Abstract
In the last decade, large-scale mass spectrometry-based phosphoproteomic studies of receptor tyrosine kinases (RTKs) have generated a compendium of signalling networks that are activated downstream of these receptors. In this article, a brief summary of previous phosphoproteomic studies on Epidermal Growth Factor Receptor (EGFR) signalling will be presented together with a perspective on the importance for the field to keep pace with new advances in RTK biology. Using examples drawn primarily from studies on the EGFR, c-Met and Flt3 receptors, areas in RTK biology which will greatly benefit from the power of phosphoproteomics will be discussed, including a. validating oncogenic RTK mutants identified in cancer genome sequencing efforts, b. spatial RTK signalling networks and c. understanding crosstalk and co-activation between members of the RTK superfamily.
Keywords: Receptor Tyrosine Kinase, Signal Transduction, Systems Biology, Phosphoproteomics, Mass Spectrometry
INTRODUCTION
Receptor Tyrosine Kinases (RTKs) are a class of transmembrane receptors that drive a wide variety of essential cellular processes in response to extracellular cues. They are characterised by three structural features comprising extracellular ligand binding domains, a transmembrane helix and a cytoplasmic region containing a tyrosine kinase domain1. Binding of RTKs to their cognate ligands initiate a cascade of phosphorylation-mediated signalling events which direct cellular programs that ultimately determine biological function and phenotype. Phosphoproteomics by mass spectrometry (MS) has emerged as a powerful tool to study these signalling networks in an unbiased, quantitative and highly sensitive manner. As a result of these efforts, the signalling community has gained a rich resource of phosphorylation sites that are catalogued in a number of publically accessible databases including Phosphosite (www.phosphosite.org) and Phosida (www.phosida.de) 2-3. In parallel with the technical developments in the field of phosphoproteomics, our knowledge of RTK biology has also significantly advanced in the last decade. While efforts in phosphoproteomics have enriched our understanding of RTK signalling networks, the field needs to keep pace with new discoveries in RTK biology. Using examples drawn from promising early phosphoproteomic studies, I will discuss new areas in RTK biology which would benefit from the power of phosphoproteomics.
CURRENT STATE-OF-THE-ART IN PHOSPHOPROTEOMICS – EGFR AS A PROTOTYPICAL EXAMPLE
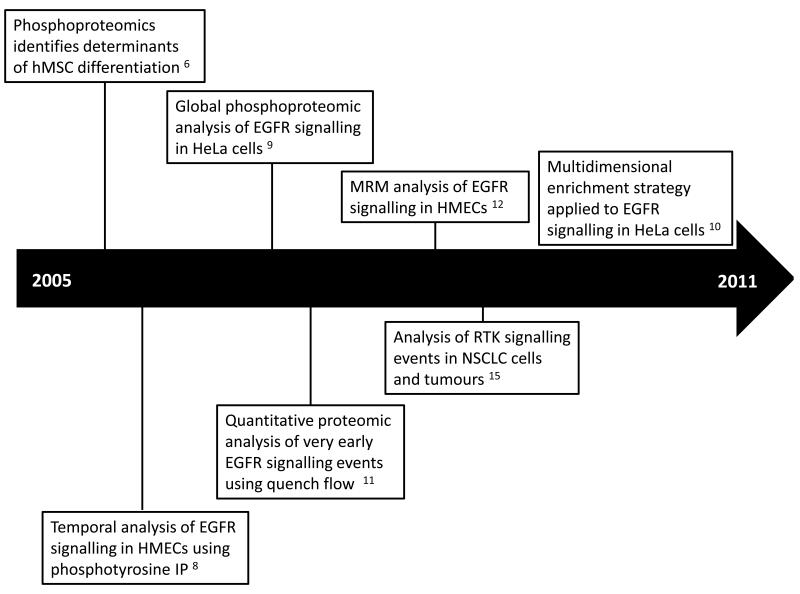
Pioneering phosphoproteomic studies have predominately focused on the prototypical member of the RTK superfamily, the Epidermal Growth Factor Receptor (EGFR). This section provides a broad summary of the published phosphoproteomics studies that have enabled new discoveries in EGFR biology (Figure 1). An in-depth description of quantitative and qualitative methods for interrogating the phosphoproteome is not within the scope of this article but the reader is directed to recent excellent reviews describing the application of phosphoproteomics to tyrosine kinases 4-5 .
