Abstract
PharmGKB is a leading resource of high quality pharmacogenomics data that provides information about how genetic variations modulate an individual's response to drugs. PharmGKB contains information about genetic variations, pharmacokinetic and pharmacodynamic pathways, and the effect of variations on drug-related phenotypes. These relationships are represented using very general terms, however, and the precise semantic relationships among drugs,and diseases are not often captured. In this paper we develop a protocol to detect and disambiguate general clinical associations between drugs and diseases using more precise annotation terms from other data sources. PharmGKB provides very detailed clinical associations between genetic variants and drug response, including genotype-specific drug dosing guidelines, and this procedure will be adding information about drug-disease relationships not found in PharmGKB. The availability of more detailed data will help investigators to conduct more precise queries, such as finding particular diseases caused or treated by a specific drug.
We first mapped drugs extracted from PharmGKB drug-disease relationships to those in the National Drug File Reference Terminology (NDF-RT) and to Structured Product Labels (SPLs). Specifically, we retrieved drug and disease role relationships describing and defining concepts according to their relationships with other concepts from NDF-RT. We also used the NCBO (National Center for Biomedical Ontology) annotator to annotate disease terms from the free text extracted from five SPL sections (indication, contraindication, ADE, precaution, and warning). Finally, we used the detailed drug and disease relationship information from NDF-RT and the SPLs to annotate and disambiguate the more general PharmGKB drug and disease associations.
Keywords: Pharmacogenomics, clinical associations, PharmGKB, NDF-RT, SPL
1. INTRODUCTION
The Pharmacogenomics Knowledge Base (PharmGKB [1]) is a publicly available internet resource for pharmacogenomics data and knowledge that provides information about genes involved in modulating the response to drugs. PharmGKB includes data about genetic variations, pharmacokinetic and pharmacodynamic pathways, and the effects of genetic variations on drug-related phenotypes. PharmGKB also provides integrated knowledge including relationships among genes, drugs, and diseases. The importance of this resource is widely recognized, and it has been used in several investigations. For example, Pathak, et al. [2] investigated pharmacodynamics (PD) and pharmacokinetics (PK) relationships between drugs and diseases in PharmGKB, and compared them with the drug-disease relationships in NDF-RT. Theobald, et al. [3] extracted the relationships between drugs, disease and genes from PubMed guided by the relationships from PharmGKB.
While PharmGKB collects, encodes, and disseminates knowledge about the impact of human genetic variations on drug response [4], the detailed semantic relationships between drugs and diseases are not often captured during the curation process. For example, PharmGKB contains a generic relationship between the drug donepezil (PA449394) and the disease dementia (PA443853), but no detailed information about that relationship is provided. The availability of more precisely annotated drug and disease associations would be invaluable to the research community, especially for drug repositioning studies.
Detailed information about drug-disease relationships would also be valuable to studies that focus on Adverse Drug Events (ADEs). Vilar et al. [5] combined data from the US Food and Drug Administration’s (FDA) Adverse Event Reporting System (AERS) with similarity measures of molecular structure, a process that achieved significant improvement in precision in computationally detecting ADEs. Benton et al. [6] conducted a study to identify potential ADEs by using medical message boards and online resources. Jiang et al. [7] developed ADEpedia by using ADE information from SPLs to create a RDF triple store. Each of these studies improve our ability to detect ADEs, which is a crucial step for identifying diseases associated with particular drugs. The detailed annotation of drug and disease associations from PharmGKB that is described in this paper adds significant value to existing resources, thereby facilitating studies like those mentioned above.
The overall goal of this study is to enrich the drug and disease associations in PharmGKB with more detailed semantic relationships. We present a method to disambiguate the clinical associations between drugs and diseases in PharmGKB. We utilized the Veterans Affairs National Drug File Reference Terminology (NDF-RT [8]) and DailyMed Structured Product Labeling (SPL [9]) as the sources of detailed information about drug-disease associations, and demonstrated how existing data sources can be used to semantically enrich each other.
2. BACKGROUND
We annotated drug and disease relationships from PharmGKB using publicly available data sources. Each source is described below, along with rationale for its use in this study.
