. Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron. 1996;17:1157–1172. doi: 10.1016/s0896-6273(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 39.Elsen GE, Choi LY, Prince VE, Ho RK. The autism susceptibility gene met regulates zebrafish cerebellar development and facial motor neuron migration. Dev Biol. 2009;335:78–92. doi: 10.1016/j.ydbio.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwall RH, Chang LY, Godowski PJ, Kahn DW, Hillan KJ, Bauer KD, Zioncheck TF. Heparin induces dimerization and confers proliferative activity onto the hepatocyte growth factor antagonists nk1 and nk2. J Cell Biol. 1996;133:709–718. doi: 10.1083/jcb.133.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakubczak JL, LaRochelle WJ, Merlino G. Nk1, a natural splice variant of hepatocyte growth factor/scatter factor, is a partial agonist in vivo. Mol Cell Biol. 1998;18:1275–1283. doi: 10.1128/mcb.18.3.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz JM, Khan SN, Ahmed ZM, Riazuddin S, Waryah AM, Chhatre D, Starost MF, Ploplis B, Buckley S, Velasquez D, Kabra M, Lee K, Hassan MJ, Ali G, Ansar M, Ghosh M, Wilcox ER, Ahmad W, Merlino G, Leal SM, Friedman TB, Morell RJ. Noncoding mutations of hgf are associated with nonsyndromic hearing loss, dfnb39. Am J Hum Genet. 2009;85:25–39. doi: 10.1016/j.ajhg.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaddy DF, Riedel MJ, Pejawar-Gaddy S, Kieffer TJ, Robbins PD. In vivo expression of hgf/nk1 and glp-1 from dsaav vectors enhances pancreatic ss-cell proliferation and improves pathology in the db/db mouse model of diabetes. Diabetes. 2010;59:3108–3116. doi: 10.2337/db09-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1cre reveals a common bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ets gene plays a critical role in 5-ht neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 47.Wu HH, Bellmunt E, Scheib JL, Venegas V, Burkert C, Reichardt LF, Zhou Z, Farinas I, Carter BD. Glial precursors clear sensory neuron corpses during development via jedi-1, an engulfment receptor. Nat Neurosci. 2009;12:1534–1541. doi: 10.1038/nn.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varela-Echavarria A, Pfaff SL, Guthrie S. Differential expression of lim homeobox genes among motor neuron subpopulations in the developing chick brain stem. Molecular and cellular neurosciences. 1996;8:242–257. doi: 10.1006/mcne.1996.0061. [DOI] [PubMed] [Google Scholar]
- 49.Troy CM, Brown K, Greene LA, Shelanski ML. Ontogeny of the neuronal intermediate filament protein, peripherin, in the mouse embryo. Neuroscience. 1990;36:217–237. doi: 10.1016/0306-4522(90)90364-a. [DOI] [PubMed] [Google Scholar]
- 50.Barclay M, Noakes PG, Ryan AF, Julien JP, Housley GD. Neuronal expression of peripherin, a type iii intermediate filament protein, in the mouse hindbrain. Histochemistry and cell biology. 2007;128:541–550. doi: 10.1007/s00418-007-0340-4. [DOI] [PubMed] [Google Scholar]
- 51.Dauger S, Pattyn A, Lofaso F, Gaultier C, Goridis C, Gallego J, Brunet JF. Phox2b controls the development of peripheral chemoreceptors and afferent visceral pathways. Development. 2003;130:6635–6642. doi: 10.1242/dev.00866. [DOI] [PubMed] [Google Scholar]
- 52.Bradley RM. Historical perspectives. In: Bradley RM, editor. The role of the nucleus of the solitary tract in gustatory processing. Boca Raton (FL): 2007. [PubMed] [Google Scholar]
- 53.Herbert H, Moga MM, Saper CB. Connections of the parabrachial nucleus with the nucleus of the solitary tract and the medullary reticular formation in the rat. The Journal of comparative neurology. 1990;293:540–580. doi: 10.1002/cne.902930404. [DOI] [PubMed] [Google Scholar]
- 54.Sieber MA, Storm R, Martinez-de-la-Torre M, Muller T, Wende H, Reuter K, Vasyutina E, Birchmeier C. Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:4902–4909. doi: 10.1523/JNEUROSCI.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cordes SP. Molecular genetics of cranial nerve development in mouse. Nature reviews Neuroscience. 2001;2:611–623. doi: 10.1038/35090039. [DOI] [PubMed] [Google Scholar]
- 56.Dahlstrom A, Fuxe K. Localization of monoamines in the lower brain stem. Experientia. 1964;20:398–399. doi: 10.1007/BF02147990. [DOI] [PubMed] [Google Scholar]
- 57.Taber E, Brodal A, Walberg F. The raphe nuclei of the brain stem in the cat. I. Normal topography and cytoarchitecture and general discussion. The Journal of comparative neurology. 1960;114:161–187. doi: 10.1002/cne.901140205. [DOI] [PubMed] [Google Scholar]
- 58.Levitt P, Moore RY. Developmental organization of raphe serotonin neuron groups in the rat. Anatomy and embryology. 1978;154:241–251. doi: 10.1007/BF00345655. [DOI] [PubMed] [Google Scholar]
