Abstract
Background: HCV has become a leading cause of liver cirrhosis and hepatocellular carcinoma and is a major health concern worldwide. To date, there is no vaccine available in the market to tackle this disease, therefore there is a strong need to develop antiviral compounds that can target all genotypes of HCV with the same efficiency. Medicinal plants have low cost and are less toxic therefore, extracts of medicinal plants can serve as important antiviral agents against HCV. This study was designed to screen phytochemicals of Accacia nilotica to find a potent drug candidate that can inhibit HCV infection effectively.
Results: Docking of NS3/4A protease and Flavonoids of Accacia nilotica revealed that most of the flavonoids bound deeply with the active site of NS3/4A protease. Compound 01 showed a high ranking on docking score. All other compounds also showed reliable docking scores and had interactions with the binding cavity of NS3/4A protease, suggesting them as a potent drug candidate to block HCV replication.
Conclusion: To recognize binding interactions of Accacia nilotica phytochemicals with NS3/4A protease, molecular docking was performed to find potential inhibitor against NS3/4A protease of HCV. After post docking analysis, important interactions were found between active compounds and active site of NS3/4A protease. It can be concluded from the study that phytochemicals of Accacia nilotica may serve as a potential drug candidate with relatively simple structural changes against HCV NS3/4A protease.
Background
Hepatitis C Virus (HCV) is a global health problem which affects 170 million people worldwide and 10 million people in Pakistan [1]. HCV causes acute and chronic hepatitis and is a major cause of liver cirrhosis and hepatocellular carcinoma [2]. HCV is a member of Flaviviridae family with a positive-sense single-stranded RNA genome which encodes three structural proteins (Core, E1, E2) and six non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A & NS5B) [3]. Among all HCV proteins, NS3/4A serine protease and NS5B polymerase are important drug targets to develop anti-HCV agents [4]. The Major role of NS3/4A is the proteolytic processing at NS4A/4B, NS4B/5A, and NS5A/5B sites, and it plays an important role in HCV replication. Because of its involvement in viral replication, it has emerged as a potential drug target for HCV. There is no vaccination available for HCV treatment and current standard of care is a combination therapy of Pegylated interferon alpha (PegIFN-α) injections with oral antiviral nucleoside analogue ribavirin (RBV) which leads to clearance of HCV in 50% genotype 1 cases and 80% of genotype 2 cases but this treatment has slow response rates and side effects in genotype 1a and 1b patients [5–7]. Current treatment is costly, less efficient and has several side effects thus, there is a strong need of developing antiviral agents that are less toxic and has potential to target all genotypes of HCV with the same efficiency. Recently, two NS3 protease inhibitors have been approved as triple therapy (PEGIFN- α , ribavirin and Boceprevir or Telaprevir) against HCV [8]. But still there is a strong need to develop specific compounds that can target important factors of the HCV life cycle [9].
Medicinal plants are being tested against HCV and many of them have proven to be beneficial as apposite substitute sources of antiviral mediators. Medicinal plants have low cost, multiple target activities, inconsequential side-effects and stumpy prospective to cause resistance therefore, are preferred over conventional treatment [10– 14]. Phytochemicals of medicinal plants such as alkaloids, organosulfur compounds, limonoids, lignans, furyl compounds, polyines, thiophenes, proteins, peptides, flavonoids, terpenoids, sulphides, polyphenolics, coumarins, saponins, chlorophyllins have functions like scavenging, antioxidant activities, hampering viral entry, DNA and RNA replication against variety of viruses [15]. Recently, our group reported that flavonoids of Acacia nilotica displayed novel inhibition of HCV titer in liver infected cells [16]. Therefore, this study was designed to screen phytochemical of Accacia nilotica against HCV NS3/4A protease using in-silico approaches.
Methodology
This study was designed to dock Accacia nilotica Flavonoids against HCV NS3/4A protein with the following communications. Intel (R) xenon (R) CPU E5620@2.40GHz system having 3.8GB RAM with the open 11.4 (X 86_64) operating platform. Protein-ligand docking was carried out using the MOE (Molecular Operating Environment) software package. Interactions between HCV NS3/4A protease and ligands were imagined using ligPlot feature of MOE software.