Figure 1.
A timeline of important phosphoprotemic studies that have enabled new discoveries in EGFR biology.
One of the earliest functional studies of EGFR signalling using quantitative phosphoproteomics was performed by Kratchmarova et al. on human mesenchymal stem cells (hMSCs) 6. By employing stable isotope labelling with amino acids in cell culture (SILAC) as a means of phosphopepetide quantification, the authors showed that despite sharing a largely overlapping subset of tyrosine phosphorylated targets (greater than 90%), only epidermal growth factor (EGF) and not platelet derived growth factor (PDGF) stimulation led to osteogenic differentiation of hMSCs. Reminiscent of the distinct phenotypic outcomes observed in PC12 cells as a result of acute or sustained Erk activation by EGF or nerve growth factor (NGF) respectively 7, the authors showed that osteogenic differentiation of hMSCs was governed exclusively by differential activation of the PI3K pathway. The PI3K pathway was shown to be activated only by PDGF and suppressing this pathway by kinase inhibition, and thereby mimicking the effect of EGF signalling, bestowed PDGF with the capability of driving osteogenic differentiation. This study was followed by a series of seminal papers that employed quantitative mass spectrometry to map the temporal phosphorylation profile of EGFR signalling networks. Using human mammary epithelial cells (HMECs), Zhang et al. coupled isobaric tag for relative and absolute quantification (iTRAQ) quantification with phosphotyrosine peptide immunoprecipitation (IP) and immobilised metal affinity chromatography (IMAC) to identify 58 proteins that were phosphorylated by EGFR activation in a time-resolved fashion 8. The authors were able to show that the EGFR signalling network could be clustered into dynamic modules using self-organising maps. Olsen et al. extended these findings by performing a global analysis of phosphorylation events downstream of EGFR activation in HeLa cells 9. Using SILAC quantification coupled to strong cation exchange (SCX) and titanium dioxide (TiO2) enrichment, the authors identified 6600 phosphorylation sites on 2,244 proteins of which a small proportion (14%) was found that be modulated by EGF. This study remains to date the largest compendium of EGFR activated phosphorylation sites. Larsen and colleagues have recently devised a multidimensional enrichment strategy that greatly increases the sensitivity of phosphoproteomic analysis 10. Using just 400μg of EGF stimulated HeLa cells, they combined sequential elution from IMAC (SIMAC), hydrophilic interaction chromatography (HILIC) and TiO2 enrichment to quantify 4700 phosphopeptides of which 636 phosphosites were modulated by EGF stimulation. Intriguingly, about two thirds of these phosphosites were not previously identified in the Olsen et al. study, suggesting that there is still a large number of EGFR signalling components that remain to be characterised and future developments in chromatographic strategies and mass spectrometry instrumentation will lead to further novel discoveries in EGFR signalling networks.
The development of new workflows has provided additional biological insights into EGFR signalling. Dengjel et al. established a continuous quench-flow system to assay the early phosphorylation events that occur within seconds of ligand engagement 11. By using peristaltic pumps, the authors rapidly mixed HeLa cells together with growth factor prior to quenching the reaction with cold ethanol after 1 to 60 seconds of ligand exposure. They termed this strategy quantitative proteomic assessment of very early signalling events (qPACE). MS analysis of the resulting lysate identified three tyrosine phosphorylation sites on EGFR that were rapidly activated within 5 seconds of EGF addition. This observation was in contrast to the phosphoserine and threonine sites on the receptor which were unaffected by EGF stimulation at these early timepoints. Interestingly, the study also showed that it was possible to resolve very early downstream signalling events and demonstrated that phosphorylation sites on phospholipase C gamma (PLC-γ) displayed a distinctly lagged activation profile compared to EGFR tyrosine phosphorylation. Although this analysis only highlighted a limited number of downstream signalling components, a combination of the qPACE technique with the aforementioned global phosphoproteomic enrichment strategies should greatly increase the depth of phosphoproteome coverage of early EGFR signalling events.