- 59.Sugaya K, Matsuyama K, Takakusaki K, Mori S. Electrical and chemical stimulations of the pontine micturition center. Neurosci Lett. 1987;80:197–201. doi: 10.1016/0304-3940(87)90653-7. [DOI] [PubMed] [Google Scholar]
- 60.Noto H, Roppolo JR, Steers WD, de Groat WC. Excitatory and inhibitory influences on bladder activity elicited by electrical stimulation in the pontine micturition center in the rat. Brain Res. 1989;492:99–115. doi: 10.1016/0006-8993(89)90893-7. [DOI] [PubMed] [Google Scholar]
- 61.Noto H, Roppolo JR, Steers WD, de Groat WC. Electrophysiological analysis of the ascending and descending components of the micturition reflex pathway in the rat. Brain Res. 1991;549:95–105. doi: 10.1016/0006-8993(91)90604-t. [DOI] [PubMed] [Google Scholar]
- 62.Cano G, Card JP, Rinaman L, Sved AF. Connections of barrington’s nucleus to the sympathetic nervous system in rats. J Auton Nerv Syst. 2000;79:117–128. doi: 10.1016/s0165-1838(99)00101-0. [DOI] [PubMed] [Google Scholar]
- 63.Rose MF, Ahmad KA, Thaller C, Zoghbi HY. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for math1. Proc Natl Acad Sci U S A. 2009;106:22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawchenko PE, Imaki T, Potter E, Kovacs K, Imaki J, Vale W. The functional neuroanatomy of corticotropin-releasing factor. Ciba Found Symp. 1993;172:5–21. doi: 10.1002/9780470514368.ch2. discussion 21-29. [DOI] [PubMed] [Google Scholar]
- 65.Sved AF, Cano G, Passerin AM, Rabin BS. The locus coeruleus, barrington’s nucleus, and neural circuits of stress. Physiol Behav. 2002;77:737–742. doi: 10.1016/s0031-9384(02)00927-7. [DOI] [PubMed] [Google Scholar]
- 66.Weidner KM, Sachs M, Birchmeier W. The met receptor tyrosine kinase transduces motility, proliferation, and morphogenic signals of scatter factor/hepatocyte growth factor in epithelial cells. J Cell Biol. 1993;121:145–154. doi: 10.1083/jcb.121.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sonnenberg E, Meyer D, Weidner KM, Birchmeier C. Scatter factor/hepatocyte growth factor and its receptor, the c-met tyrosine kinase, can mediate a signal exchange between mesenchyme and epithelia during mouse development. J Cell Biol. 1993;123:223–235. doi: 10.1083/jcb.123.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stenman J, Yu RT, Evans RM, Campbell K. Tlx and pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development. 2003;130:1113–1122. doi: 10.1242/dev.00328. [DOI] [PubMed] [Google Scholar]
- 69.Chan AM, Rubin JS, Bottaro DP, Hirschfield DW, Chedid M, Aaronson SA. Identification of a competitive hgf antagonist encoded by an alternative transcript. Science. 1991;254:1382–1385. doi: 10.1126/science.1720571. [DOI] [PubMed] [Google Scholar]
- 70.Lamballe F, Genestine M, Caruso N, Arce V, Richelme S, Helmbacher F, Maina F. Pool-specific regulation of motor neuron survival by neurotrophic support. J Neurosci. 2011;31:11144–11158. doi: 10.1523/JNEUROSCI.2198-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorrindo P, Williams KC, Lee EB, Walker LS, McGrew SG, Levitt P. Gastrointestinal dysfunction in autism: Parental report, clinical evaluation, and associated factors. Autism Res. 2012;5:101–108. doi: 10.1002/aur.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gascon E, Gaillard S, Malapert P, Liu Y, Rodat-Despoix L, Samokhvalov IM, Delmas P, Helmbacher F, Maina F, Moqrich A. Hepatocyte growth factor-met signaling is required for runx1 extinction and peptidergic differentiation in primary nociceptive neurons. J Neurosci. 2010;30:12414–12423. doi: 10.1523/JNEUROSCI.3135-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waselus M, Valentino RJ, Van Bockstaele EJ. Collateralized dorsal raphe nucleus projections: A mechanism for the integration of diverse functions during stress. Journal of chemical neuroanatomy. 2011;41:266–280. doi: 10.1016/j.jchemneu.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Bockstaele EJ, Biswas A, Pickel VM. Topography of serotonin neurons in the dorsal raphe nucleus that send axon collaterals to the rat prefrontal cortex and nucleus accumbens. Brain research. 1993;624:188–198. doi: 10.1016/0006-8993(93)90077-z. [DOI] [PubMed] [Google Scholar]
- 75.Fischer OM, Giordano S, Comoglio PM, Ullrich A. Reactive oxygen species mediate met receptor transactivation by g protein-coupled receptors and the epidermal growth factor receptor in human carcinoma cells. J Biol Chem. 2004;279:28970–28978. doi: 10.1074/jbc.M402508200. [DOI] [PubMed] [Google Scholar]