Ligand preparation
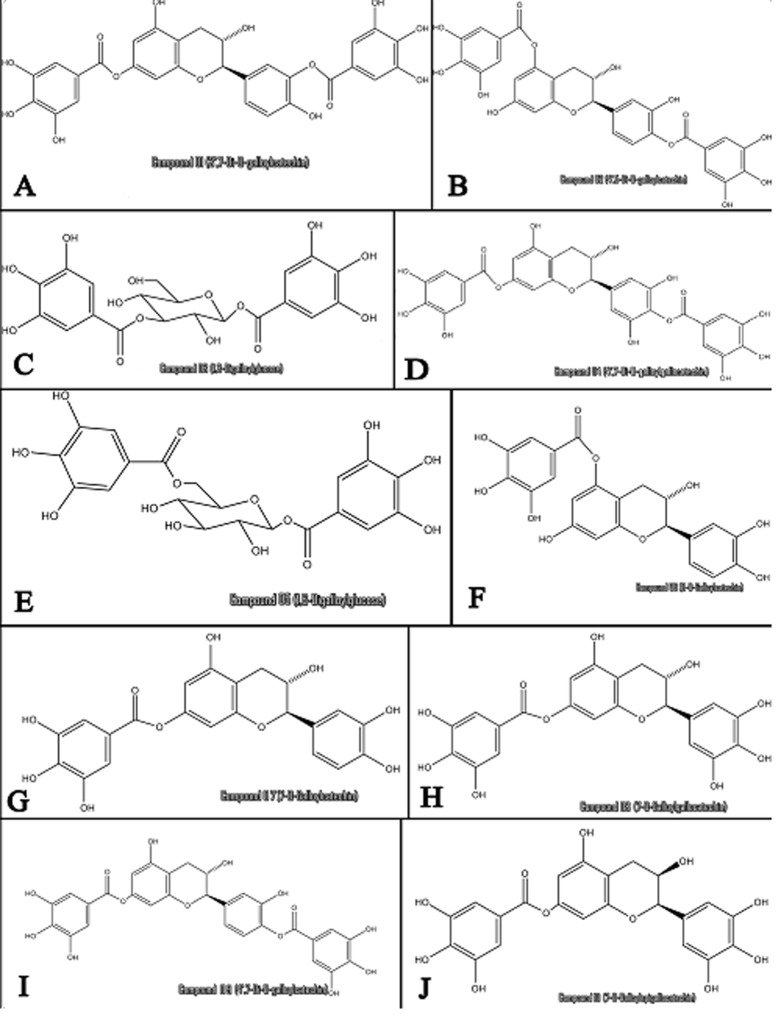
A literature search was performed to find Accacia nilotica Flavonoid Structures. Chemical structures of Favonoids were constructed using ChemDraw software (Figure 1) and were converted into their respective 3D structures. 3D structures were optimized by adding hydrogens through MOE software package. Energies of selected molecules were minimized with parameters (gradient: 0.05, Force Field: MMFF94X, Chiral Constraint and Current Geometry). Molecules were then saved in mdb format and were used as input file for MOE-Docking.
Figure 1.
Chemical Structure of Accacia nilotica phytochmicals constructed using Chemdraw Software.
Preparation of Receptor Protein
HCV NS3/4A protease structure was retrieved from Protein Data Bank using PDB ID: 2FM2 and was optimized by removing water molecules, 3D protonation and Energy minimization. Energy minimization was carried out using parameters (gradient: 0.05, Force Field: MMFF94X+Solvation and Chiral Constraint: Current Geometry). Energy minimization was terminated when the root mean square gradient falls below the 0.05. The minimized structure was used as the receptor protein for Docking.
Molecular Docking
MOE docking program with default parameters was used to bind the selected ligands with receptor protein and to find the correct conformation (with the rotation of bonds, structure of molecule is not rigid) of the ligand so as to obtain minimum energy structure. After docking, best conformations were analyzed for hydrogen bonding/π-π interactions.