One of the major challenges associated with discovery-based MS approaches is its inherent inability to acquire reproducible data across biological/technical replicates or between multiple cellular conditions. This problem occurs as a result of the automated selection of precursor ions for fragmentation by information dependent acquisition (IDA) workflows. To overcome this problem, Wolf-yadlin et al. developed a targeted quantitative MS strategy to generate a high-resolution temporal map of EGFR signalling across seven time-points 12. Using multiple reaction monitoring (MRM) in a hybrid triple quadrupole/linear ion trap mass spectrometer to monitor the levels of 222 phosphopeptides, the authors were able to demonstrate that this method was superior over existing IDA approaches in improving data reproducibility. The MRM strategy had an 88% overlap of phosphopeptides across four analyses while the IDA approach only generated a 34% overlap. This study is the first application of MRM to quantify signal transduction and has the exciting potential to be broadly applied to other RTK signalling networks. These targeted approaches are particularly beneficial for the acquisition of reproducible data for systems biology applications where high density phosphoproteomic measurements across multiple conditions are essential for the development of predictive computational models of EGFR signalling networks 13-14.
All of the described approaches have primarily been confined to the characterisation of in vitro EGFR signalling in cell line models. Using a semi-quantitative MS strategy, Rikova et al. screened a panel of 41 non-small cell lung cancer (NSCLC) cell lines and 150 tumours for their levels of phosphotyrosine-mediated signalling proteins 15. Remarkably, the authors showed that the spectrum of activated RTKs in NSCLC cell lines were distinct from those found in tumours. While EGFR, c-Met and EphA2 receptors had the highest levels of RTK phosphorylation in NSCLC cell lines, the discoidin domain receptors (DDR1 and 2) and EGFR formed the largest component of phosphorylated RTKs in tumour specimens. These findings indicate that cancer cell lines only represent tractable model systems for signalling studies and further characterisation of tissue and tumours samples are required for a better mechanistic understanding of disease processes. With the recent development and application of isotopic-based MS quantification approaches to animal models of disease and patient specimens 16-18, it is anticipated that forthcoming phosphoproteomic analyses will reveal further insights into in vivo EGFR signalling networks.
VALIDATING ONCOGENIC RTK MUTANTS
The advent of next generation sequencing technologies has revolutionized the cost and speed at which whole genome sequencing can be performed 19. Cancer sequencing projects driven by The Cancer Genome Atlas project (TCGA) and The International Cancer Genome Consortium (ICGC) have uncovered a large number of RTK mutations and translocations in human tumours 20. While some of these aberrations are found in mutational ‘hotpots’, for instance in the ectodomain and kinase domain of EGFR, and are fairly well characterized 21, many of the identified mutations are novel and their functional contribution to RTK activity and signalling specificity is unknown. In tandem with the deluge of sequencing data that is being generated, bioinformatic approaches to distinguish driver from passenger mutations have been developed to prioritize the functional annotation of RTK mutations 22-24. Nonetheless, functional validation of these driver mutations represents the greatest bottleneck in the ultimate goal of translating the cancer genome into a mechanistic understanding of the disease with the accompanying development of patient-specific therapies for cancer.