2.1 PharmGKB
PharmGKB contains genomic, phenotype and clinical information collected from pharmacogenomics studies. More specifically, PharmGKB provides information regarding variant annotations, drug-centered pathway, pharmacogene summaries, clinical annotations, pharmacogenomics based drug-dosing guidelines, drug labels with pharmacogenomics information. PharmGKB also provides to registered research network members tools to browse, query, download, submit, edit and process the information [4].
2.2 Veterans Affairs National Drug File Reference Terminology (NDF-RT)
NDF-RT is produced and maintained by the U.S. Department of Veterans Affairs (VA). It is used for modeling drug characteristics including ingredients, chemical structure, dose form, physiologic effect, mechanism of action, pharmacokinetics, and related diseases [10]. NDF-RT concepts are grouped into several general categories, including drug and disease. The drug category includes VA classifications of medications, generic ingredient preparations used in medications, and orderable (clinical) VA drug products. The disease category includes pathophysiologic data as well as certain non-disease physiologic states that are treated, prevented, or diagnosed by an ingredient or drug product. In addition, the disease category may also describe contraindications.
Although PharmGKB provides mappings with ATC (Anatomical Therapeutic Chemical) and DrugBank, we used NDF-RT in this study for several reasons: 1) ATC is a proprietary terminology; 2) neither ATC nor DrugBank provide information about drug and disease associations; 3) NDF-RT has been integrated into RxNorm, which is specified by U.S. Meaningful Use regulations; 4) NDF-RT is updated weekly.
2.3 Structured Product Labeling (SPL)
The SPL standard has been adopted by the FDA for exchanging information about drugs and drug ingredients, including dosage, strength, usage, and known ADEs [10]. SPL documents contain structured sections that in turn contain unstructured content that comprise the product label (including all text, tables and figures). SPL defines the content of human prescription drug labeling in an XML format that are organized by section headings; 76 section headings have been defined and coded by LOINC [11].
SPLs were used in this study as a data source that compliments NDF-RT. Other studies have used this resource in a similar manner, which allows us to extend our current work with other resources and applications.
2.4 National Center for Biomedical Ontology (NCBO) Annotator
The NCBO annotator is an ontology-based web service for annotating textual biomedical data with concepts from more than 200 ontologies, which are part of two important repositories of biomedical ontologies and terminologies: the UMLS Metathesaurus and NCBO BioPortal [12]. The NCBO annotator provides the capability for detecting and annotating disease terms within the SPL free text used in this study.
3. Methods
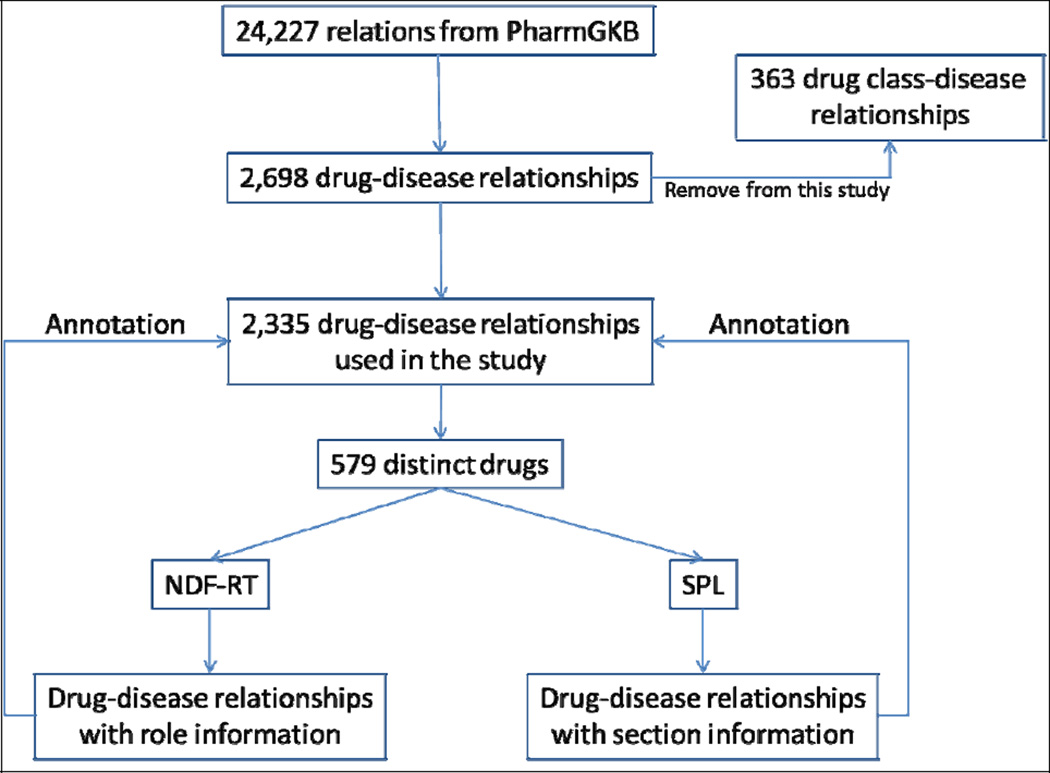
