Abstract
Peptidylglycine monooxygenase is a copper-containing enzyme which catalyzes the amidation of neuropeptide hormones, the first step of which is the conversion of a glycine-extended pro-peptide to its α-hydroxyglcine intermediate. The enzyme contains two mononuclear Cu centers termed CuM (ligated to imidazole nitrogens of H242, H244 and the thioether S of M314) and CuH (ligated to imidazole nitrogens of H107, H108 and H172) with a Cu-Cu separation of 11 Å. During catalysis, the M site binds oxygen and substrate and the H site donates the second electron required for hydroxylation. The WT enzyme shows maximum catalytic activity at pH 5.8, and undergoes loss of activity at lower pHs due to a protonation event with a pKA of 4.6. Low pH also causes a unique structural transition in which a new S ligand coordinates to copper with an identical pKA, manifest by a large increase in Cu-S intensity in the XAS. In previous work (Bauman, A. T., Broers, B. A., Kline, C. D., and Blackburn, N. J. (2011) Biochemistry 50, 10819–10828) we tentitively assigned the new Cu-S interaction to binding of M109 to the H-site (part of an HHM conserved motif common to all but one member of the family). Here we follow up on these findings via studies on the catalytic activity, pH-activity profiles, and spectroscopic (EPR, XAS and FTIR) properties of a number of H-site variants, including H107A, H108A, H172A and M109I. Our results establish that M109 is indeed the coordinating ligand, and confirm the prediction that the low pH structural transition with associated loss of activity is abrogated when the M109 thioether is absent. The histidine mutants show more complex behavior, but the almost complete lack of activity in all three variants coupled with only minor differences in their spectroscopic properties suggests that unique structural elements at H are critical for functionality. The data suggest a more general utility for the HHM motif as a copper- and pH-dependent conformational switch.
Introduction
Mononuclear copper monooxygenases represent a small but important group of metalloenzymes involved in neurotransmitter and peptide hormone biosynthesis. They include the enzymes dopamine β-monooxygenase (DBM)1 (1) and tyramine β-monooxygenase (TBM) (2) involved in catecholamine biosynthesis, and peptidylglycine monooxygenase (PHM) which catalyzes the amidation of neuropeptides hormones, the first step of which is the conversion of a glycine-extended pro-peptide to its α-hydroxyglcine intermediate (3). The active sites of these enzymes appear to be homologous as determined by a combination of crystallographic (4–8), spectroscopic (9–13), kinetic (1, 2, 14, 15) and computational studies (8, 16–18), but PHM remains the only member of the group for which crystal structures are available. The two copper centers termed CuM and CuH are mononuclear, and are separated by 11 Å of solvent-filled channel, in contrast to the better characterized dinuclear centers in hemocyanins, tyrosinases (17, 19–21) and oxygen activating models (22, 23) in which the Cu-Cu distance is 3–4 Å. A CuM-superoxo intermediate has been suggested on the basis of additional crystallographic (24) and biochemical data (14, 25) while in silico studies (16, 17) have validated Cu(II)-O2•− as a probable reactive oxygen species. It has been further suggested that the large spatial separation of the Cu centers in PHM prevents immediate formation of the peroxide, and thus allows the potent electrophilic reactivity of the mononuclear Cu(II)-superoxo species to be fully expressed in the form of H-atom abstraction from the substrate (14, 16, 17) to form a mononuclear hydroperoxo species at CuM and a substrate radical.
The M-site is considered to be the catalytic locus and is coordinated by H242, H244 and solvent ligands in the oxidized form with a weak EXAFS-indetectable interaction with the thioether of M314; on reduction the solvent ligands dissociate and the thioether S from M314 binds to the Cu(I) (12, 13, 26). A structure of reduced PHM co-crystalized with a slow substrate has allowed the visualization of a “pre-catalytic complex” involving a dioxygen molecule bound at CuM, the bond length of which is consistent with a Cu(II)-superoxo species. The O-O bond is oriented away from the C–H bond of the substrate which binds nearby, but a facile rotation about the Cu-O bond could bring the distal O and the substrate C–H bond into alignment (24). The M314 ligand plays a critical role in optimizing the M-site for catalysis since mutation to His, Cys or Asp results in ~95% loss in activity (2, 27). Whereas the M-site is the catalytic center, the H-site is believed to be an electron transfer center responsible for supplying the second electron necessary to complete the monooxygenation reaction. In the resting oxidized protein CuH is unremarkable with a [(His)3(OH2)] ligand set, but reduction again induces loss of solvent, and generates a Cu(I) site with Cu-N(His) distances more typical of a 2-coordinate system (1.88 Å) (26, 28, 29). The similarity of the EXAFS of the reduced protein in the WT and H172A derivatives suggests that of the three copper-coordinating His residues (107,108, and 172), H172 is only weakly bound in the reduced protein. Nevertheless, mutation to alanine has a dramatic effect on catalysis with the kcat decreasing by three orders of magnitude (15). Furthermore, crystallographic analysis reveals a structural interaction between the M and H sites, with the M314I inducing dissociation of the H107 ligand from the H-center, some 11 Å distant (6). H172 forms a stacking interaction with the conserved Y79 residue, and it has been suggested from studies on the related enzyme TBM (30), that the H172 ligand might form the exit pathway for the electron as it transfers from H to M using Y79 and oriented water molecules as additional elements of the ET pathway (31).
WT PHM shows maximum catalytic activity at pH 5.8, and undergoes loss of activity at lower pHs due to a protonation event with a pKA of 4.6. Low pH also causes a unique structural transition in which a new S ligand coordinates to copper with an identical pKA, manifest by a large increase in Cu-S intensity in the XAS. In previous work we tentitatively assigned the new Cu-S interaction to binding of M109 to the H-site (part of the HHM conserved motif common to all but one member of the family), induced by protonation of one of the H-site histidine residues (27). These data suggested that the H-site is also conformationally mobile and hint at allosteric gating of ET via long-range structural perturbations. In the present paper we follow up on these findings via studies on the catalytic activity, pH-activity profiles, and spectroscopic properties of a number of H-site variants, including H107A, H108A, H172A and M109I. Our results establish that M109 is indeed the coordinating ligand, and confirm the prediction that this mutant should show no decrease in activity at low pH. The histidine mutants show more complex behavior, but the almost complete lack of activity in all three variants coupled with only minor changes in spectroscopic properties suggests that unique structural elements at H are critical for functionality.
Materials and Methods
Buffers and ascorbate were obtained from Sigma-Aldrich at a minimum purity of 99%. Beef liver catalase was acquired from Roche. Substrates Ac-Tyr-Val-Gly (Ac-YVG) and dansyl-Tyr-Val-Gly (dansyl-YVG) were purchased from Peptide International and American Peptide Co, respectively.
Construction of PHMcc CuH site Mutants
WT, H172A, and H107A PHMcc were constructed as previously reported (32, 33). PHMcc mutants (H108A and M109I) were individually introduced into pBS.∆ProPHM382s (obtained as a gift from Betty A. Eipper and Richard E. Mains) using Splicing by Overlap Extension (SOEing) (32, 34). Sense and antisense oligonucleotide primers (Table S1) encoding about 15 bases downstream and upstream of the mutation were used for site-directed mutagenesis and paired with primers upstream and downstream of two restriction enzyme sites, ClaI & XbaI. PCR products were purified on agarose gels. Final PCR products were Phenol-Chloroform extracted, digested using restriction enzymes (NEB), fractionated on agarose gels, purified via Qiagen PCR kit, and then ligated into the pCIS.2CXXNH expression vector (also a gift from Betty A. Eipper and Richard E. Mains). Sequence analysis was performed on mutant clones, and Qiagen midi prep was used to ensure 20 ug/20 uL of recombinant DNA for transfection.
Screening PHMcc CuH Site Mutants
CHO DG44 cells were transfected with the recombinant DNA using Lipofectamine 2000 (Invitrogen). The transfected cells were subsequently selected for Dhfr cell lines in α-minimum Eagle’s medium containing 10% dialyzed fetal bovine serum (33, 35). Only those cells that retained the Dhfr gene (co-located with PHM on the plasmid) were capable of growth under these conditions. Monoclonal cell lines were created by serial dilution into 96- well plates, in order to select for wells which contained single-cell colonies. These were passed individually into a fresh 96 well, grown to confluence, and screened via Western blot for PHMcc production under similar conditions. The strongest producers were inoculated into a Hollow Fiber Bioreactor with 5 MWCO (Fibercell Systems, Inc).
Western Blot Analysis
CHO DG44 cells were incubated in DMEM/F12 containing 0.5% Fetal Clone II (FCII, Fisher) for at least 24 hours before a sample was collected which was then combined with SDS, and heated for five minutes at 100° C. Each sample was separated by 8–25% SDS-PAGE, and then transferred to an Immobilon P membrane (Millipore) using the PhastSystem. PHM proteins were visualized using rabbit antibody 246 [rPAM(116–13l)] (36) diluted 1:1500, and secondary antibody-anti-rabbit IgG (Sigma) diluted 1:1000, followed by an AP Conjugate Substrate Kit (Bio-Rad Laboratories).
PHMcc Expression and Purification
Variant cell-lines WT, H107A, and H172A (kindly provided to us by Richard E. Mains and Betty A. Eipper), and H108A and M109I (constructed in house) were grown as described previously (26, 37). Briefly, the stably transfected cell lines were thawed from freezer stock into a T75 flask with 20 mL of DMEM/F12 medium containing 10% FCII serum (Fisher). At 80 percent confluence the cells were passed into five NUNC triple flasks (500 cm2 area per flask) which were also grown to confluence. Cells were trypsinized and resuspended in 50 mL medium with 10% FCII serum prior to inoculation into the extracapillary space (ECS) of a Hollow Fiber Bioreactor (Fibercell Systems 4300-C2008, MWCO 5 kD, 3000 cm2 surface area) precultured with 2 L of 50 mM PBS pH 7.35 and 2 L of DMEM/F12 10% FCII serum (26, 37, 38).
Individual bioreactors containing each of the variants were fed with DMEM/F12/10% FCII serum for a month, after which the serum level was reduced to 0.5% FCII serum (38). At this point, the bioreactors were fed with 0.5% serum-containing medium every other day and spent medium (20 mL) from the ECS was collected and frozen at −20 °C for later purification. About a month worth of bioreactor harvest (300 mL) for each variant was purified as previously described (38).
PHMcc Copper Reconstitution
Purified enzyme was dialyzed against 20 mM sodium phosphate buffer, pH 8.0 and then reconstitution with cupric sulfate by slow addition of 2.5 molar equivalents Cu(II) per protein followed by two cycles of dialysis to remove unbound cupric ions. Concentrations were determined using OD280(1%) = 0.980 on a Cary 50 spectrophotometer. Copper concentrations were determined using a Perkin-Elmer Optima 2000 DV inductively coupled plasma optical emission spectrometer.