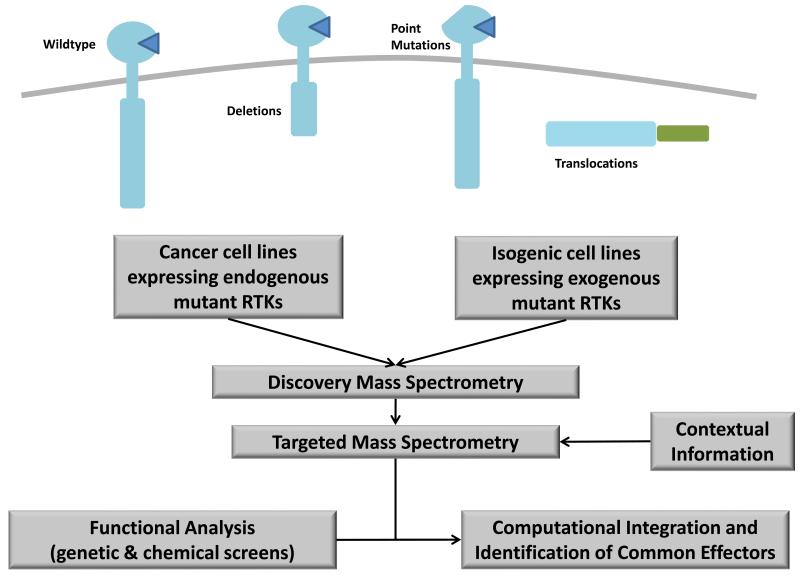
Quantitative phosphoproteomics is ideally positioned to tackle this challenge. Studies performed on oncogenic mutants of EGFR have shown that mass spectrometry is capable of accurately mapping the signalling network changes that occur upon acquisition of RTK mutations 25-26. For instance, Guo and co-workers used a semi-quantitative MS approach to profile the phosphotyrosine status of NSCLC cells endogenously expressing EGFR mutants that are sensitive to the tyrosine kinase inhibitor gefitinib 27. The authors showed that mutant EGFR cell lines generally displayed higher phosphorylation levels of downstream adaptor proteins such as Grb2-associated binding protein 1 (Gab1) and SH2 domain containing transforming protein 1 (SHC1) compared to cells expressing wildtype (WT) EGFR. The authors have recently extended this analysis to identify AKT-RSK-S6 kinase substrates downstream of these mutant receptors by using motif-specific antibodies to enrich for phosphopeptides that contain the characteristic AKT substrate motif (RxRxxpS/pT) 28. However, such analyses are generally confounded by the heterogeneous genetic background of different NSCLC lines which make it challenging to establish if the observed signalling alterations are indeed the consequence of EGFR mutations. To overcome this problem, Guha et al. used isogenic immortalised human bronchial epithelial cells (HBECs) that were transduced with either WT or two mutants of EGFR (Del E746-A750 and L858R) 29. Using SILAC combined with phosphotyrosine IP, the study demonstrated that cells expressing the two EGFR mutants exhibited much higher baseline tyrosine phosphorylation levels than WT EGFR. More importantly, they observed receptor phosphorylation differences between the two EGFR mutants, with the Del E746-A750 mutant exhibiting a five-fold increase in phosphorylation at the Y727 site compared to the L858R mutant. This finding indicates that different EGFR mutants activate unique downstream signalling pathways as a result of differential receptor phosphorylation. Subsequent phosphoproteomic studies of the EGFR mutants EGFRvIII and EGFRvIV in glioblastoma further reinforce this observation 26, 30. The knowledge gleaned from such phosphoproteomics analyses can also be used to design ‘network-based’ therapeutic strategies to overcome chemoresistance driven by mutant RTKs. We have previously employed quantitative MS to identify crosstalk between EGFRvIII and the c-Met receptor in glioblastoma cells 26, 31. This interaction was exploited to design a novel combinatorial strategy to overcome EGFRvIII-driven glioblastoma cell growth. Co-inhibition of both the EGFRvIII and c-Met receptor resulted in additive cancer cell death and conferred sensitivity to EGFR inhibitor monotherapy and DNA damaging agents. These studies cumulatively highlight that mass spectrometry is a promising tool for resolving signalling features that occur downstream of RTK mutants and has the potential to inform the selection of signalling candidates for targeted therapy in patients with heterogeneous tumours that may contain a diverse spectrum of RTK mutations (Figure 2).
Figure 2.
Strategy for the analysis of oncogenic RTK mutants in cancer. Either cancer cell lines containing endogenous mutant RTKs or isogenic cell lines transduced to express mutant RTKs can be analysed using discovery-based mass spectrometry. Contextual information can then be assessed using targeted mass spectrometry methods such as MRM. Finally in combination with functional analyses, computational approaches can be used to identify common effectors of mutant RTK signalling.