Results & Discussion
Docking Analysis
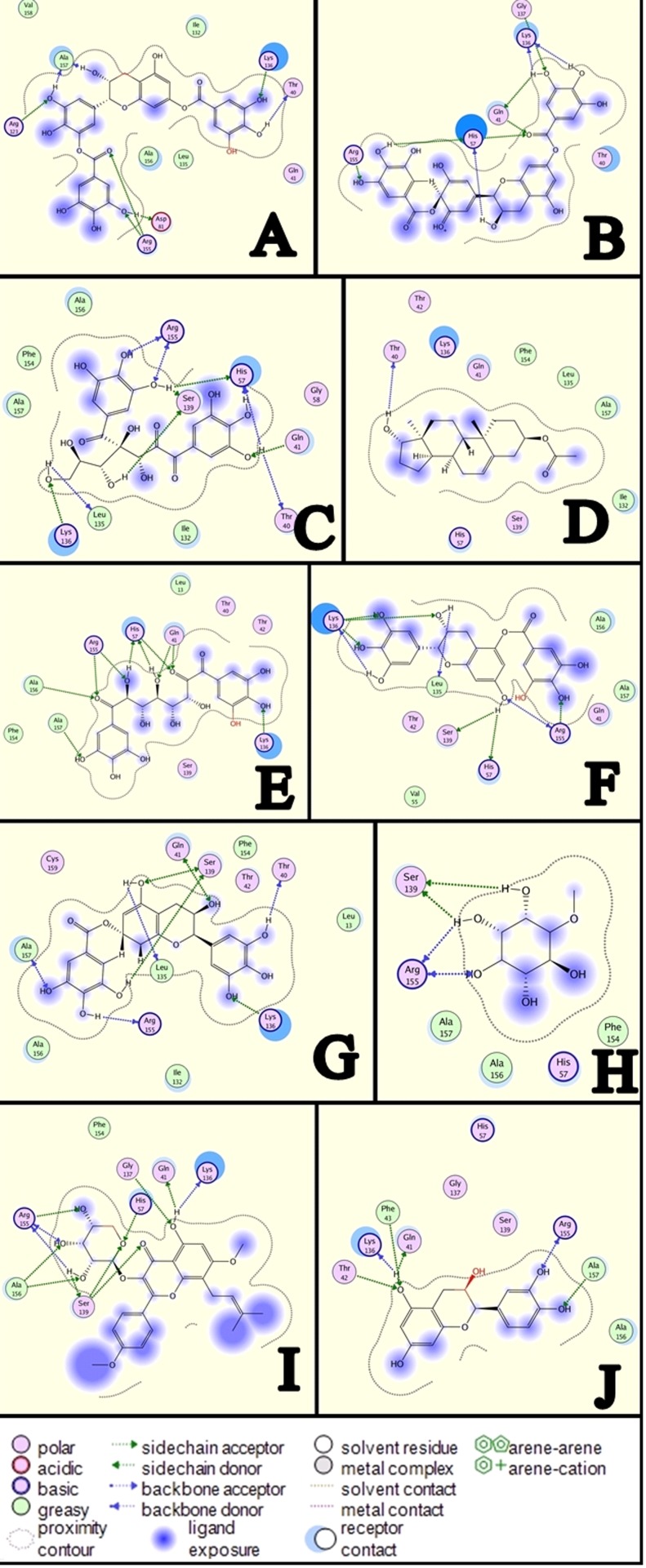
All ligands were docked with the active site of NS3/4A protease enzyme and top ranked conformations of each ligand were saved in a separate database. Compound 01 was ranked as top docked molecule on the basis of S-Score obtained from MOE docking algorithm. Thus, it can be inferred that this compound can serve as potential inhibitor against NS3/4A protease. All other compounds were having S-Score close to each other. All compounds were studied in detail to obtain their interaction information that can be important for the inhibition of NS3/4A protease enzyme. Interaction diagrams were obtained using MOE ligand interaction analysis feature.
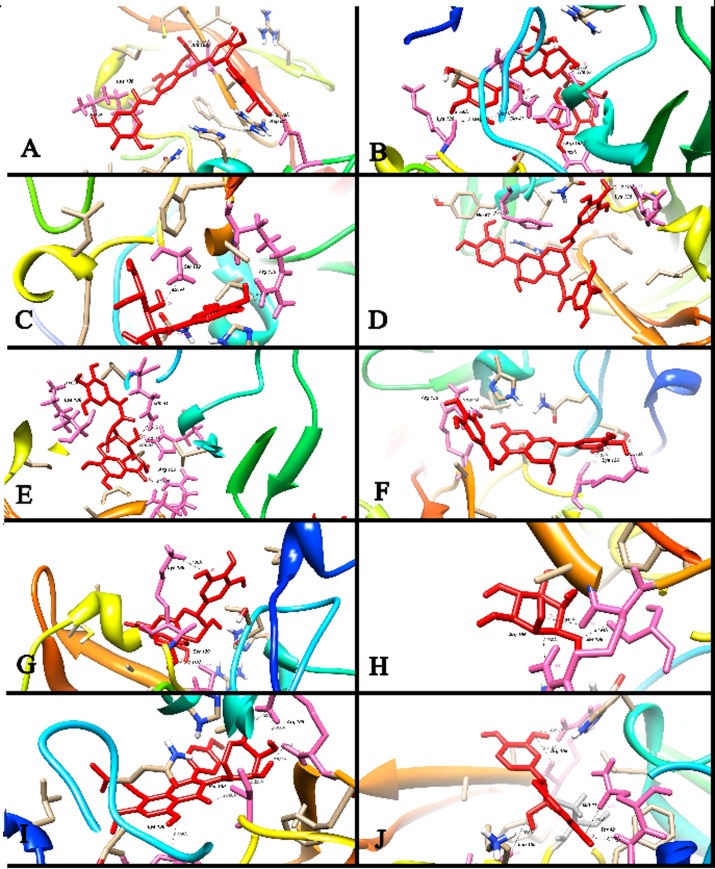
Binding interactions of Ligands and Protein
The most active Compound 01 was ranked as first on the docking score. It is clear from (Figure 2 & Figure 3A) that this compound was bound deep into the binding cavity of NS3 protease making interactions with the residues Lys 136 and Asp 81. On the basis of docking score Compound 02, Compound 03, Compound 04, Compound 05, Compound 06, Compound 07, Compound 08, Compound 09, Compound 10, and Compound 11 were ranked close to each other as shown in Table 1 (see supplementary material). Among all these compounds, Compound 02, Compound 03, and Compound 04 showed three interactions with the residues of the binding cavity of protein. The compound 02 can be seen from (Figure 2 & Figure 3B) that it was docked tightly into the binding site of protein. There were some five prominent interactions observed between the Compound 02 and the binding site residues Gln 41 and Arg 155 and His 57. The solvent contact was found almost all over the ligand. The compounds Compound 03 and Compound 04 showed almost similar interactions with the protein. Both compounds were having interactions with Gln 41, Ser 139 and Arg 155. The Compound 03 can be seen clearly in (Figure 2 & Figure 3C) making four interactions with the residues Gln 41, Ser 139 and Arg 155. The compound Compound 04 is having three interactions with binding site residues Gln 41, Ser 139 and Arg 155 of protein (Figure 2 & Figure 3D).