Specific Activity Measurements
Enzymatic activity was measured by monitoring oxygen consumption in a Rank Brother’s oxygen electrode at 37° C, as previously reported (39). Each reaction was performed in a water-jacketed vessel in 2 mL total volume containing 100 mM MES pH 5.5, 200 µL of a 6 mg/mL catalase solution (47,000 units per mg), 100 µL of 100 µM Cu(II) solution, 10 µL of 2 M stock ascorbate, and 80 µM dansyl-YVG substrate. In some cases various concentrations of imidazole up to 10 mM were added in an attempt to rescue activity. The reaction was allowed to equilibrate for approximately 1 minute, the reaction vessel was capped, and a baseline was measured for 50 seconds prior to initiation of the reaction. The reaction was initiated by addition of 10 to 20 µL of enzyme (concentrations varied depending on the activity of the particular variant) through the cap using a Hamilton syringe. The oxygen consumption was monitored and analyzed as previously reported (27, 39). Steady state kinetic measurements were performed as above, varying concentrations of dansyl-YVG between 2.5 and 400 µM. Kinetic constants were determined by fitting raw data to the Michaelis-Menten equation using nonlinear regression. Similar assay conditions were used for the measurement of pH-activity profiles in the pH range 3 – 9, except that a mixed buffer system was employed containing equal volumes of 100 mM each of MES, HEPES, CHES, and formic acid adjusted to the desired pH with sodium hydroxide.
Coupling of oxygen and product
Coupling of oxygen consumption to product formation was determined for each mutant using HPLC to determine substrate consumed, and the O2-electrode to determine oxygen consumed at 37° C. A reverse phase HPLC Varian Pro Star solvent delivery module was utilized to separate and quantify substrate consumption and product accumulation as previously reported (27). Reactions were performed in a water-jacketed glass reaction vessel, under similar reaction conditions as for the specific activity measurements (substrate and enzyme concentrations were adjusted for each experiment). The reaction was allowed to equilibrate for 1 minute prior to initiating the reaction with enzyme. An aliquot of 200 µL from the reaction vessel was removed and quenched with 20 µL of 20% TFA after 150–200 seconds of reaction time. (In the case of M109I, an aliquot of 200 µL from the reaction vessel was removed and quenched with 200 µL of 20% TCA in order to quench the reaction). Substrate and product were separated via HPLC, and concentrations were determined using a standard curve of 10–250 µM dansyl-YVG run under the same conditions (27, 39). Micromoles oxygen consumed were determined by subtracting the O2 concentrations at the time of sampling from the value immediately before reaction initiation.
XAS Samples
Oxidized samples were prepared in a single step by 5-fold dilution of 2 mM protein in 20 mM phosphate pH 8.0 (4 mM in Cu (II)) with the appropriate mixed buffer containing 20% ethylene glycol (27). Reduced protein samples were prepared under anaerobic conditions by 5-fold dilution of a 2 mM protein (4 mM in Cu(II) sample of the oxidized enzyme with the appropriate buffer containing 5 mM ascorbate and 20% ethylene glycol. Samples were transferred to an XAS cuvette via a syringe and flash frozen in liquid nitrogen. Final PHMcc copper concentrations ranged from 600 to 1200 µM.
Collection and Analysis of XAS Data
Copper K-edge (8.9 keV) extended X-ray absorption fine structure (EXAFS) and X-ray absorption near edge structure (XANES) data were collected at the Stanford Synchrotron Radiation Lightsource operating at 3 GeV with currents between 300 and 450 mA maintained by continuous top-up. Samples were measured on beamline 7–3 using a Si[220] monochromator and a Rh-coated mirror upstream with 13 keV energy cutoff in order to reject harmonics. Data were collected in fluorescence mode using a highcount rate Canberra 30-element Ge array detector with maximum count rates per array element less than 120 kHz. A Z-1 nickel oxide filter and Soller slit assembly inserted in front of the detector was used to reduce elastic scattering relative to the Cu Kα fluorescence. Four to six scans of a sample containing only buffer were averaged and subtracted from the averaged data for each protein sample to remove the Ni Kβ fluorescence and produce a flat pre-edge baseline. Samples were measured as aqueous glasses in 20 percent ethylene glycol at 10 K. Output from each detector channel was inspected for glitches and dropouts before inclusion in the final average.
Data reduction and background subtractions were performed using the program modules of EXAFSPAK (40). Spectral simulation was carried out using EXCURVE version 9.2 (41–43) as described previously (26). Simulations of the EXAFS data used a mixed-shell model consisting of imidazole from histidines residues and S (Met) coordination. The threshold energy, E0, was chosen at 8985 eV and refinement of structural parameters included distances (R), coordination numbers (N), and Debye-Waller factors (2σ2), and included multiple scattering contributions from outer-shell atoms of imidazole rings.
CO Binding
Purified PHMcc was concentrated to approximately 2 mM (4 mM in copper) in 20 mM phosphate pH 8.0, and pH-adjusted with four volumes of 50 mM mixed buffer MES/HEPES/CHES/Formate at either pH 3.5 or 7.5 in a septum-sealed conical vial. Samples were purged with CO before the addition of a 5-fold excess (5 mM) of anaerobic buffered ascorbate, and then incubated under an atmosphere of pure CO for 10 – 15 minutes.
The carbonylated protein solutions were loaded into the IR cell at a final concentration of 500 µM (1 mM in copper). After the protein data were collected, the cell was flushed with buffer and remeasured to collect a baseline. FTIR data were recorded on a Bruker Tensor 27 FTIR spectrometer at room temperature with a sample chamber that was continuously purged with CO2-free dry air. Samples were equilibrated inside the instrument sample chamber for 15 minutes to allow purging of water vapor and CO2 prior to data collection. One thousand scans were collected for each sample and buffer spectrum from 2250–1900 cm−1 at a nominal resolution of 2 cm−1. Baseline subtraction and spectral analysis were performed using the GRAMS AI Spectroscopy Software (Thermo).
Results
Steady State Kinetics
The catalytic activity of all three variants (H107A, H108A, and M109I) was measured under saturating conditions of ascorbate and atmospheric O2, as a function of peptidylglycine substrate (dansyl-YVG), and the data fit by non-linear regression to a standard Michelis-Menton equation. Kinetic constants are compared with data for the WT enzyme in Table 1. The H107A and H108A have low activity which can be seen to be primarily the result of a large decrease in kcat. The effect on KM is different for the two mutants, with H108A binding the peptide substrate more tightly than WT, and H107A binding three times weaker. Given the fact that the substrate binds in the vicinity of the M center, the effects on KM induced by His to Ala mutation at the H center are intriguing. At the pH optimum for catalysis (5.8), the M109I substitution is not expected to have any effect on H-site copper coordination. However we consistently observed the somewhat puzzling result of a significant decrease in specific activity, with the major effect on kcat. Addition of imidazole to the His to Ala mutants up to a concentration of 10 mM was unable to rescue catalytic activity.
Table 1.
Kinetic parameters, oxygen coupling ratios and copper binding stoichiometry of PHM H-site variants compared with the wild-type enzyme. Estimated errors in kinetic constants are ± 4 percent. Values for oxygen coupling in the WT protein are from reference 15.
| Variant | Km (uM) | kcat (s−1) | Specific Activity |
[Dansyl YVG] per [O2] |
Cu:PHM |
|---|---|---|---|---|---|
| WT | 8.2 | 13.8 | 25.7 | 0.90 ± 0.08 | 2.02 ± 0.15 |
| M109I | 11.8 | 4.6 | 13.7 | 1.01 ± 0.19 | 2.08 ± 0.14 |
| H107A | 3.3 | 0.08 | 0.25 | 0.97 ± 0.05 | 2.01 ± 0.01 |
| H108A | 19.4 | 0.11 | 0.38 | 0.97 ± 0.13 | 2.04 ± 0.23 |
Copper Binding
One possibility for the dramatic decrease in catalytic rate of the H107A and H108A variants would be a loss of copper due to the loss of a critical histidine residue. Measurement of copper binding stoichiometry showed that this was not the origin of the loss of activity, since both His to Ala mutants bound Cu(II) at a ratio of close to 2:1 (Table 1). This result is comparable to that for the H172A mutant which bound Cu(II) with a ratio between 1 and 2 (15, 28). The data suggests that loss of either H107 or H108 can be compensated by coordination of a solvent to complete the expected 4-coordinate geometry for a cupric ion. Nevertheless, the higher binding ratios for the H107A and H108A mutants relative to H172A, may indicate that H172 is more important for stabilizing the H-site structure. The contiguous positioning of H107 and H108 on the same β-strand constrains these ligands to coordinate via their Nδ donor atoms, which may introduce strain into the 4-coordinate (His)3(OH2) ligand set in the WT. Therefore, replacement of either H107 or H108 with a solvent ligand may result in a lower energy structure than a similar substitution at H172. The non-coordinating M109I variant reconstitutes with 2 Cu(II) per protein as predicted for the presence of all three coordinating His residues.
Characterization of the Cu(II) centers by XAS and EPR
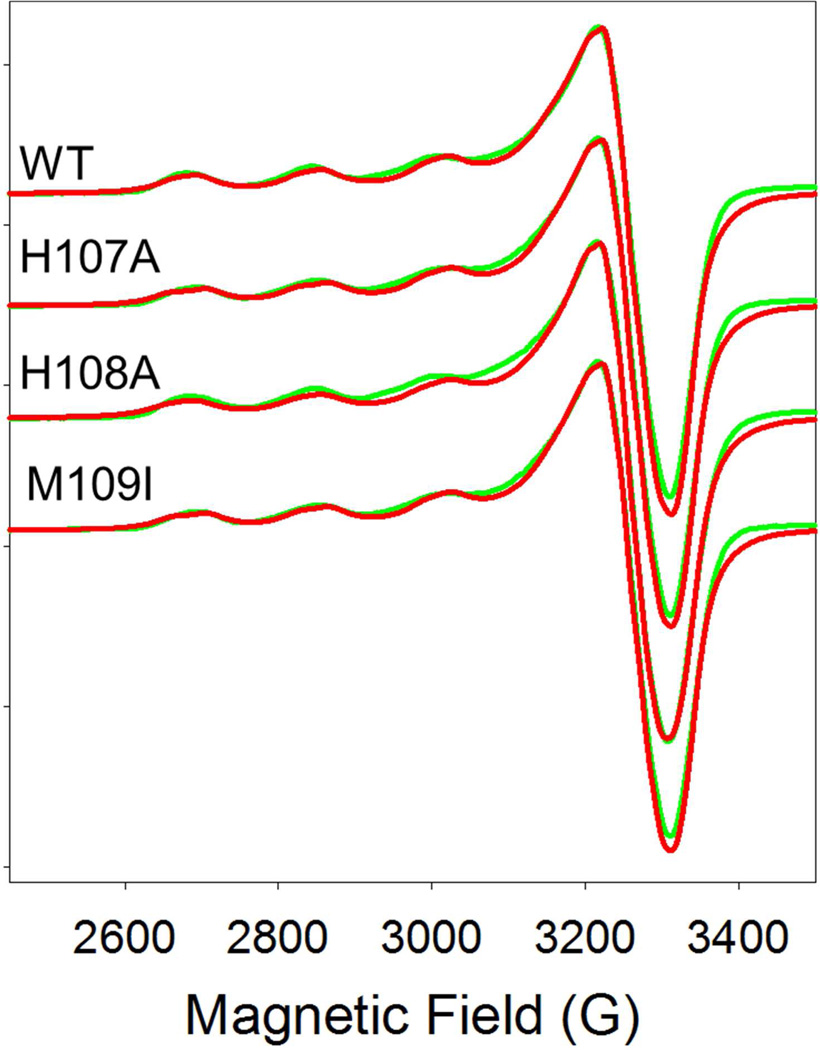
To gain further insight into the effects of the substitutions, we carried out EPR and XAS studies on the oxidized forms. X-band EPR spectra for WT, H107A, H108A and M109I at pH 5.5 are shown in Figure 2. All four spectra are extremely similar and despite the 11 Å separation, are typical of isolated mononuclear cupric centers with little or no dipolar coupling as found previously for members of this family of enzymes (9, 12, 13, 44). Simulations using the program SIMPIP (45–47) gave the best fits when two axially symmetric sites were included in 1:1 ratio, as expected for the two non-equivalent H and M sites in PHM. The g- and A-values for each site are listed in Table 2. The two sites differ slightly, with site 1 being more axial and having higher gz and Az values than site 2. Site 1 is most likely assigned to the M-center as the higher g- and A-values suggest more O-donor ligands (solvent) and fewer N-donors (His) than site 2. In line with this assignment, site 1 for the H107A and H108A variants does not change significantly, while the gz and Az values for site 2 increase slightly, as predicted by the substitution of a histidine by solvent. Alternatively, the vacant position which results from the His to Ala substitution could be occupied by an endogenous protein ligand such as a main-chain amide O group. Notwithstanding these subtle changes suggested by the simulations, the EPR parameters for the His to Ala variants are remarkably similar to WT, and rule out large changes in coordination geometry as the result of histidine removal at the H-center.