There are issues that complicate the routine use of phosphoproteomics in the functional validation of mutant RTK signalling networks. Signalling processes are heavily influenced by cellular context, for instance the presence of additional mutations in other oncogenes and tumour suppressors within the same cell could dramatically alter the networks activated by RTKs. Similarly, extracellular stimuli such as matrix and stromal interactions have been shown to modulate RTK signalling processes 32. Is the identification of context-specific changes in cellular signalling networks at the genome-wide scale a tractable problem? Previous studies by Lauffenburger and colleagues combining signal transduction pathway analysis with computational modelling have suggested that cancer cells share a common effector signalling program 33. While contextual information may result in differences in kinase-mediated signalling networks, the fundamental components or effectors of signalling are likely to be common to most cells. By taking advantage of this principle, it may be possible to simplify the problem of context-specific RTK signalling by focusing on the modulating effects of RTK mutations on components of the common effector machinery. Through the use of targeted MS-based approaches such as MRM to map these common effectors 12, 34, one can now test this principle by collecting large-scale datasets of mutant RTK signalling networks in different contexts (e.g. genetic background). Analysis of these datasets in combination with functional phenotypic studies, such as RNA interference and chemical genetic screens, using computational modelling approaches should allow one to identify the common effectors that are critical for distinguishing mutant RTK oncogenic phenotypes 13-14, which may then serve as potential candidates for therapeutic development or biomarker validation (Figure 2).
SPATIAL RTK SIGNALLING NETWORKS
There is an increasing appreciation that the activation of an RTK at the plasma membrane represents only one facet of its complex biology. Upon ligand engagement, many RTKs are internalised into endosomal compartments, prior to signal degradation and receptor recycling 35. Previous studies have demonstrated that spatial compartmentalisation is a mechanism by which cells diversify the signalling networks activated by RTKs. For instance, Kermorgant and co-workers have shown that c-Met endocytosis is required for the phosphorylation and nuclear accumulation of both Erk1/2 and STAT3 36. Interestingly, translocation of c-Met from the peripheral endosomal compartment to the perinuclear endosomal compartment is required only for STAT3 nuclear localisation but not for Erk1/2, suggesting that signalling specificity is conferred by compartmentalisation of c-Met. In a subsequent study, the same group demonstrated that accumulation of receptor levels in the endosomal compartment is a mechanism by which mutant forms of c-Met conferred in vivo oncogenicity and metastasis 37. Similarly, Haugh et al. have observed that while both surfaced localised EGFR and endosomal EGFR can activate the Ras pathway, only the surface localised receptor is able to activate PLC-γ 38-39. In addition to the endosomal compartments, it has been reported that EGFR also translocates to both the mitochondrial and nuclear compartments and play important roles in activating signalling components present in these organelles 40-42. Identifying the signalling differences that arise as a result of spatial organisation of RTKs will be critical in establishing the functional relevance of receptor compartmentalisation. The ability of phosphoproteomics to quantitatively map signalling networks in an unbiased fashion makes it an attractive approach for addressing some of the pressing questions in this burgeoning field of spatial signalling networks.
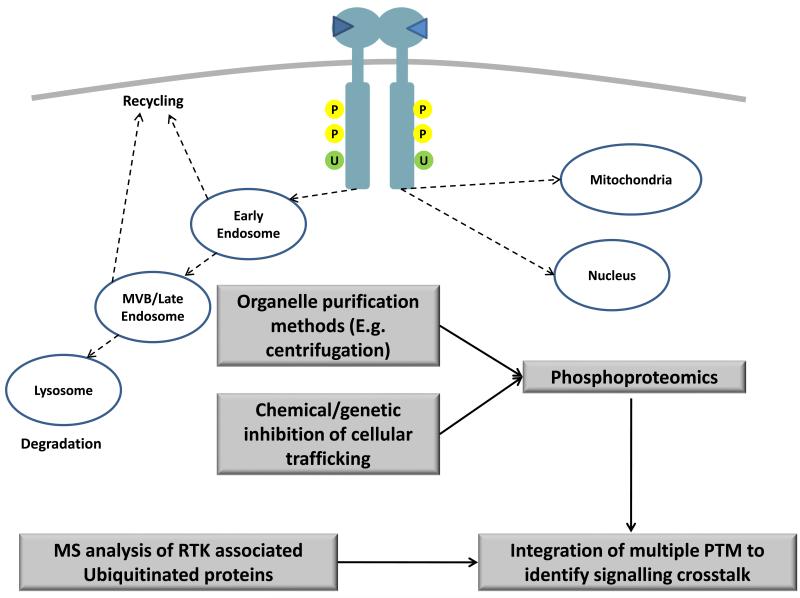
Studies on protein compartmentalisation have historically been performed using organelle isolation techniques such as centrifugation (Figure 3). For example, the global phosphoproteomic study of EGFR signalling by Olsen et al. was done using HeLa cells subjected to cytoplasmic and nuclear separation through centrifugation 9. In this manner, the authors were able to show that phosphorylated STAT5 translocated from the cytoplasm to the nucleus upon temporal stimulation with EGF. However this technique requires cells to be subjected to multiple and often lengthy fractionation steps which may affect the integrity of the biological sample. Furthermore, the use of organelles separation would itself preclude the investigation of the signalling interactions that occur in an inter-compartmental manner, e.g. interactions between endosomal bound RTK and cytoplasmic adaptor proteins. An alternative approach to organelle fractionation is to employ chemical or genetic means to disrupt components of the cellular trafficking machinery to probe for spatial RTK signalling networks in cells. An example is the use of dynasore (a GTPase inhibitor of dynamin) or dominant negative and temperature sensitive mutants of dynamin to block clathrin-mediated endocytosis in cells and prevent the accumulation of RTKs in the endosomal compartment 37. Similarly, brefeldin A (BFA) is a chemical inhibitor that targets the guanine nucleotide exchange factors that regulate the Arf GTPases and prevents surface expression of RTKs while retaining the receptors in the endoplasmic reticulum (ER). Using BFA in combination with quantitative global phosphoproteomics, Choudhary and colleagues were able to show that an oncogenic mutant of fms-like tyrosine kinase receptor 3 (Flt3) that contains internal tandem duplications (Flt3-ITD) activated distinct kinases when localised to the ER versus the plasma membrane (PM) 43. Flt3-ITD in the ER appeared to enrich for Pim1 kinase motifs while Flt3-ITD in the PM preferentially activated proteins containing the motifs for AKT and MAPKs. Intriguingly, distinct Flt3-ITD phosphorylation sites were shown to be activated between the two compartments, suggesting that the observed signalling changes may be the result of altered phosphorylation at the level of the receptor. While the chemical approach allows for the study of protein compartmentalisation in intact cells, many of these compounds inhibit general cellular processes and would exert pleiotropic effects in cells. Moving forward, it is likely that both organelle isolation and chemical inhibitor strategies will need to be employed in a complementary manner to obtain a more accurate phosphoproteomic profile of spatial RTK signalling.
Figure 3.
Analysis of spatial signalling networks using mass spectrometry. After ligand binding, RTKs undergo endocytosis to different compartments, including the early endosome, multivesicular bodies (MVBs) and the mitochondria. Spatial signalling networks can be analysed either using purification methods to enrich for specific organelle components (such as centrifugation) or by employing chemical or genetic approaches to inhibit cellule trafficking processes. It is also important to incorporate other post-translational modifications (PTMs) such as ubiquitination (U) together with phosphorylation (P) to identify crosstalk mechanisms that occur between different PTMs in the regulation of spatial signalling networks.
The internalisation and intracellular trafficking of many RTKs are driven by a complex interplay between protein phosphorylation and both ubiquitin (Ub)-dependent and –independent mechanisms 44-45. Ubiquitination is a reversible protein modification that is added to RTKs by E3 ubiquitin ligases in response to receptor phosphorylation. This complexity is compounded by the ability of Ub itself to undergo further rounds of Ub addition on lysine residues to form polyubiquitin chains. These monoubiquitin and polyubiquitin modifications act as molecular tags for recruiting components of the endosomal sorting and lysosomal degradation machinery to the receptor46. In the case of EGFR, phosphorylation at Y1045 leads to the recruitment of the E3 ubiquitin ligase, Cbl which in turn ubiquitinates the receptor at lysine residues within the tyrosine kinase domain 47. Additionally, Cbl can also indirectly bind EGFR through the Grb2 adaptor protein at the Y1068 and Y1086 sites of the receptor 48. Proteins containing ubiquitin binding domains (UBDs) then interact with the ubiquitinated receptor to drive endosomal sorting and trafficking. Two recent studies have used quantitative MS approaches to identify the players involved in the ubiquitination regulated network downstream of EGFR 49-50. Argenzio et al. used a Ub specific antibody to immunopurify ubiquitinated proteins in EGF stimulated HeLa cells 49. They also performed a tandem affinity purification (TAP) using TAP-tagged Ub exogenously transfected into the B82L mouse fibroblast cell line to identify additional ubiquitinated targets. Using a complementary approach, Akimov et al. used GST-tagged UBDs from endocytic the adaptor proteins Epsin-1 and Eps15 to enrich for proteins that are ubiquitinated upon activation of EGFR in HeLa cells 50. There was significant overlap between the EGF-responsive ubiquitinated proteins in both studies. More importantly, many of the enriched proteins have previously been shown to be phosphorylated within similar timescales, suggesting potential crosstalk between the two forms of posttranslational modifications (PTMs). Future efforts in the computational integration of proteomic datasets of disparate but interconnected forms of PTMs will be required to build a more comprehensive understanding of spatial regulation of RTK signalling (Figure 3).