Data sets were retrieved from the PharmGKB July 16th, 2011 snapshot. Of the 24,227 relations in the dataset, 2,698 drug-disease relationships were extracted, including 363 relationships between a drug class and a disease. The drug class-disease relationships were excluded from the analysis since NDF-RT only provides relationships between individual drugs and diseases. Therefore, 2,335 drug-disease relationships from PharmGKB were annotated in this study. These relationships included 579 distinct drugs (called “PharmGKB drug” in this report), and 444 distinct diseases. The annotation process is illustrated in Figure 1.
Figure 1.
PharmGKB drug-disease annotation process diagram
We mapped PharmGKB drugs to records in the NDF-RT and SPL data sets, then used the disease information obtained from these two resources to more precisely annotate the PharmGKB drug-disease relationships. The annotation workflow is shown in Figure 2. The steps involved are described in the following sections.
Figure 2.
Annotation workflow
3.1 NDF-RT extraction
NDF-RT provides role relationships that describe and define concepts according to their relationships with other concepts. Each role has a domain (the kind of concept whose definition may use the role) and a range (the kind of concept to which the role can refer) [10]. In this study, we focused on the role relationships between NDF-RT concepts that are “Generic Ingredients or Combinations” or “Disease”, as shown in Table 1.
Table 1.
Definition of role relationships from NDF-RT
| Role | Definition |
|---|---|
| may_treat | therapeutic use or indication of a generic ingredient preparation or drug |
| may_prevent | preventative use or indication of a generic ingredient preparation or drug |
| may_diagnose | diagnostic use or indication of a generic ingredient preparation or drug |
| induces | therapeutic effect or state caused by a generic ingredient preparation or drug |
| CI_with | therapeutic or co-morbid contraindication of a generic ingredient preparation or drug |
The NDF-RT API [13] was used to retrieve information for the 579 PharmGKB drugs, using the drug name as an input parameter (Figure 1). The query results included the NDF-RT drug identifier (NUI) and its corresponding role relationships (Table 1).
To ensure the accuracy of the mappings between PharmGKB and NDF-RT, we manually reviewed all mappings in which the NDF-RT concept names did not exactly match the PharmGKB drug names. The review process confirmed that all non-exact mappings were due only to differences in spelling, use of synonymous terms, and differences in drug representation format, and therefore they were retained for further analysis.
3.2 SPL Extraction
SPLs provide high quality information for marketed drugs, which includes generic names, ingredients, dosage forms, routes of administration, and usage of the drug. While the SPLs are only semi-structured, they are a valuable resource that can be used to enrich the annotation for PharmGKB drug and disease pairs, and therefore we utilized them in this study.
The SPL data set was downloaded from DailyMed on 11/4/2011, and stored in a local database. We extracted the disease information from the five SPL sections listed in Table 2 as detailed below, and used this information to annotate PharmGKB drug and disease relationships..
Table 2.