Figure 2.
Comparison of the EPR spectra of WT PHM with its H-site variants H107A, H108A, and M109I. Red lines represent experimental data, green lines simulated data using SIMPIP. Parameters used in the fits are listed in Table 2.
Table 2.
EPR parameters derived from simulations to the X-band EPR spectra of WT, H107A, H108A and M109I at pH 5.5. Instrumental parameters were as follows: Microwave frequency 9.400, modulation amplitude 4 Gauss, microwave power 2 mW, Temperature 100 K. No account was taken of differences in nuclear magnetic moments of 63- and 65-Cu isotopes. Line shapes were Lorenzian in all cases.
| gx | gy | gz | A(x,y) | A(z) | W(x,y) | W(z) | |
|---|---|---|---|---|---|---|---|
| WT- Site 1 | 2.051 | 2.069 | 2.310 | 18 | 533 | 49 | 106 |
| Site 2 | 2.042 | 2.083 | 2.279 | 25 | 510 | 49 | 94 |
| H107A- site 1 | 2.051 | 2.069 | 2.309 | 18 | 526 | 49 | 105 |
| Site 2 | 2.044 | 2.082 | 2.289 | 25 | 521 | 49 | 93 |
| H108A-Site 1 | 2.052 | 2.070 | 2.312 | 18 | 536 | 49 | 128 |
| Site 2 | 2.043 | 2.085 | 2.283 | 25 | 528 | 49 | 110 |
| M109I- Site 1 | 2.051 | 2.069 | 2.308 | 18 | 531 | 49 | 107 |
| Site 2 | 2.041 | 2.082 | 2.279 | 25 | 510 | 49 | 96 |
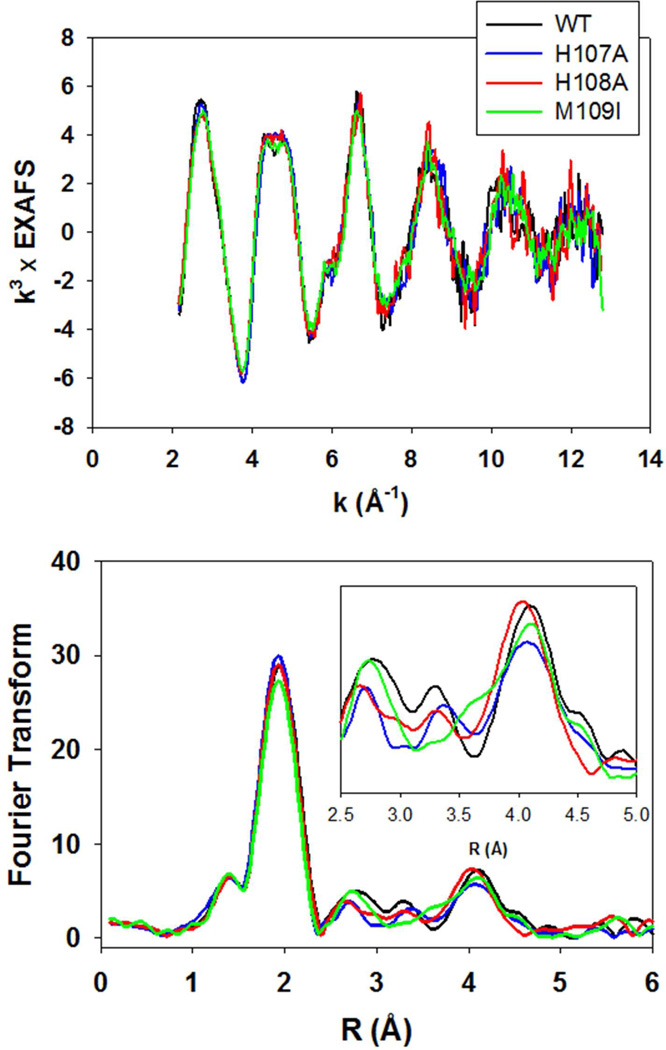
The copper coordination was also explored using X-ray absorption spectroscopy (XAS). Fig. 3 (top) shows a co-plot of the EXAFS of the WT and all three variants. The spectra overlay exactly, with differences less than the level of noise in the data. Simulations of the spectra (Table S1) confirm the result obtained by inspection of the four datasets, namely that they give rise to almost identical parameters, and correspond to the average coordination of 4N/O ligands per Cu(II) center, reported previously for DBM (10), PHM (26) and TBM (2). This is not unexpected since distinguishing features arising from the substitution of one histidine in five (averaged over both copper centers) would only be observable in the shape and/or intensity of the outer-shell features at R=2.8–4.3 Å, which are the signatures of imidazole coordination. Fig 3 (bottom panel) shows a comparison of the Fourier transforms of the WT protein with the three variants, from which it is evident that the intensities of the shell around 3 Å do appear to correlate with the loss of imidazole intensity in the H107A and H108A mutants. However, the trend is much less obvious in the 4 Å shell, where multiple scattering contributions dominate (48), and small differences in imidazole orientation can result in greater intensity shifts than coordination numbers themselves. The EXAFS data therefore confirm conclusions derived from EPR, that the H-site His residue in H107A and H108A is replaced by coordinated solvent and does not perturb the coordination geometry of the site in an observable fashion.
Figure 3.
Top, overlay of the experimental EXAFS of oxidized PHM variants. Black trace, WT; blue trace, H107A; red trace H108A; green trace M109I. Bottom, overlay of experimental Fourier transforms color-coded as for the EXAFS above. The inset shows an expanded view of the imidazole outer-shell region of the transform.
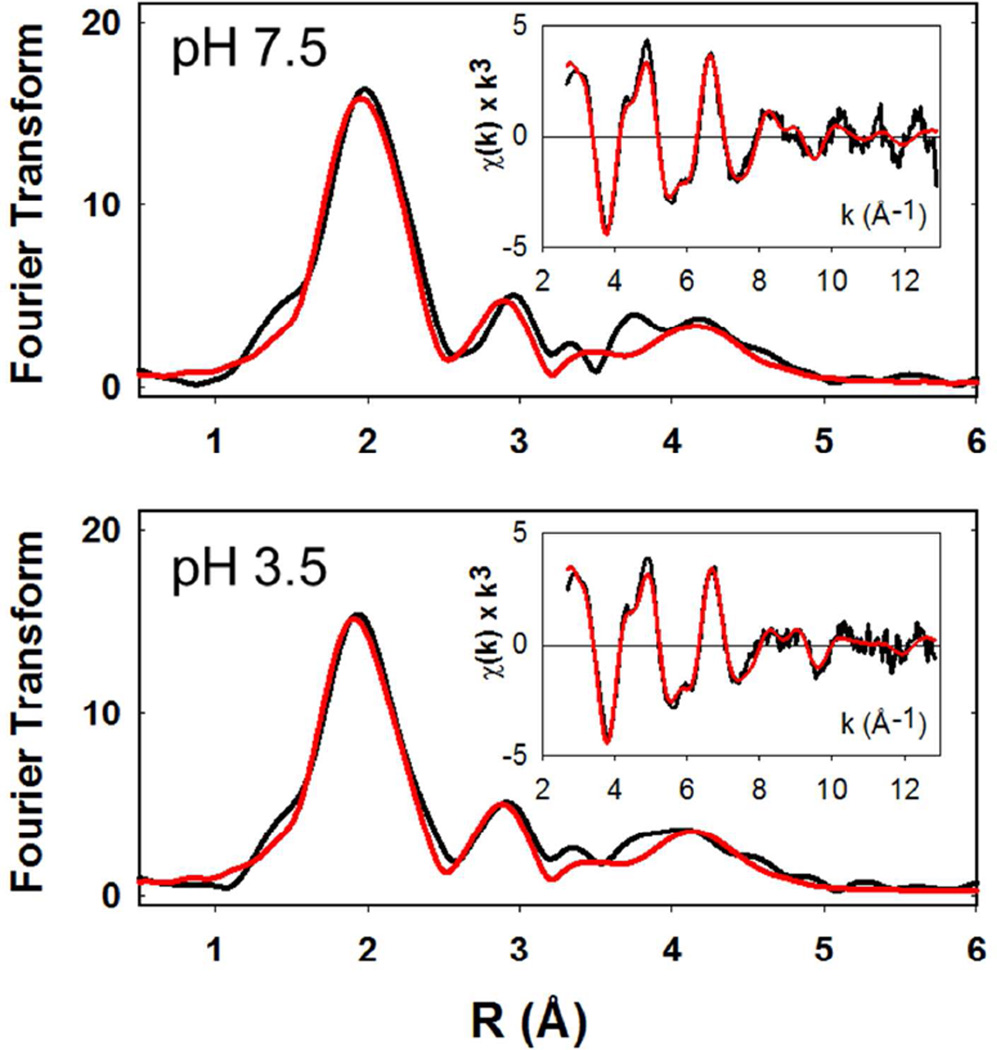
XAS studies on the Reduced Proteins
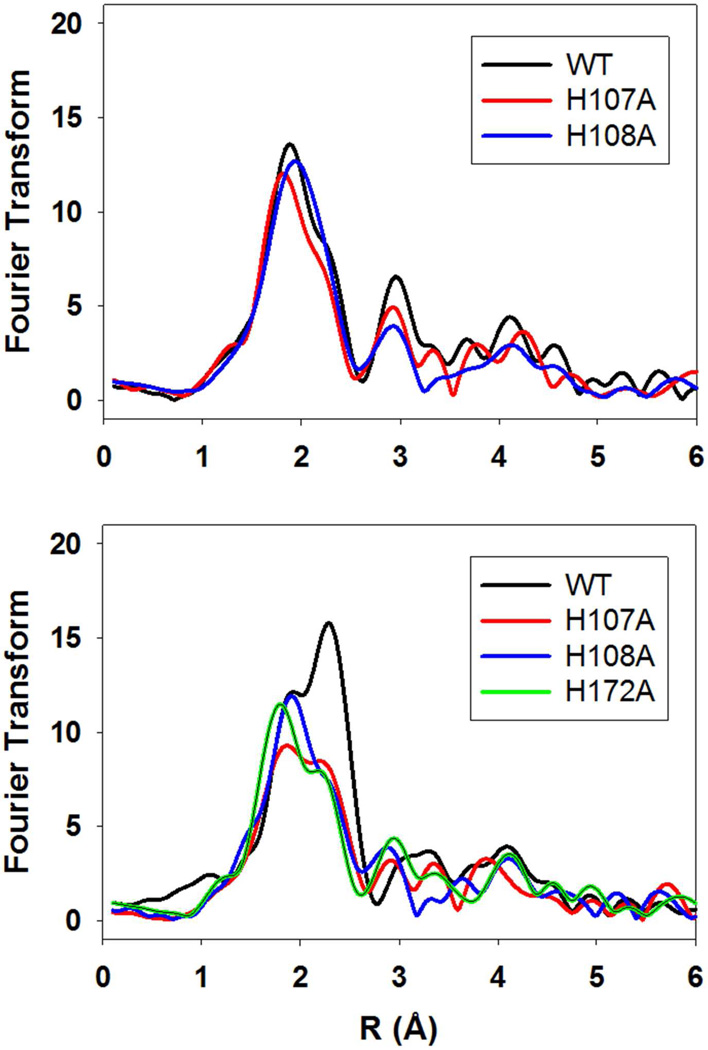
Copper coordination in the reduced proteins was probed by XAS. Figure 4 (top panel) compares the Fourier transforms for WT, H107A, and H108A at pH 7.5. The data show more complex behavior than predicted solely on the basis of histidine shell occupancy with both intensities and peak positions changing, albeit with shell occupancy decreasing by less than the predicted 20 percent. These data can be simulated (Table 3) with the expected histidine coordination numbers, and Cu-N bond lengths ranging between 1.95 Å for the WT and 1.88 Å for the H108A variant, and are broadly consistent with the trend towards a 2-coordinate site at CuH. A similar trend was observed previously in a study of the H172A variant (28) (included in Table 3 for comparison) where the average Cu-N(His) bond length also decreased towards the value (1.87 – 1.89 Å) expected for a 2-coordinate bis-imidazole Cu(I) complex (29). However, in contrast to H172A, the absorption edges of the H107A and H108A (Figure 5) do not show the expected increase in intensity of the 8983 eV edge feature associated with a linear 2-coordinate Cu(I) complex (29, 49–51) suggesting that the 2-coordinate H-centers in H107A and H108A are significantly distorted from linearity. The data all show the presence of 0.5 Cu-S due to M314 coordination at the M-center, but interestingly, the Cu-S distance appears to decrease by ~0.04 Å in H107A and H108A relative to the WT protein. This may suggest that intersite cross-talk may influence H and M individual site structure in subtle ways that are difficult to extract from the average coordination as determined by EXAFS analysis. Notwithstanding these uncertainties, it is clear that all the H-site single His variants adopt a 2-coordinate configuration with variable degrees of distortion from linearity. Fits to the EXAFS and FTs of these variants at pH 7.5 are given in Table S2 (Supporting Information).
Figure 4.
Comparison of the Fourier transforms of ascorbate-reduced PHM H-site variants. Top panel pH 7.5; bottom panel pH 3.5. Spectra are color coded as follows: black, WT; red, H107A; blue H108A; green H172A.
Table 3.
Parameters used in the simulation of the EXAFS of the H-site variants of PHM
| Fa | Nob | R(Å)c | DW(Å2) | Nob | R(Å)c | DW(Å2) | Nob | R(Å)c | DW(Å2) | -E0 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cu-N(His)d | Cu-O/Ne | Cu-S | |||||||||
| Oxidized Proteins pH 7.5 | |||||||||||
| WT | 0.318 | 2.5 | 1.97 | 0.0123 | 1.5 | 1.97 | 0.0123 | 4.69 | |||
| H107A | 0.316 | 2.0 | 1.96 | 0.0123 | 2.0 | 1.96 | 0.0123 | 5.21 | |||
| H108A | 0.447 | 2.0 | 1.97 | 0.0129 | 2.0 | 1.97 | 0.0129 | 4.95 | |||
| H172Af | 0.167 | 2.0 | 1.96 | 0.0120 | 2.0 | 1.96 | 0.0120 | ||||
| M109I | 0.287 | 2.5 | 1.97 | 0.0137 | 1.5 | 1.97 | 0.0137 | −4.30 | |||
| Reduced Proteins pH 7.5 | |||||||||||
| WT | 0.373 | 2.5 | 1.92 | 0.0175 | 0.5 | 2.24 | 0.0123 | 0.34 | |||
| H107A | 0.487 | 2.0 | 1.88 | 0.0156 | 0.5 | 2.20 | 0.0123 | −0.57 | |||
| H108A | 0.360 | 2.0 | 1.91 | 0.0152 | 0.5 | 2.20 | 0.0101 | −0.03 | |||
| H172Af | 0.379 | 2.0 | 1.90 | 0.0130 | 0.5 | 2.23 | 0.0200 | ||||
| M109I | 0.373 | 2.5 | 1.95 | 0.0150 | 0.5 | 2.21 | 0.0094 | 0.26 | |||
| Reduced Proteins pH 3.5 | |||||||||||
| WT | 0.327 | 2.5 | 1.95 | 0.0182 | 1.0 | 2.26 | 0.0102 | −0.66 | |||
| H107A | 0.860 | 2.0 | 1.91 | 0.0194 | 0.6 | 2.23 | 0.0120 | −0.26 | |||
| H108A | 0.629 | 2.0 | 1.91 | 0.0147 | 0.5 | 2.22 | 0.0120 | 0.24 | |||
| H172A | 0.659 | 2.0 | 1.88 | 0.0165 | 0.6 | 2.22 | 0.0124 | −0.67 | |||
| M109I | 0.280 | 2.5 | 1.93 | 0.0159 | 0.5 | 2.21 | 0.0139 | −0.15 | |||
F is a least-squares fitting parameter defined as
Coordination numbers are generally considered accurate to ± 25%
In any one fit, the statistical error in bond-lengths is ±0.005 Å. However, when errors due to imperfect background subtraction, phase-shift calculations, and noise in the data are compounded, the actual error is closer to ±0.02 Å.
Fits modeled histidine coordination by an imidazole ring, which included single and multiple scattering contributions from the second shell (C2/C5) and third shell (C3/N4) atoms respectively. The Cu-N-Cx angles were as follows: Cu-N-C2 126°, Cu-N-C3 −126°, Cu-N-N4 163°, Cu-N-C5 −163°.
Distances of the Cu-N(His) and Cu-N/O (non-His) shells were constrained to be equal in fits to the oxidized proteins
Data from reference (28)
Figure 5.
Comparison of absorption edges of ascorbate-reduced PHM H-site variants at neutral and low pH.
For the WT protein, M109 does not coordinate at pH 7.5 so that the H-site of M109I is expected to be similar to that of WT at this pH. The EXAFS and FT of M109I at pH 7.5 is shown in the top panel to Figure 6, and the spectral parameters extracted from simulations are listed in Table 3. These data confirm that M109I can be simulated with all three His ligands coordinated at the H-site (average 2.5 over both copper centers), with a longer Cu-N(His) bond length of 1.95 Å similar to the WT protein. However, the Cu-S bond length has decreased to 2.21 which may signal some perturbation at the M-center, as the result of removal of the methionine side chain. While the decrease in RCu-S is close to the limit of detection, we note that this intriguing result could be related to the unexpected ca 40% decrease in catalytic activity also observed for this variant.
Figure 6.
pH dependence of the EXAFS and Fourier transforms of the M109I variant. Black traces are experimental data, red traces are simulated data. The top panel represents data collected at pH 7.5, the bottom panel is data collected at pH 3.5.