Figure 2.
The 2D pictures of the docked conformations of most active compounds.
Figure 3.
A) Interactions between Compound 01 and active site of NS3/4A; B) Interactions between Compound 02 and active site of NS3/4, C) Interactions between Compound 03 and active site of NS3/4, D) Interactions between Compound 04 and active site of NS3/4A. , E) Interactions between Compound 05 and active site of NS3/4A; F) Interactions between Compound 06 and active site of NS3/4A; G) Interactions between Compound 07 and active site of NS3/4A; H) Interactions between Compound 08 and active site of NS3/4A; I) Interactions between Compound 09 and active site of NS3/4A; J) Interactions between Compound 10 and active site of NS3.
Conclusion
To recognize binding interactions of Accacia nilotica phytochemicals with NS3/4A protease, molecular docking was performed to find a potential inhibitor against NS3/4A protease of HCV. After post docking analysis, important interactions were found between active compounds and active site of NS3/4A protease. Thus, molecular docking can be helpful to find the relative binding affinities of active compounds prior to synthesis and biochemical testing of new inhibitors. It can be concluded from the study that phytochemical of Accacia nilotica may serve as a potential drug candidate with relatively simple structural changes against HCV NS3/4A protease.
Supplementary material
Acknowledgments
We are thankful to Zaheer-ul-Haq Qasmi for providing MOE software
Footnotes
Citation:Khan et al, Bioinformation 9(14): 710-714 (2013)
References
- 1.Naggie S. Top Antivir Med. 2012;20:154. [PMC free article] [PubMed] [Google Scholar]
- 2.Ashfaq UA, et al. Virol J. 2011;8:161. doi: 10.1186/1743-422X-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, et al. J Virol. 1994;68:5045. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashfaq UA, et al. Genet Vaccines Ther. 2011;9:7. doi: 10.1186/1479-0556-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poynard T, et al. Hepatology. 2003;38:75. doi: 10.1053/jhep.2003.50267. [DOI] [PubMed] [Google Scholar]
- 6.Feld JJ, Hoofnagle JH. Nature. 2005;436:967. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 7.Raychoudhuri A, et al. J Virol. 2010;84:10991. doi: 10.1128/JVI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghany MG, et al. Hepatology. 2011;54:1433. doi: 10.1002/hep.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Labrador FX, et al. Recent Pat Antiinfect Drug Discov. 2008;3:157. doi: 10.2174/157489108786242369. [DOI] [PubMed] [Google Scholar]
- 10.Vlietinck AJ, Vanden Berghe DA. J Ethnopharmacol. 1991;32:141. doi: 10.1016/0378-8741(91)90112-q. [DOI] [PubMed] [Google Scholar]
- 11.Cowan MM, et al. Clin Microbiol Rev. 1999;12:564. doi: 10.1128/cmr.12.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briskin DP. Plant Physiol. 2000;124:507. doi: 10.1104/pp.124.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JE. Altern Med Rev. 2001;6:567. [PubMed] [Google Scholar]
- 14.Jassim SA, Naji MA. J Appl Microbiol. 2003;95:412. doi: 10.1046/j.1365-2672.2003.02026.x. [DOI] [PubMed] [Google Scholar]
- 15.Naithani R, et al. Mini Rev Med Chem. 2008;8:1106. doi: 10.2174/138955708784567368. [DOI] [PubMed] [Google Scholar]
- 16.Rehman S, et al. Virol J. 2011;8:220. doi: 10.1186/1743-422X-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.