SPL sections used in this study
| SPL Sections | LOINC CODE |
|---|---|
| INDICATION AND USAGE | 34067-9 |
| ADVERSE REACTIONS | 34084-4 |
| PRECAUTIONS | 42232-9 |
| WARNINGS | 34071-1 |
| CONTRAINDICATIONS | 34070-3 |
3.3 SPL disease information extraction
For each PharmGKB drug, SPL documents were located in our local database and disease information was extracted from the sections listed in Table 2. To identify SPLs for a given drug, the RxCUI was used as an intermediary identifier to retrieve the SPL setId from the RxNav REST API [14]. Two separate calls to the API were required: one to obtain the RxCUI for a given PharmGKB drug name and another to retrieve the SPL setId for a given RxCUI. Once SPL documents were identified using the setId, unstructured text was extracted using SAXParser. The text was submitted to the NCBO annotator to obtain disease terms.
3.4 Annotating SPL free text
Using the NCBO annotator, the free text from the five SPL sections listed in Table 2 was annotated with terms from standardized terminologies. To increase the specificity of the annotations, the annotated concepts were filtered by UMLS semantic type [14]; terms that mapped to one of the disease-related types listed in Table 3 were retained for further evaluation. For example, the phrase “ARICEPT is indicated for the treatment of dementia of the Alzheimers type” was obtained from the”INDICATION” section of one SPL document. As shown in Figure 3, the NCBO annotator successfully identified the term “dementia” and mapped it to a term from the OMIM ontology; that term has a semantic type of “Disease or Syndrome” (semantic type T047). Similar results were obtained for all text extracted from the SPL documents.
Table 3.
UMLS sematic types used in this study
| UMLS semantic types | Names |
|---|---|
| T047 | Disease or Syndrome |
| T191 | Neoplastic Process |
| T046 | Pathologic Function |
| T033 | Finding |
| T184 | Sign or Symptom |
| T048 | Mental or Behavioral Dysfunction |
Figure 3.
An example of NCBO annotator output in XML.
Due to the conceptual overlap among sections (see Table 2), it is not uncommon to find statements related to indications and contraindications in the same section. For example, the statement “VICOPROFEN is not indicated for the treatment of such conditions as osteoarthritis or rheumatoid arthritis” expresses a contraindication as a negated indication. Automatically detecting and annotating this type of statement may be possible using a tuned natural language processing (NLP) tool, but since a validated NLP system was not available we employed a hybrid approach. Specifically, we utilized OpenNLP [15] to split the content from the “INDICATION” section into individual sentences and used a list of negative words (e.g., “not”, “neither”, “unable”) to identify potential contraindication statements. If a negative word was found in a sentence, we flagged that sentence for manual review. This approach, while fairly primitive, was an efficient method for identifying statements that may generate inaccurate annotations.
3.5 Annotating PharmGKB drug-disease associations
The results from the previous steps yielded precise semantic relationships between drugs and diseases, which were used to disambiguate the general associations listed in PharmGKB. We compared each PharmGKB drug–disease relationship to annotation data for the corresponding drug-disease pairs from NDF-RT and SPL (see Figure 2). Known synonyms for drug and disease terms were used to maximize the number of successful matches. In cases where matches were not found by term identifier for diseases, substring matching was employed. Figure 4 shows an example of PharmGKB drug-disease annotation results.
Figure 4.
Example of PharmGKB drug-disease annotation result
4. Results
4.1 Annotation results from NDF-RT
The NDF-RT API was used to obtain mappings between 579 PharmGKB drug names and NDF-RT unique identifiers (NUIs). Mappings were obtained for 464 PharmGKB drug names, whereas 115 PharmGKB drugs failed to map to NDF-RT concepts (the reason for the failure will be addressed in the Discussion section). In most cases where mappings were obtained, the NDF-RT concept name exactly matched the PharmGKB drug name. Manual review was conducted for the 14 non-exact matches that were returned (Table 4). All 14 matches were found to be correct after accounting for differences in spelling, use of synonymous terms, and drug representation format. Therefore, those 14 mappings were included in the subsequent analysis.
Table 4.