pH Dependence and the Role of M109 in the Low-pH Transition
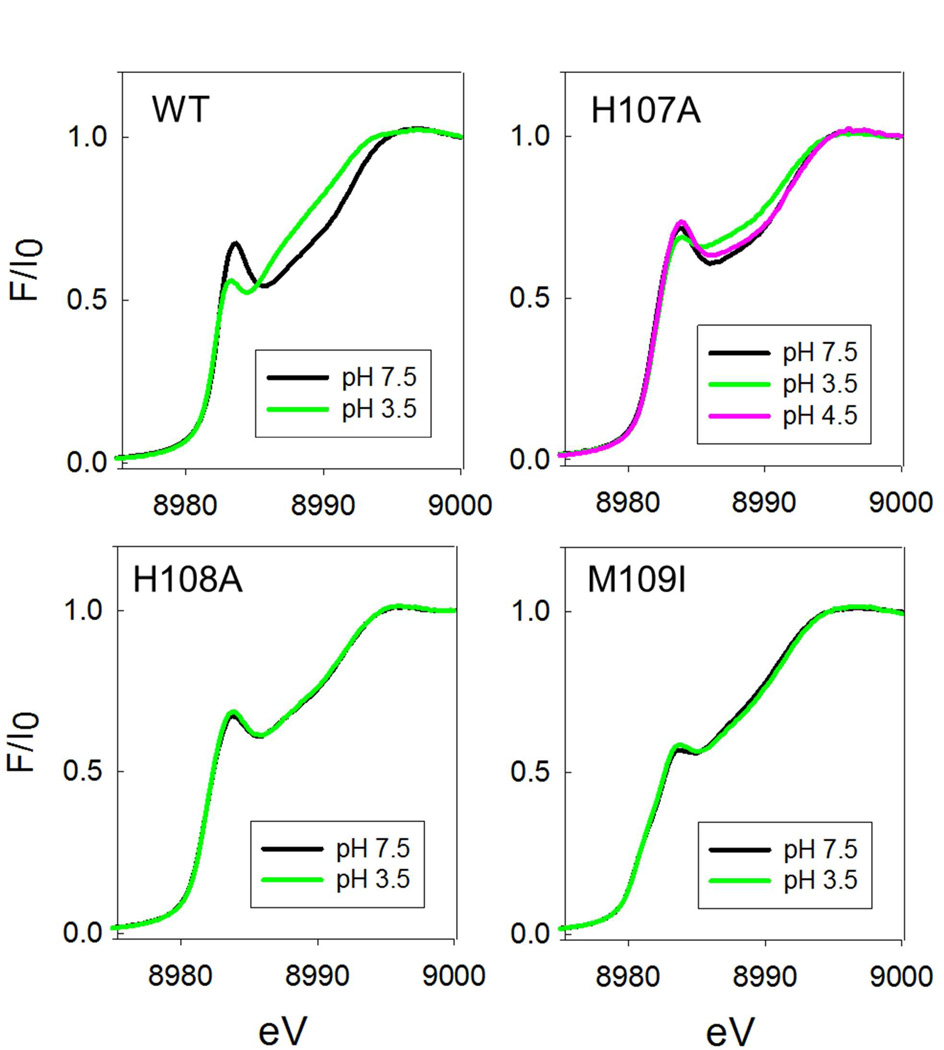
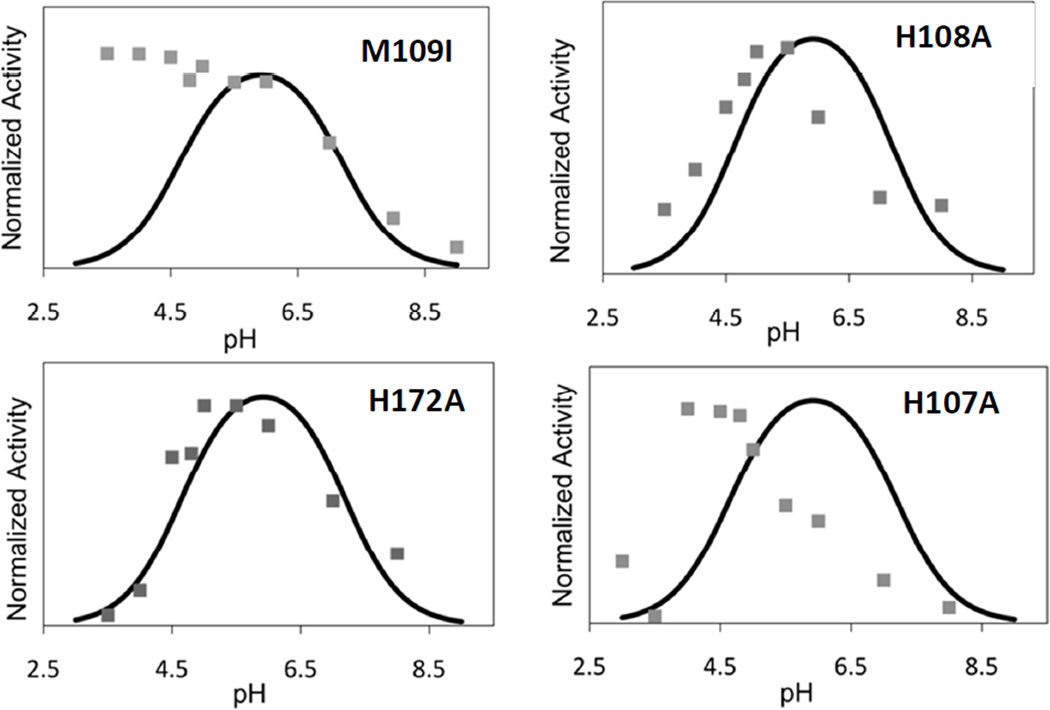
Our previous studies have suggested that the decrease in activity at low-pH is due to a conformational change induced by a protonation event with pKA of 4.6–4.7 which results in the coordination of an additional Met ligand at one or other of the two coppers (27). Based on observation of similar behavior in the homologue TBM, and sequence comparisons between PHM, TBM, and DBM we proposed that M109 was the likely origin of the low-pH Met ligand, and that the conformational change was initiated by protonation of one of the His ligands at the H-center. The hypothesis leads to two predictions (i) M109I should show no decrease in catalytic activity at low pH, and (ii) the Met-off to Met-on transition should be absent in M109I. These predictions were tested by measuring the pH dependence of both the catalytic activity and the EXAFS-derived H-site coordination of the M109I variant.
Figure 7(a) compares the pH-activity profile of M109I with that of the WT enzyme. Differences in catalytic rate were factored out by normalizing the rate to unity at the pH optimum of the WT enzyme so that changes in pH-dependence of the rate profile were directly comparable. In the Figure, the data for WT are represented by the solid black line which corresponds to the simulation of the WT rate versus pH data published previously (27). The data provides a dramatic confirmation of the prediction, viz that in M109I the rate remains high as the pH decreases below 5.5, and may actually increase in the pH range 5.5–3.0. In a second set of experiments, we compared the EXAFS of M109I at pH 7.5 and 3.5 as shown in Figure 6. The spectrum at pH 3.5 (Fig. 6 bottom panel) is identical to that at pH 7.5, and lacks the increased intensity at 2.3 Å due to the additional Cu-S(Met) ligand, which is the hallmark of the low-pH structural transition (see Fig. 4 bottom panel and reference (27)). These data confirm our second prediction, namely that the low-pH Cu-S(Met) interaction is eliminated in the M109I variant. Therefore, we can state with confidence that M109 coordinates via its thioether S atom in the low-pH form.
Figure 7.
pH-rate profiles for PHM H-site variants M109I, H108A, H172A and H107A. Rates were measured using the standard assay conditions at the different pHs as described in the text. To aid in comparison to the WT protein, all rate data have been normalized to unity, despite large differences in activity (as shown in Table 1). The solid trace in each panel is the simulated rate profile for the WT enzyme as determined in reference (27).
An unanswered question is the origin of the group which protonates. Previously we argued that a pKA of 4.6 was consistent with protonation of the coordinated Nδ/Nε of the imidazole side chain of a histidine ligand. This in turn leads to the prediction that mutation of the protonatable His residue might also induce the conformational change, and result in a species which (i) exhibited the met-on form at all pHs. The EXAFS of H107A and H108A at pH 7.5 (Fig. 4 top panel) do not show this behavior. Perhaps surprisingly, the mutations also abrogate M109 coordination at pH 3.5 as show in Fig. 4 (bottom) and Table 3. Additionally, it might be anticipated that mutation of the protonatable His residue would also abrogate the decrease in activity at low pH, producing instead an enzyme form with minimal activity over the entire pH range. Data on the pH-activity profiles of the H-site His to Ala variants are shown in Fig. 7. H172A shows behavior almost identical to WT, while H108A shows a slight shift in pH optimum to lower pH values. H107A, on the other hand shows a pH-rate profile which more closely resembles that of M109I. This would imply that H107 is the ligand that protonates since its absence abrogates M109 coordination and the associated decrease in catalytic rate, although it is still unclear why the other His variants do not show M109 Cu-S interaction in their low-pH EXAFS spectra.
CO binding to WT and M109I at low pH
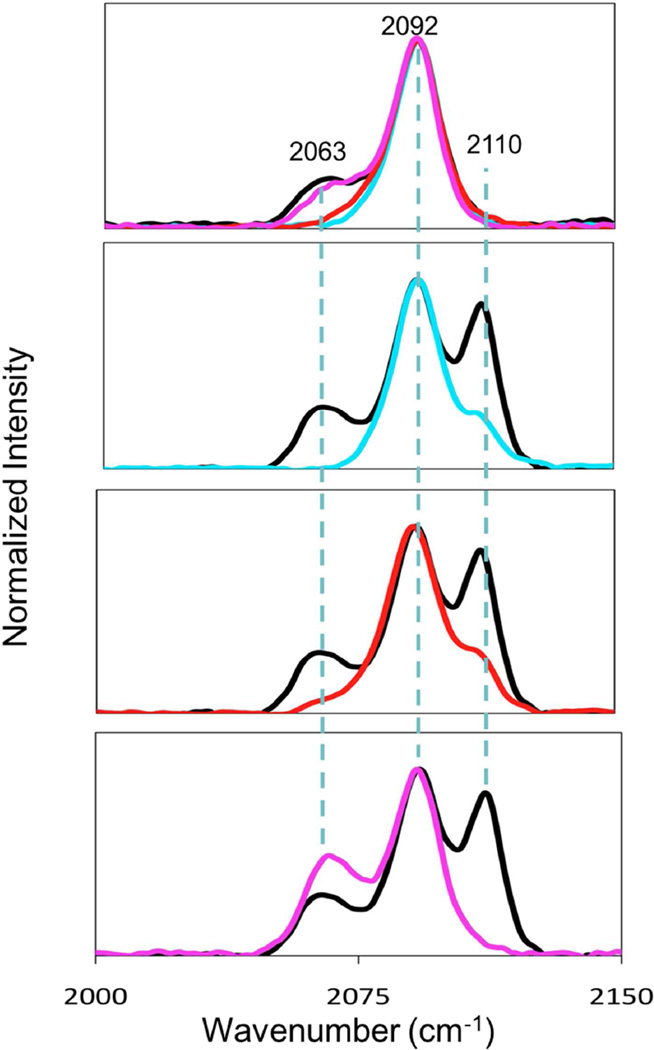
CO binds to WT enzyme at pH 5.5 and above to generate an M-site carbonyl complex which has been characterized by FTIR (11, 37) and crystallography (7). The C≡O stretching frequency is 2092 cm−1 which is consistent with a binding site comprised of 2 His, one S(Met) and CO (11, 12, 37). To gain further insight into the coordination changes at low-pH, we used FTIR to compare the CO-binding chemistry of the WT and the H-site variants at pH 7.5 and pH 3.5. These data are shown in Figure 8. As expected, all proteins show the 2092 cm−1 band associated with the M-site carbonyl at both pHs. However, a new band is observed at 2110 cm−1 in the WT protein that is absent in M109I, and we therefore assign this band to an H-site carbonyl coordinated by the thioether of M109, two His residues and the CO. This ligand set is identical to that of the M-site CO complex, yet its ν(CO) is 20 cm−1 higher, suggestive of weaker back-bonding into the π* orbitals on the CO ligand. At present we have no explanation for the differences in frequency, but the low-pH carbonyl and it’s absence in M109I adds confidence to the assignment of M109 as the coordinating ligand in the low-pH H-site structure. The IR of the H107A and H108A variants at pH 3.5 also shows some intensity at 2110 cm−1, but of much lower intensity than the WT protein. This may suggest that a small population of H107A and H108A molecules exist in the thioether-bound conformation, or that CO induces a small shift towards this conformation.
Figure 8.
FTIR spectra of the CO complexes of WT PHM and its H-site variants in the C≡O stretching region. (a) Comparison of IR spectra at pH 7.5 for WT (black), H107A (cyan), H108A (red) and M109I (magenta); (b) – (d) comparison of IR spectra at pH 3.5 for WT (black) and H107A (cyan) (panel (b)); WT (black) and H108A (red) (panel (c)); WT (black) and M109I (magenta) (panel (d)). Spectra are normalized to the intensity of the WT 2092 cm−1 band.
Discussion
Our data establish that mutation of any one of the three His residues at the H-center of PHM reduces activity to detectable yet extremely low levels. Most of the loss is associated with kcat, although KM varies by a factor of 3 with H107A having the highest affinity and H108A the lowest. This loss of activity is not due to inability to bind Cu in either the oxidized or reduced states. Furthermore, the structures as visualized by EPR and EXAFS spectroscopy appear similar in all cases to those observed in the WT fully active enzyme. In the oxidized mutant proteins, solvent appears competent to bind in place of imidazole to generate 4-or 5-coordinate H-site structures, whereas in the reduced proteins any two histidine residues appear competent to bind Cu(I), albeit with some indication from absorption edge intensities of different degrees of distortion from linearity. Since H107 and H108 are contiguous residues on the same β-strand they are constrained to bind in a trans -configuration via their Nδ imidazole N atoms and hence are likely most stable in linear 2-coordinate geometry (49). This was previously noted in an earlier study of the H172A variant where a significant increase in absorption edge intensity (characteristic of 2-coordinate linear geometry (29, 50)) was observed. For the H107A and H108A variants the 8983 eV intensity was not significantly increased above that found in the WT protein, suggesting that these variants may be unable to reorient so as to adopt the thermodynamically preferred linear structure. This observation, coupled to the almost complete loss of activity in single H-site variants, may suggest that the H-site is built on a fairly rigid scaffold where precise orientation of His ligands is an essential element of function.