- 76.Tsuboi Y, Kakimoto K, Akatsu H, Daikuhara Y, Yamada T. Hepatocyte growth factor in cerebrospinal fluid in neurologic disease. Acta Neurol Scand. 2002;106:99–103. doi: 10.1034/j.1600-0404.2002.01125.x. [DOI] [PubMed] [Google Scholar]
- 77.Kern MA, Bamborschke S, Nekic M, Schubert D, Rydin C, Lindholm D, Schirmacher P. Concentrations of hepatocyte growth factor in cerebrospinal fluid under normal and different pathological conditions. Cytokine. 2001;14:170–176. doi: 10.1006/cyto.2001.0875. [DOI] [PubMed] [Google Scholar]
- 78.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 79.Miyazawa K, Kitamura A, Naka D, Kitamura N. An alternatively processed mrna generated from human hepatocyte growth factor gene. Eur J Biochem. 1991;197:15–22. doi: 10.1111/j.1432-1033.1991.tb15876.x. [DOI] [PubMed] [Google Scholar]
- 80.Lokker NA, Mark MR, Luis EA, Bennett GL, Robbins KA, Baker JB, Godowski PJ. Structure-function analysis of hepatocyte growth factor: Identification of variants that lack mitogenic activity yet retain high affinity receptor binding. EMBO J. 1992;11:2503–2510. doi: 10.1002/j.1460-2075.1992.tb05315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cioce V, Csaky KG, Chan AM, Bottaro DP, Taylor WG, Jensen R, Aaronson SA, Rubin JS. Hepatocyte growth factor (hgf)/nk1 is a naturally occurring hgf/scatter factor variant with partial agonist/antagonist activity. J Biol Chem. 1996;271:13110–13115. doi: 10.1074/jbc.271.22.13110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental fig. 1 – Comparison of the localization of Met and Isl1 in the brainstem at E13. a-c, Isl1 riboprobes were used to detect Isl1 expression on adjacent sections of those used in fig. 1. The Isl1+ structures were compared with those in Chemoarchitectonic Atlas of the Developing Mouse Brain (Jacobowitz and Abbott 1997). d-f are the same sections shown in fig. 1 a-c. Arrows denote Met-expressing neurons that are positioned comparably to the Isl1+ motor neurons. Scale bar = 30 μm. Corresponding planes of sections from an E13 brain in sagittal view are shown on the cartoon drawing on the right. Section 1 = a and d; section 2 = b and e; section 3 = c and e. g, Very weak signal for the Met transcript was detected in a limited number of neurons in the area containing the nucleus of trochlear nerve (nIV). Scale bar = 30 μm and 10 μm for the box.
Supplemental fig. 2 – Peripherin expression overlaps with tdTomato-labeled cranial motor nuclei in Isl1cre/Rosa-tdTomatofx/+. Immunostaining using an antibody against peripherin was performed on sections adjacent to those used in fig. 2. a, d, and g are tdTomato fluorescence expressed under the control of Isl1cre in 3 successive sections. The level of the section is indicated on the bottom in a cartoon showing a sagittal view of the E16 mouse brain. a is at plane 1; d at plane 2; g at plane 3. b, e, and h are immunostaining using anti-peripherin. c, f, and i are overlapping images of tdTomato fluorescence and anti-peripherine. Peripherin immunoreactivity was seen overlapping with tdTomato+ cranial motor nuclei at E16. Scale bar = 20 μm.
Supplemental fig. 3 – No detectable MET immunoreactivity in dtTomato labeled neurons residing in nIII, nVI, nV, and nVI. Micrographs show MET immunofluorescence in a series of coronal sections harvested from Isl1cre/Rosa-tdTomatofx/+ mice at E13. (a, d, and g), endogenous tdTomato fluorescence, expressed under the control of Isl1cre at 3 successive levels; (b, e, and h), MET immunostaining on the same sections; (c, f, and i), overlay of tdTomato and MET immunofluorescence. Arrowhead in “a” indicates the spinal trigeminal nerve (V) labeled with tdTomato. Arrowhead in b points to the same region depicted in a, showing no MET immunoreactivity. Arrows indicate tdTomato+ neurons that are MET− within the boundaries of nVI, nIV, and nIII. Doubled arrowheads denote a MET-expressing region in superior colliculus that was also seen by Met in situ hybridization (fig. 1c, asterisk). Scale bar=30 μm.
Supplemental fig. 4 – Pax2 expression in some MET+ neurons. a, double immunofluorescence staining with anti-MET and anti-Pax2 on E13 coronal sections. V4, 4th ventricle. The approximate location of the section shown is indicated on the cartoon on the right. b, higher magnification of boxed region in a. Arrows denote some cells that are both Pax2+ and MET+. Scale bar, A = 10 μm; B = 5 μm.
Supplemental fig. 5 – Expression of Hgf in E10 limb bud. a, schematic drawing of the relative locations of two Hgf DIG-riboprobes used for staining. Identical labeling patterns were found with both probes. For consistency, we used probe #1 in this report. b, as a positive control, the ISH probes detect Hgf mRNA in the E10 limb buds.