Non-exact string matches between PharmGKB drug and NDF-RT
| PharmGKB Drug Name | NDF-RT Concept Name | RxNorm Name |
|---|---|---|
| abacavir | avacavir | abacavir |
| carmustine | camustine | Carmustine |
| colchicine | colchcine | Colchicine |
| valproic acid | divalproex | Valproic Acid |
| copper sulfate | cupric sulfate | Copper Sulfate |
| copper sulfate | copper (as cupric sulfate) | Copper Sulfate |
| copper sulfate | cupric sulfate, anhydrous | Copper Sulfate |
| ethanol | alcohol | Ethanol |
| ethanol | alcohol,ethyl | Ethanol |
| drotrecogin alfa | drotrecogin alfa (activated) | drotrecogin alfa |
| epoetin alfa | epoetin alfa, recombinant | Epoetin Alfa |
| estradiol | estradiol 17-beta | Estradiol |
| certolizumab pegol | certolizumab | certolizumab pegol |
| beta carotene | carotene, beta | Beta Carotene |
Using the RxNav API, disease (role) information was obtained for each of the 464 distinct PhramGKB drugs for which NUIs were available. Unfortunately, not all NDF-RT concepts had associated disease information (e.g., hydrocortisone) and 84 drugs were excluded from further analysis for this reason. The remaining 380 distinct drugs were related to 672 distinct diseases through 3148 drug-disease role pairs. Table 5 shows the number of relations by NDF-RT role type (defined in Table 1). For example, there are 1703 relations that describe drugs that “may_treat” a given disease. The 1703 "may treat" relations consisted of 368 distinct drugs and 510 distinct diseases.
Table 5.
Mapping PharmGKB drugs to NDF-RT
| Role type | # of relations | # of distinct drugs | # of distinct diseases |
|---|---|---|---|
| CI_With | 1230 | 375 | 241 |
| induces | 19 | 10 | 13 |
| may_diagnose | 6 | 5 | 6 |
| may_prevent | 190 | 98 | 74 |
| may_treat | 1703 | 368 | 510 |
| Total | 3148 | 380 | 672 |
4.2 Annotation results from SPL
To create drug-disease associations from the SPL resource, the section names were used to define the type of association between the drug referenced by the SPL document and the disease term(s) that were found within the section. These associations were then used to disambiguate the drug-disease relationships in PharmGKB.
RxCUI generation
To identify SPLs for a given drug, the RxCUI was used as an intermediary identifier to retrieve the SPL setId from the RxNav REST API. Of the 579 PharmGKB drug names, 459 were mapped to 17,426 RxCUIs (14,726 unique RxCUIs) (mappings were not found for 120 drugs using this method). Another 18 distinct PharmGKB drugs resulted in 808 non-exact mappings between PharmGKB and RxNorm (see, examples in Table 6). By manually reviewing the literature (see the “Reference” column in Table 6), we verified that the 808 non-exact mappings were caused by synonymous representations used by PharmGKB and RxNorm. Therefore, those mappings were included in the subsequent analysis.
Table 6.
Examples of non-exact mapping in drug name between PharmGKB and RxNorm
| PharmGKB Drug Name |
RxNorm Concept Name |
Reference |
|---|---|---|
| glibenclamide | glyburide 1.25 mg oral tablet [micronase] | Glibenclamide (INN), also known as glyburide (USAN), is an antidiabetic drug in a class of medications known as sulfonylureas, closely related to sulfa drugs [16]. |
| fenofibrate | fenofibric acid 35 mg oral tablet [fibricor] | Fenofibric acid, an active form of fenofibrate, increases apolipoprotein A-I-mediated high-density lipoprotein biogenesis by enhancing transcription of ATP-binding cassette transporter A1 gene in a liver X receptor-dependent manner [17] |
| cyanocobalamin | vitamin b 12 0.1 mg/ml injectable solution [crystal b-12] | Cyanocobalamin is an especially common vitamer of the vitamin B12 family [18] |
SPL extraction
The 14,726 distinct RxCUIs generated in the above step were used to retrieve SPL setIds via the RxNAV API. A total of 12,549 setIds were returned, and text from the sections listed in Table 2 was extracted from each SPL document, which was then annotated with disease terms. The extraction and annotation results for each section are listed in Table 7. In this table, we list unique numbers of SPL sections, annotated drug-disease pairs (e.g. drug-indication, drug-ADE, etc.), relevant drugs, and relevant diseases.
Table 7.