It is puzzling that solvent readily substitutes for the missing imidazole side chain, yet exogenous ligands do not appear to bind at H. Azide, nitrite and peroxide bind exclusively to the M-center in the oxidized protein while CO binds to M in the reduced form (7, 8, 11–13,37). Likewise addition of imidazole to any of the H-site His to Ala mutants fails to rescue activity (ref (28) and this work). This may be due to a lack of reactivity (similar to azide), but it may also support the hypothesis advanced above that H-site reactivity is intimately involved with the connectivity of the His ligands to the protein scaffold, which either completes ET circuitry, or organizes other key elements of structure. It is noteworthy that the x-ray crystal structure of the M-site M314I mutant shows a highly perturbed H-site structure with H107 completely dissociated (6). The cross-talk between H and M implied by this structure is also manifest in changes in the KM for substrate binding of H-site mutants, even though the substrate binds at a site many angstroms distant. Thus subtle changes in H-site structure may propagate through the scaffold to M, where they could inhibit the enzyme from achieving critical conformations necessary for H-tunneling (52, 53).
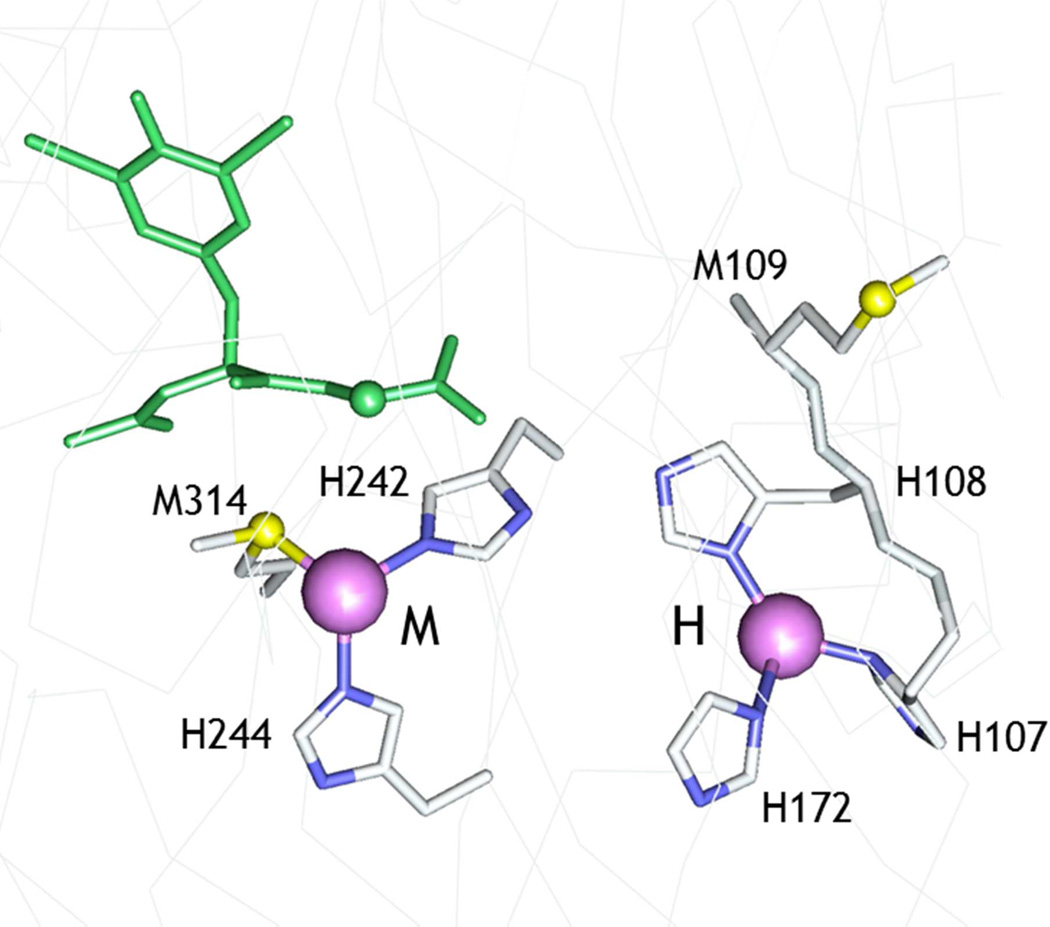
The pH dependence of catalytic activity gives further insight into protonation/ deprotonation events that interconvert active and inactive states. In this study, we have focused on the protonation event that generates an inactive state with a pKA of 4.6 (27, 39). Our earlier work advanced the hypothesis that pH induced a conformational switch between a catalytically competent active state of the H-center and an inactive state containing a new Cu-S ligand. We further speculated that the new S residue was derived from the side chain of M109 which is part of the H-site conserved HHM motif but points away from CuH on the opposite side of the β-strand (Fig. 1). The hypothesis allowed us to make two predictions (i) that the absence of a thioether at residue 109 would prevent the M109 S(Met) coordination thereby attenuating the driving force for this conformational switch, and (ii) that its absence would therefore eliminate the loss of activity at low pH. Both of these predictions were borne out by the data. The M109I variant showed a small increase in activity in the pH range 5.5 – 3.0, and lacked the high-intensity Cu-S interaction characteristic of the low-pH state of the WT enzyme. This allows us to conclude with confidence that in the WT enzyme, the low-activity state has undergone a conformational switch which flips the β-sheet, repositioning the coordinating ligands such that M109 is in a favorable orientation to bind to CuH.
Figure 1.
Active site structure of oxidized WT PHM (pdb file 1OPM). The substrate di-iodo-tyrosyl-glycine (I-YG) is shown in green. The H-site shows the relative orientation of the H107H108M109 motif in the active conformation, with the Met residue pointing away from the copper.
The observed pKa for the catalytic transition of 4.6, is within the range expected for protonation of histidine residues coordinated to Cu(I). Well established case of this behavior include the reduced forms of cupredoxins (54, 55), and Cu/Zn superoxide dismutases (56–58), where protonation is coupled to addition of an electron so as to keep the overall charge constant. As noted previously (27), the C-terminal histidine ligand of the cupredoxin site is located in a loop of sequence between the Cys112 and Met121 (azurin numbering), and the pKa for histidine protonation is sensitive to both the identity and length of the sequence (59) with values ranging from 2 to 6. For example replacement of the native loop of azurin (C112TFPGH117SALM, pKaH117<2) with shorter loops from amicyanin or plastocyanin produces chimeras with pKAs for protonation of the corresponding histidine of 5.5 and 4.3 respectively, while for plastocyanin, the pKAs of the native protein and the chimera in which the native loop (C84SPH87QGAGM92) is replaced with the azurin loop-sequence are 4.7 and 4.9. We reasoned that if protonation of an H-site coordinating histidine was responsible for the conformational switch, then its mutation to alanine should either eliminate, or at least strongly perturb both the pH-rate profile, and the structural transition leading to S(Met) binding. The data show that H172A exhibits WT pH-rate profile, while H108A has a rate profile only slightly shifted to lower pH. H107A on the other hand has a strongly perturbed rate profile which approximates to that of M109I showing an increase in rate between 5.5 and 4, below which the activity crashes to zero. Additionally, close inspection of the EXAFS data suggests an increase in Cu-S site occupancy for H107A to 0.65 at pH 3.5, while FTIR shows evidence for the S-coordinated H-site carbonyl (2010 cm−1). These observations may suggest an equilibrium between M109-on and -off states in H107A, and leads us to propose that H107 is the residue which protonates. The inability of the H107A mutant to induce a complete conformational switch was at first puzzling, as the prediction was that the absence of the protonating residue would generate the Met109-on state at all pHs. However, further analysis suggests that the switch may be driven by the replacement of the coordinating histidine by its bulky non-coordinating protonated form, and is induced by a combination of S(M109) coordination and relief of steric crowding. In this model, the hole created by the Ala substitution would create no steric restrictions, and could therefore be a stable entity at all pHs. We also note that H107A does not appear to protonate as readily in the M109I variant, as no decrease in activity is observed with M109I between pH 6 and 3. This observation implies that the pKA for His protonation is coupled to the ability of the Met ligand to coordinate: without the driving force for S ligation, Cu(I) out-competes the proton for histidine binding.
The M109-on state of the enzyme is catalytically incompetent, and the obvious next question is why? Binding of CO to the low pH inactive (S-bound) form of the WT enzyme induces a new band at 2110 cm−1, absent in the M109I variant, which we may logically assign to a 4-coordinate H-site carbonyl with two histidines, one methionine and CO. On the other hand the active state of the H-site does not form a CO complex. These observations give hints to the possible geometrical differences between active and inactive states. Cu(I) carbonyls are generally formed from 3-coordinate precursors to give predominately 4-coordinate tetrahedral complexes (51, 60, 61) and react poorly if at all with 2-coordinate Cu(I) complexes. A recent study of an H-site PHM model peptide containing the HH motif confirmed this chemistry: the 2-coordinate Cu(I)-Nδ,Nδ-bis-imidazole complex reacted sluggishly with CO to generate a weak 3-coordinate CO complex with a low-intensity ν(C≡O) between 2105 and 2110 cm−1. However, in the presence of an additional mole equivalent of imidazole, the 4-coordinate Cu(I)(His)3CO species was formed stoichiometrically, and exhibited a strong ν(C≡O) at lower frequency (2075 cm−1) as expected (29). With regards to the low pH WT PHM spectrum, it is possible that the 2110 cm−1 is due to a Cu(I)(His)2CO structure, but the strong intensity of the band together with its absence in the M109I variant suggests that the 4-coordinate Cu(I)(His)2S(Met)CO species is more likely. We believe that the assignment is reasonable since the thioether ligand is a poor donor, and hence may have little impact on the extent of back bonding.
The question remains, if the low activity S-bound H-site readily forms a carbonyl, why not the 3-coordinate high activity site? The answer must lie in the ability of lack thereof of the 3-coordinate precursor to rearrange on CO binding to a tetrahedral coordination. This again suggests that a rigid protein scaffold associated with the H-site active form may be required for electron transfer.