SPL extraction and annotation results
| Indications | Adverse Reactions |
Contraindications | Precautions | Warnings | |
|---|---|---|---|---|---|
| # Unique sections | 4,678 | 5,179 | 3,481 | 4,400 | 4,932 |
| # unique drugs | 430 | 430 | 401 | 334 | 427 |
| # unique diseases | 3,688 | 8,850 | 1,917 | 5,632 | 5,510 |
| # Unique drug-disease relations | 17,350 | 145,391 | 10,278 | 54,544 | 60,570 |
4.3 Evaluation of NCBO annotation
To evaluate the accuracy of the NCBO annotation, the annotations from a random set of 60 PharmGKB drugs were manually reviewed. From this set of 60 PharmGKB drugs, 98 SPL labels were obtained. We manually reviewed the free text from the indications section of these SPLs and compared them to the 1023 corresponding annotations from the NCBO annotator. The evaluation results are listed in Table 8. The false positives were instances where non-disease terms such as, “pressure” or “stomach” were mapped to a term that was classified as one of the disease-related semantic types (Table 3). In addition, 13 false negatives were found that were not identified by NCBO annotator, such as “Prophylaxis of Organ Rejection in Renal Transplantation”.
Table 8.
Evaluation results for NCBO annotation
| # Indications | True positive | False positive | False negative | Precision | Recall | F-measure |
|---|---|---|---|---|---|---|
| 1023 | 816 | 194 | 13 | 80.8% | 98.4% | 88.7% |
4.4 Review of Negated Statements
A total 80 statements from SPL sections were flagged for review due to the presence of a negative word. Manual review of each statement confirmed that 20 of them were true positives (an expression of a contraindication). The remaining 60 statements were false positives (e.g., “children under 2 years of age, do not use, consult a doctor”), which were re-annotated and included in the subsequent analysis for the drug and disease pairs.
4.5 PharmGKB drug-disease relationships annotation
Using the annotations and mappings from the NDF-RT and SPL data sets, 1,222 distinct PharmGKB drug-disease relationships were able to be semantically disambiguated using this approach, out of the 2,335 relationships that were present in PharmGKB at the time of this study. These relationships included 394 distinct drugs and 275 distinct diseases (Tables 9 and 10). These results will be made publicly available and will be contributed to PharmGKB.
Table 9.
Annotation results for PharmGKB drug-disease relationships by NDF-RT
| Role/section types | # unique Drug-disease pairs |
|---|---|
| may_diagnose | 1 |
| may_prevent | 54 |
| may_treat | 313 |
| CI_With | 61 |
| induces | 1 |
Table 10.
Annotation results for PharmGKB drug-disease relationships by SPL
| Role/section types | # unique Drug-disease pairs |
|---|---|
| Indication | 620 |
| ADE | 833 |
| Warnings | 722 |
| Precautions | 564 |
| Contraindications | 188 |
5. Discussion
In this study, we employed two different approaches for extracting detailed semantic annotations for drug-disease pairs from the NDF-RT and SPL resources, and subsequently used them to disambiguate the general drug-disease relationships in PharmGKB. For NDF-RT, the mapping process was very straightforward due to two aspects: the content was available in a structured format (XML) and its existing drug-disease associations reduced the need for additional curation. While this approach was relatively simple computationally, NDF-RT yielded fewer drug-disease associations than the SPL dataset (Tables 9 and 10).
The SPL resource required the extraction and annotation of drug and disease relations from free text. To accomplish this, we explored an approach that utilized OpenNLP and the NCBO annotator to extract and annotate disease information, and we used manual annotation to evaluate the accuracy of the annotations. While the development of this process was not the main focus of this work, it was a necessary step toward accomplishing the primary goal of obtaining semantically rich drug-disease associations. The SPL resource would be much more valuable to the research community if the information was available in a structured and codified format. We support ongoing efforts to mine, annotate, and connect information contained in SPLs with that in other resources. An example of these efforts is the LinkedSPLs project [19], which provides links between SPLs and terminologies such as DrugBank and RxNorm.
Overall, 52% (1,222 of 2,335) of PharmGKB drug-disease relationships were successfully disambiguated using one or more precise relationships extracted from the NDF-RT and SPL resources. This project not only provides tangible results in the form of detailed drug-disease associations, but also demonstrates how resources such as NDF-RT and SPL can be used to add value to already high-quality human-curated repositories such as PharmGKB. In most cases when annotations were available from both NDF-RT and SPL, the annotations were consistent with each other. For example, NDF-RT defines anemia as a contraindication for the administration of hydroxyurea (i.e., the role type of the association is “CI_with”). Similarly, “anemia” is present within the #x0201C;CONTRAINDICATION” section of an SPL for hydroxyurea. Some annotations from these two resources were different, however. For example, NDF-RT defines an association between norepinephrine and hypotension with the role type “CI_with” (i.e., specifying a contraindication), whereas the opposite relationship ("INDICATION") was extracted from SPL. When this was observed, the conflict was resolved by consulting DailyMed, which in this example supported the latter association (an indication).
A significant fraction (130 of 579, or 22%) of the drug names listed in PharmGKB could not be mapped to NDF-RT and/or RxNorm. Specifically, 115 PharmGKB drugs could not be mapped to a NUI and 103 failed to map to an RxCUI. Many failed mappings were due to PharmGKB's use of chemical IUPAC names, which were not included in NDF-RT or RxNorm. For example, the PharmGKB drug entry “3-beta-hydroxy-5-androsten-17-one” failed to map to the terminologies using the methods described in this project. However, a synonym for this compound is “Dehydroepiandrosterone”, which has a NUI (N0000148779). To address this issue, additional data sources such as PubChem [20] and DrugBank [21], which contain IUPAC names, could be used to identify additional synonyms. Currently, only PubChem has an API that could be used to query for this information; DrugBank does not have an API but the data could be downloaded to a local repository and queried directly.
PharmGKB also contains some entries for compounds that are not used for treatment, and therefore are not included in NDF-RT or RxNorm. For example, PharmGKB associates "calcein" with four diseases (“Leukemia”, “Leukemia, Myeloid”, “Leukemia, Myeloid, Acute” and “neoplasms”), but this compound is a cytoplasmic fluorescent dye used for diagnosis rather than for pharmaceutical treatment [22].
Finally, we failed to find mappings for some drug-disease associations due to the use of general disease terms within PharmGKB. For example, the PharmGKB dataset used for this study included terms such as “Transplantation” (N=22), “Drug Toxicity” (N=93), and “Neoplasms” (N= 108). Other terms, such as "Death" (N=55) were also included as diseases. While some of these terms might be included as an upper-level term in the NDF-RT or NxNorm terminologies, it is possible that other ontologies might be needed to express these concepts.
6. Conclusion
We present a method for disambiguating PharmGKB drug and disease associations that utilizes the NDF-RT and SPL resources. This work focused on individual drugs; future work could include the annotation of drug classes using concepts from NDF-RT and/or ATC.
While the results of this study yielded meaningful results that can be of immediate use to the research community, more work is required to maximize the potential of these data sources. For instance, automatic annotation methods could be improved but ideally the primary data sources (e.g., SPL) would contain structured and codified content that is computationally accessible. Also, new terms could be added to existing terminologies and ontologies but perhaps more important are the additions of cross-references and links among those resources. Both of these approaches will add value to these data sets, facilitating drug-repositioning studies and efforts to detect ADEs.
Highlights.
We disambiguate PharmGKB drug and disease associations by NDF-RT and SPL.
Detailed clinical associations are clearly represented in PharmGKB.
The work helps to understand drug and disease relations in details from PharmGKB.
Reveals standardized drug information will accelerate clinical drug integration.
ACKNOWLEDGMENTS
This work was supported by the NIH/NIGMS (U19 GM61388; the Pharmacogenomic Research Network).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Qian Zhu, Email: zhu.qian@mayo.edu, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA.
Robert R Freimuth, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA.
Jyotishman Pathak, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA.
Matthew J Durski, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA.
Christopher G Chute, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA.