Most cuproproteins including PHM and DBM are packaged into vesicles in the TGN as folded apo-proteins that still require metalation. This function is performed by copper transporting ATPases (62–65) which are members of the P1B family of heavy metal transporters found in all forms of life from bacteria to mammals where they function in copper export across membranes. Mammalian ATP7A resides in the TGN or vesicular membranes and pumps copper from the cytosolic to the lumenal side of the membrane where it is believed to transfer copper directly to PHM without the intermediacy of a intravesicular copper chaperone (66). ATP7A contains a lumenal loop rich in His and Met residues (MDHHFATLHHNQNMSKEEMINLHSSM) (67, 68) which has been shown to bind Cu(I) at pH 8 with 2-coordinate (His)2 ligation and weak additional interaction with a Met residue (69). Based on in vitro data, it was postulated that that this loop binds copper as it exits the membrane channel, and passes it on to cuproproteins such as PHM which are also associated with the lumenal membrane (62). As the vesicles mature, their pH drops as the result of H+ import, resulting in an internal pH of ~5.5, close to the pH optimum of the monooxygenases with the consequence that the ATP7A lumenal loop is subject to the same pH constraints as its putative PHM partner. At present, the effect of pH on the Cu(I)-binding properties of the loop has not been investigated but it is intriguing that similar HM signals appear to be present in both systems, suggesting that HHXnM motifs may have more general utility as Cu-dependent conformational switches that could modulate coordination in response to pH. Further studies are underway to explore this and other aspects of HHM chemistry.
Supplementary Material
Acknowledgement
We thanks Drs. Betty Eipper and Richard Mains for the gift of cell lines for the WT and H107A PHM proteins.
Footnotes
This work was supported by a grant from the National Institutes of Health (R01 NS027583) to N.J.B. We gratefully acknowledge the use of facilities at the Stanford Synchrotron Radiation Lightsource which is supported by the National Institutes of Health Biomedical Research and Technology Program Division of Research Resources, and by the US Department of Energy Office of Biological and Environmental Research.
Abbreviations used: MES, 2-(N-morpholino)ethanesulfonic acid; HEPES, 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid; CHES, N-cyclohexyl-2-aminoethanesulfonic acid; dansyl-YVG, dansyl- Tyr-Val-Gly; PHM, peptidylglycine monooxygenase; EXAFS, extended X-ray absorption fine structure; XAS, X-ray absorption spectroscopy; HPLC, high pressure liquid chromatography; ICP-OES, inductively coupled plasma- optical emissions spectrometry; DBM, dopamine β-monooxygenase; TBM, tyramine β-monooxygenase; TFA, trifluoroacetic acid; TCA, trichloroacetatic acid; WT, wild-type;; Dhfr, dyhydrofolate reductase gene; CHO, chinese hamster ovary; DMEM F-12, Dulbecco’s modified Eagle’s medium; FBS FCII, fetal clone II; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; PBS, phosphate buffer saline; ECS, extra capillary space; MWCO, molecular weight cut off.
Supporting information available. Supporting information (two figures and one table) is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Klinman JP. The copper-enzyme family of dopamine b -monooxygenase and peptidylglycine a -hydroxylating monooxygenase: Resolving the chemical pathway for substrate hydroxylation. J. Biol. Chem. 2006;281:3013–3016. doi: 10.1074/jbc.R500011200. [DOI] [PubMed] [Google Scholar]
- 2.Hess CR, Klinman JP, Blackburn NJ. The copper centers of tyramine beta-monooxygenase and its catalytic-site methionine variants: an X-ray absorption study. J. Biol. Inorg. Chem. 2010;15:1195–1207. doi: 10.1007/s00775-010-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prigge ST, Mains RE, Eipper BA, Amzel LM. New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell. Mol. Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: the structure of peptidylglycine a-hydroxylating monooxygenase. Science. 1997;278:1300–1305. doi: 10.1126/science.278.5341.1300. [DOI] [PubMed] [Google Scholar]
- 5.Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Substrate-mediated electron transfer in peptidylglycine a-hydroxylating monooxygenase. Nature Struct. Biol. 1999;6:976–983. doi: 10.1038/13351. [DOI] [PubMed] [Google Scholar]
- 6.Siebert X, Eipper BA, Mains RE, Prigge ST, Blackburn NJ, Amzel LM. The catalytic copper of peptidylglycine alpha-hydroxylating monooxygenase also plays a critical structural role. Biophys. J. 2005;89:3312–3319. doi: 10.1529/biophysj.105.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chufan EE, Prigge ST, Siebert X, Eipper BA, Mains RE, Amzel LM. Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase. J. Am. Chem. Soc. 2010;132:15565–15572. doi: 10.1021/ja103117r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudzka K, Moreno DM, Eipper B, Mains R, Estrin DA, Amzel LM. Coordination of peroxide to the Cu(M) center of peptidylglycine a-hydroxylating monooxygenase (PHM): structural and computational study. J. Biol. Inorg. Chem. 2013;18:223–232. doi: 10.1007/s00775-012-0967-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess CR, Wu Z, Ng A, Gray EE, McGuirl MA, Klinman JP. Hydroxylase activity of Met471Cys tyramine b-monooxygenase. J. Am. Chem. Soc. 2008;130:11939–11944. doi: 10.1021/ja800408h. [DOI] [PubMed] [Google Scholar]
- 10.Blackburn NJ, Hasnain SS, Pettingill TM, Strange RW. Copper K-EXAFS studies of oxidized and reduced dopamine-b-hydroxylase: confirmation of a sulfur ligand to Cu(I) in the reduced enzyme. J. Biol. Chem. 1991;266:23120–23127. [PubMed] [Google Scholar]
- 11.Pettingill TM, Strange RW, Blackburn NJ. Carbonmonoxy dopamine-b-hydroxylase: structural characterization by FTIR, fluorescence and XAS spectroscopy. J. Biol. Chem. 1991;266:16996–17003. [PubMed] [Google Scholar]
- 12.Boswell JS, Reedy BJ, Kulathila R, Merkler DJ, Blackburn NJ. Structural investigations on the coordination environment of the active-site copper centers of recombinant bifunctional peptidylglycine a-amidating enzyme. Biochemistry. 1996;35:12241–12250. doi: 10.1021/bi960742y. [DOI] [PubMed] [Google Scholar]
- 13.Chen P, Bell J, Eipper BA, Solomon EI. Oxygen activation by the noncoupled binuclear copper site in peptidylglycine a-hydroxylating monooxygenase. Spectroscopic definition of the resting sites and the putative CuIIM-OOH intermediate. Biochemistry. 2004;43:5735–5747. doi: 10.1021/bi0362830. [DOI] [PubMed] [Google Scholar]
- 14.Evans JP, Ahn K, Klinman JP. Evidence that dioxygen and substrate activation are tightly coupled in dopamine b -monooxygenase: Implications for oxygen activation. J. Biol. Chem. 2003;278:49691–49698. doi: 10.1074/jbc.M300797200. [DOI] [PubMed] [Google Scholar]
- 15.Evans JP, Blackburn NJ, Klinman JP. The catalytic role of the copper ligand H172 of peptidylglycine a-hydroxylating monooxygenase: a kinetic study of the H172A mutant. Biochemistry. 2006;45:15419–15429. doi: 10.1021/bi061734c. [DOI] [PubMed] [Google Scholar]
- 16.Chen P, Solomon EI. Oxygen activation by the noncoupled binuclear copper site in peptidylglycine a-hydroxylating monooxygenase. Reaction mechanism and role of the noncoupled nature of the active site. J. Am. Chem. Soc. 2004;126:4991–5000. doi: 10.1021/ja031564g. [DOI] [PubMed] [Google Scholar]
- 17.Chen P, Solomon EI. O2 activation by binuclear Cu sites: noncoupled versus exchange coupled reaction mechanisms. Proc. Natl. Acad. Sci. U S A. 2004;101:13105–13110. doi: 10.1073/pnas.0402114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crespo A, Marti MA, Roitberg AE, Amzel LM, Estrin DA. The catalytic mechanism of peptidylglycine a-hydroxylating monooxygenase investigated by computer simulation. J. Am. Chem. Soc. 2006;128:12817–12828. doi: 10.1021/ja062876x. [DOI] [PubMed] [Google Scholar]
- 19.Magnus KA, Hazes B, Ton-That H, Bonaventura C, Bonaventura J, Hol WGJ. Crystallographic analysis of oxygenated and deoxygenated states of arthropod hemocyanin shows unusual differences. Proteins Struct. Funct. Genet. 1994;19:302–309. doi: 10.1002/prot.340190405. [DOI] [PubMed] [Google Scholar]
- 20.Gerdemann C, Eicken C, Krebs B. The crystal structure of catechol oxidase: new insight into the function of type-3 copper proteins. Acc. Chem. Res. 2002;35:183–191. doi: 10.1021/ar990019a. [DOI] [PubMed] [Google Scholar]
- 21.Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J. Biol. Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- 22.Lewis EA, Tolman WB. Reactivity of dioxygen-copper systems. Chem. Rev. 2004;104:1047–1076. doi: 10.1021/cr020633r. [DOI] [PubMed] [Google Scholar]
- 23.Citek C, Lyons CT, Wasinger EC, Stack TD. Self-assembly of the oxy-tyrosinase core and the fundamental components of phenolic hydroxylation. Nat Chem. 2012;4:317–322. doi: 10.1038/nchem.1284. [DOI] [PubMed] [Google Scholar]
- 24.Prigge ST, Eipper BA, Mains RE, Amzel M. Dioxygen binds end-on to mononuclear copper in a precatalytic enzyme complex. Science. 2004;304:864–867. doi: 10.1126/science.1094583. [DOI] [PubMed] [Google Scholar]
- 25.Bauman AT, Yukl ET, Alkevich K, McCormack AL, Blackburn NJ. The hydrogen peroxide reactivity of peptidylglycine monooxygenase supports a Cu(II)-superoxo catalytic intermediate. J. Biol. Chem. 2006;281:4190–4198. doi: 10.1074/jbc.M511199200. [DOI] [PubMed] [Google Scholar]
- 26.Blackburn NJ, Rhames FC, Ralle M, Jaron S. Major changes in copper coordination accompany reduction of peptidylglycine monooxygenase. J. Biol. Inorg. Chem. 2000;5:341–353. doi: 10.1007/pl00010663. [DOI] [PubMed] [Google Scholar]
- 27.Bauman AT, Broers BA, Kline CD, Blackburn NJ. A Copper-Methionine Interaction Controls the pH-Dependent Activation of Peptidylglycine Monooxygenase. Biochemistry. 2011;50:10819–10828. doi: 10.1021/bi201193j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaron S, Mains RE, Eipper BA, Blackburn NJ. The catalytic role of the copper ligand H172 of peptidylglycine a -hydroxylating monooxygenase (PHM): a spectroscopic study of the H172A mutant. Biochemistry. 2002;41:13274–13282. doi: 10.1021/bi020404z. [DOI] [PubMed] [Google Scholar]