REFERENCES
- 1.Hewett MO, Rubin DE, Easton DL, Stuart KL, Altman JM, Klein RB. T E: PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30(1):163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pathak J, Weiss LC, Durski MJ, Zhu Q, Freimuth RR, Chute CG. Integrating VAs NDF-RT Drug Terminology with PharmGKB: Preliminary Results; Pacific Symposium on Biocomputing (PSB); 2012. [PMC free article] [PubMed] [Google Scholar]
- 3.Theobald M, Shah N, Shrager J. Extraction of conditional probabilities of the relationships between drugs, diseases and genes from PubMed guided by relationships in PharmGKB; Proceedings of the American Medical Informatics Associated Symposium; 2009. pp. 124–128. [PMC free article] [PubMed] [Google Scholar]
- 4.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. Pharmacogenomics Knowledge for Personalized Medicine. [Last accessed by April. 11, 2013];Clinical Pharmacology & Therapeutics. 2012 92(4):414–417. doi: 10.1038/clpt.2012.96. PharmGKB, www.pharmgkb.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vilar S, Harpaz R, Chase HS, Costanzi S, Rabadan R, Friedman C. Facilitating adverse drug event detection in pharmacovigilance databases using molecular structure similarity: application to rhabdomyolysis. JAMIA. 2011 Dec;18(Suppl 1):i73–i80. doi: 10.1136/amiajnl-2011-000417. Epub 2011 Sep 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benton A, Ungar L, Hill S, Hennessy S, Mao J, Chung A, Leonard CE, Holmes JH. Identifying potential adverse effects using the web: A new approach to medical hypothesis generation. J Biomed Inform. 2011 Dec;44(6):989–996. doi: 10.1016/j.jbi.2011.07.005. Epub 2011 Jul 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang G, Solbrig HR, Chute CG. ADEpedia: A Scalable and Standardized Knowledge Base of Adverse Drug Events Using Semantic Web Technology. AMIA Annu Symp Proc. 2011;2011:607–616. Epub 2011 Oct 22. [PMC free article] [PubMed] [Google Scholar]
- 8. [Last accessed by April. 11 2013];VA National Drug File Reference Terminology. http://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/NDFRT.
- 9. [Last accessed by April. 11 2013];The FDA Structured Product Labeling URL. http://www.fda.gov/ForIndustry/DataStandards/StructuredProductLabeling/default.htm.
- 10. [Last accessed by April. 11 2013];DailyMed. http://dailymed.nlm.nih.gov/dailymed/about.cfm.
- 11.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49:624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 12.Jonquet C, Shah NH, Youn CH, Callendar C, Storey MA. NCBO Annotator: Semantic Annotation of Biomedical Data. ISWC. 2009:170–173. [Google Scholar]
- 13. [Last accessed by April. 11 2013];RxNav API. http://rxnav.nlm.nih.gov/RxNormRestAPI.html.
- 14. [Last accessed by April. 11, 2013];UMLS Semantic Types. http://www.nlm.nih.gov/research/umls/META3_current_semantic_types.html.
- 15. [Last accessed by April. 11, 2013];OpenNLP. http://opennlp.apache.org/ [Google Scholar]
- 16.Glibenclamide. http://en.wikipedia.org/wiki/Glibenclamide. [Google Scholar]
- 17.Arakawa R, Tamehiro N, Nishimaki-Mogami T, Ueda K, Yokoyama S. Fenofibric acid, an active form of fenofibrate, increases apolipoprotein A-I-mediated high-density lipoprotein biogenesis by enhancing transcription of ATP-binding cassette transporter A1 gene in a liver X receptor-dependent manner. Arterioscler. Thromb. Vasc. Biol. 2005;25:1193–1197. doi: 10.1161/01.ATV.0000163844.07815.c4. [DOI] [PubMed] [Google Scholar]
- 18.Cyanocobalamin. http://en.wikipedia.org/wiki/Cyanocobalamin. [Google Scholar]
- 19.Hassanzadeh O, Zhu Q, Freimuth R, Boyce R. Extending the "Web of Drug Identity" with Knowledge Extracted from United States Product Labels. AMIA Summit on Clinical Research Informatics. 2013 submitted to. [PMC free article] [PubMed] [Google Scholar]
- 20. [Last accessed by April. 11, 2013];PubChem. http://pubchem.ncbi.nlm.nih.gov/
- 21. [Last accessed by April. 11, 2013];DrugBank. www.drugbank.ca. [Google Scholar]
- 22.Parish CR. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol. Cell Biol. 1999;77:499–508. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]