- 29.Himes RA, Park YG, Barry AN, Blackburn NJ, Karlin KD. Synthesis and X-ray absorption spectroscopy structural studies of Cu(I) complexes of histidylhistidine peptides: The predominance of linear 2-coordinate geometry. J. Am. Chem. Soc. 2007;129:5352–5353. doi: 10.1021/ja0708013. [DOI] [PubMed] [Google Scholar]
- 30.Osborne RL, Zhu H, Iavarone AT, Blackburn NJ, Klinman JP. Interdomain Long-Range Electron Transfer Becomes Rate-Limiting in the Y216A Variant of Tyramine beta-Monooxygenase. Biochemistry. 2013;52:1179–1191. doi: 10.1021/bi3013609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardenas DJ, Cuerva JM, Alias M, Bunuel E, Campana AG. Water-based hydrogen-atom wires as mediators in long-range proton-coupled electron transfer in enzymes: a new twist on water reactivity. Chemistry. 2011;17:8318–8323. doi: 10.1002/chem.201100964. [DOI] [PubMed] [Google Scholar]
- 32.Eipper BA, Quon ASW, Mains RE, Boswell JS, Blackburn NJ. The catalytic core of peptidylglycine a-hydroxylating monooxygenase: investigation by site-directed mutagenesis, Cu x-ray absorption spectroscopy, and electron paramagnetic resonance. Biochemistry. 1995;34:2857–2865. doi: 10.1021/bi00009a016. [DOI] [PubMed] [Google Scholar]
- 33.Kolhekar AS, Keutman HT, Mains RE, Quon ASW, Eipper BA. Peptidylglycine a-hydroxylating monooxygenase: active site residues, disulfide linkages, and a two-domain model of the catalytuc core. Biochemistry. 1997;36:10901–10909. doi: 10.1021/bi9708747. [DOI] [PubMed] [Google Scholar]
- 34.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 35.Oyarce AM, Steveson TC, Jin L, Eipper BA. Dopamine beta-monooxygenase signal/anchor sequence alters trafficking of peptidylglycine alpha-hydroxylating monooxygenase. J. Biol. Chem. 2001;276:33265–33272. doi: 10.1074/jbc.M101088200. [DOI] [PubMed] [Google Scholar]
- 36.Husten EJ, Eipper BA. The membrane-bound bifunctional peptidylglycine a-amidating monooxygenase protein. Exploration of its domain structure through limited proteolysis. J. Biol. Chem. 1991;266:17004–17010. [PubMed] [Google Scholar]
- 37.Jaron S, Blackburn NJ. Does superoxide channel between the copper centers in peptidylglycine monooxygenase? A new mechanism based on carbon monoxide reactivity. Biochemistry. 1999;38:15086–15096. doi: 10.1021/bi991341w. [DOI] [PubMed] [Google Scholar]
- 38.Bauman AT, Ralle M, Blackburn N. Large scale production of the copper enzyme peptidylglycine monooxygenase using an automated bioreactor. Protein Expression and Purification. 2007;51:34–38. doi: 10.1016/j.pep.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 39.Bauman AT, Jaron S, Yukl ET, Burchfiel JR, Blackburn N. pH dependence of peptidylglycine monooxygenase. Mechanistic implications for Cumethionine binding dynamics. Biochemistry. 2006;45:11140–11150. doi: 10.1021/bi060905a. [DOI] [PubMed] [Google Scholar]
- 40.George GN. EXAFSPAK. Menlo Park, CA: Stanford Synchrotron Radiation Laboratory; 1995. [Google Scholar]
- 41.Binsted N, Hasnain SS. State of the art analysis of whole X-ray absorption spectra. J. Synchrotron Rad. 1996;3:185–196. doi: 10.1107/S0909049596005651. [DOI] [PubMed] [Google Scholar]
- 42.Gurman SJ, Binsted N, Ross I. A rapid, exact, curved-wave theory for EXAFS calculations. J. Phys. C. 1984;17:143–151. [Google Scholar]
- 43.Gurman SJ, Binsted N, Ross I. A rapid, exact, curved-wave theory for EXAFS calculations. II. The multiple-scattering contributions. J. Phys. C. 1986;19:1845–1861. [Google Scholar]
- 44.Blackburn NJ, Concannon M, Khosrow Shahiyan S, Mabbs FE, Collison D. The active site of dopamine-b-hydroxylase. Comparison of derivatives containing four and eight coppers per tetramer using potentiometry and EPR spectroscopy. Biochemistry. 1988;27:6001–6008. doi: 10.1021/bi00416a026. [DOI] [PubMed] [Google Scholar]
- 45.Nilges MJ. SIMPIP EPR simulation program. Urbana-Champaign: Illinois EPR Research Center (IERC), University of Illinois; 1979. [Google Scholar]
- 46.Nilges MJ, Matteson K, Belford RL. A software package for the simulation of ESR powder-type spectra. In: Hemminga MA, Berliner LJ, editors. ESR Spectroscopy in Membrane Biophysics. New York: Springer; 2006. [Google Scholar]
- 47.Siluvai GS, Mayfield M, Nilges MJ, DeBeer George S, Blackburn NJ. Anatomy of a red copper center: Spectroscopic identification and reactivity of the copper centers of Bacillus subtilis Sco and Its Cys-to-Ala variants. J. Am. Chem. Soc. 2010;132:5215–5226. doi: 10.1021/ja910759v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strange RW, Blackburn NJ, Knowles PF, Hasnain SS. X-ray absorption spectroscopy of metal-histidine coordination in metalloproteins. Exact simulation of the EXAFS of tetraimidazole-copper(II) nitrate and other copper-imidazole complexes by the use of a multiple scattering treatment. J. Am. Chem. Soc. 1987;109:7157–7162. [Google Scholar]
- 49.Himes RA, Park GY, Siluvai GS, Blackburn NJ, Karlin KD. Structural studies of copper(I) complexes of amyloid-beta peptide fragments: formation of two-coordinate bis(histidine) complexes. Angew Chem Int Ed Engl. 2008;47:9084–9087. doi: 10.1002/anie.200803908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pickering IJ, George GN, Dameron CT, Kurz B, Winge DR, Dance IG. X-ray absorption spectroscopy of cuprous-thiolate clusters in proteins and model systems. J. Am. Chem. Soc. 1993;115:9498–9505. [Google Scholar]
- 51.Sanyal I, Karlin KD, Strange RW, Blackburn NJ. Chemistry and structural studies on the dioxygen-binding copper-1,2 dimethylimidazole system. J. Am. Chem. Soc. 1993;115:11259–11270. [Google Scholar]
- 52.Francisco WA, Knapp MJ, Blackburn NJ, Klinman JP. Hydrogen tunneling in peptidylglycine a-hydroxylating monooxygenase. J. Am. Chem. Soc. 2002;124:8194–8195. doi: 10.1021/ja025758s. [DOI] [PubMed] [Google Scholar]
- 53.Klinman JP. The role of tunneling in enzyme catalysis of C-H activation. Biochim. Biophys. Acta. 2006;1757:981–987. doi: 10.1016/j.bbabio.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Guss JM, Harrowell PR, Murata M, Norris VA, Freeman HC. Crystal structure analyses of reduced (CuI) poplar plastocyanin at six pH values. J. Mol. Biol. 1986;192:361–387. doi: 10.1016/0022-2836(86)90371-2. [DOI] [PubMed] [Google Scholar]
- 55.Li C, Sato K, Monari S, Salard I, Sola M, Banfield MJ, Dennison C. Metal-binding loop length Is a determinant of the pKa of a histidine ligand at a type 1 copper site. Inorg. Chem. 2011;50:482–488. doi: 10.1021/ic101413e. [DOI] [PubMed] [Google Scholar]
- 56.Bertini I, Luchinat C, Monnanni R. Evidence of the breaking of the copper-imidazolate bridge in copper/cobalt-substituted superoxide dismutase upon reduction of the Cu(II) centers. J. Am. Chem. Soc. 1985;107:2178–2179. [Google Scholar]
- 57.Blackburn NJ, Hasnain SS, Binsted N, Diakun GP, Garner CD, Knowles PF. An extended-x-ray-absorption-fine-structure study of bovine erythrocyte superoxide dismutase in aqueous solution. Direct evidence for three-coordinate Cu(I) in the reduced enzyme. Biochem. J. 1984;219:985–990. doi: 10.1042/bj2190985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. The solution structure of reduced dimeric copper zinc superoxide dismutase. The structural effects of dimerization. Eur. J. Biochem. 2002;269:1905–1915. doi: 10.1046/j.1432-1033.2002.02840.x. [DOI] [PubMed] [Google Scholar]
- 59.Li C, Banfield MJ, Dennison C. Engineering copper sites in proteins: loops confer native structures and properties to chimeric cupredoxins. J. Am. Chem. Soc. 2007;129:709–718. doi: 10.1021/ja0661562. [DOI] [PubMed] [Google Scholar]
- 60.Pasquali M, Floriani C. Cu(I)-carbon monoxide chemistry: recent advances and perspectives. In: Karlin KD, Zubieta J, editors. Copper Coordination Chemistry, Biochemical and Inorganic Perspectives. New York: Adenine Press; 1984. pp. 311–330. [Google Scholar]
- 61.Blackburn NJ, Strange RW, Farooq A, Haka MS, Karlin KD. X-ray absorption studies of three-coordinate dicopper(I) complexes and their dioxygen adducts. J. Am. Chem. Soc. 1988;110:4263–4272. [Google Scholar]
- 62.Steveson TC, Ciccotosto GD, Ma X-M, Mueller GP, Mains RE, Eipper BA. Menkes protein contributes to the function of peptidylglycine a-amidating monooxygenase. Endocrinology. 2003;144:188–200. doi: 10.1210/en.2002-220716. [DOI] [PubMed] [Google Scholar]
- 63.Petris MJ, Strausak D, Mercer JF. The Menkes copper transporter is required for the activation of tyrosinase. Hum. Mol. Genet. 2000;9:2845–2851. doi: 10.1093/hmg/9.19.2845. [DOI] [PubMed] [Google Scholar]
- 64.Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008;454:1142–1146. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin Z, Itoh S, Jeney V, Ushio-Fukai M, Fukai T. Essential role for the Menkes ATPase in activation of extracellular superoxide dismutase: implication for vascular oxidative stress. FASEB J. 2006;20:334–336. doi: 10.1096/fj.05-4564fje. [DOI] [PubMed] [Google Scholar]
- 66.El Meskini R, Culotta VC, Mains RE, Eipper BA. Supplying copper to the cuproenzyme peptidylglycine a-amidating monooxygenase. J. Biol. Chem. 2003;278:12278–12284. doi: 10.1074/jbc.M211413200. [DOI] [PubMed] [Google Scholar]
- 67.Pope CR, Flores AG, Kaplan JH, Unger VM. Structure and function of copper uptake transporters. Current topics in membranes. 2012;69:97–112. doi: 10.1016/B978-0-12-394390-3.00004-5. [DOI] [PubMed] [Google Scholar]
- 68.Barry AN, Otoikhian A, Bhatt S, Shinde U, Tsivkovskii R, Blackburn NJ, Lutsenko S. The Lumenal Loop Met672-Pro707 of Copper-transporting ATPase ATP7A Binds Metals and Facilitates Copper Release from the Intramembrane Sites. J. Biol. Chem. 2011;286:26585–26594. doi: 10.1074/jbc.M111.229039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otoikhian A, Barry AN, Mayfield M, Nilges M, Huang Y, Lutsenko S, Blackburn NJ. Lumenal loop M672-P707 of the Menkes protein (ATP7A) transfers copper to peptidylglycine monooxygenase. J. Am. Chem. Soc. 2012;134:10458–10468. doi: 10.1021/ja301221s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.