RTK COACTIVATION NETWORKS
The availability of a diverse range of phosphoproteomic technologies, both MS-based and alternative formats, has resulted in the widespread use of these tools for screening purposes, including the routine screening of cancer cell lines and tumours 51-54. One of the unanticipated outcomes of these screening efforts is the revelation that RTKs rarely act in isolation but rather cooperate in a web of coactivated RTK networks 54. For instance, the A431 epidermoid carcinoma cell line has been used in the past two decades as a workhorse for studying EGFR signalling pathways since it expresses abnormally high levels of the receptor. Ciaccio et al. used microwestern arrays to investigate the phosphorylation-mediated signalling networks activated in A431 cells upon stimulation with EGF in a dose and time-dependent manner 55. By integrating this data with Bayesian network modelling, the authors were able to show that stimulation of EGFR resulted in the ordered activation of a hierarchical cascade of multiple downstream RTKs, including PDGFR, ErbB2, ErbB4, c-Kit, c-Met, FGFR1 and IGF1R. A431 cells display distinct phenotypic behaviour in response to different concentrations of EGF. Stimulation at a low dose of EGF promotes cell proliferation while higher doses of the ligand results in growth inhibition and terminal differentiation 56-57. It is plausible that this biphasic phenomenon may be the effect of different combinations of RTKs being coactivated simultaneously at distinct EGF concentrations rather than the consequence of EGFR receptor activation alone. There is currently a poor understanding of the functional relevance of RTK coactivation and more work needs to be done on elucidating the integrated signalling networks downstream of coactivated RTKs and their resulting effects on cellular behaviour. This endeavour has been particularly challenging since many RTKs share common downstream signalling pathways and require highly sensitive and reproducible detection methods to delineate the individual pathways activated by RTK coactivation.
Phosphoproteomics by MS has yet to be employed in the systematic study of RTK coactivation networks. The goal of such a study would be to determine the signalling contribution of the individual receptors to integrated RTK coactivation signals. A previous large-scale study of receptor crosstalk in macrophages has shed light on the design criteria that would be important for such an experiment. Ranganathan and co-workers treated RAW 264.7 macrophage cells with 22 receptor-specific ligands singly or in all possible pairwise combinations 58. They then measured the effect of these ligand combinations on cyclic adenosine monophosphate (cAMP) synthesis, Ca2+ mobilisation, cytokine secretion and the phosphorylation status of downstream signalling proteins, including AKT, p38, STAT3, Erk1/2 and RSK. In performing these experiments, they looked for ligand combinations that exhibited non-additive effects, i.e. Effects that cannot simply be accounted for by a linear addition of the contributing single ligand responses. The authors found that there were multiple instances where cellular responses were silent under single ligand addition but became uncovered upon simultaneous activation by multiple ligands. They termed this effect as context-dependence where the responses driven by specific ligands are not independent but are largely reliant on the effects of others. Similar to the common effector machinery described earlier 33, the authors made a second observation that despite the large number of ligand combinations tested, the downstream signalling components converge onto a small set of interactions that are able integrate and regulate cellular behaviour. These two important findings are likely to also apply to RTK coactivation networks. Previous studies have shown that distinct RTKs phosphorylate common downstream adaptor proteins and kinases, albeit activating them to differing degrees 59. One of the open questions in the field is whether RTK coactivation leads to a simple linear extrapolation of the individual signalling profiles of contributing RTKs or if there is cooperativity and amplification as a result of the coactivation event.
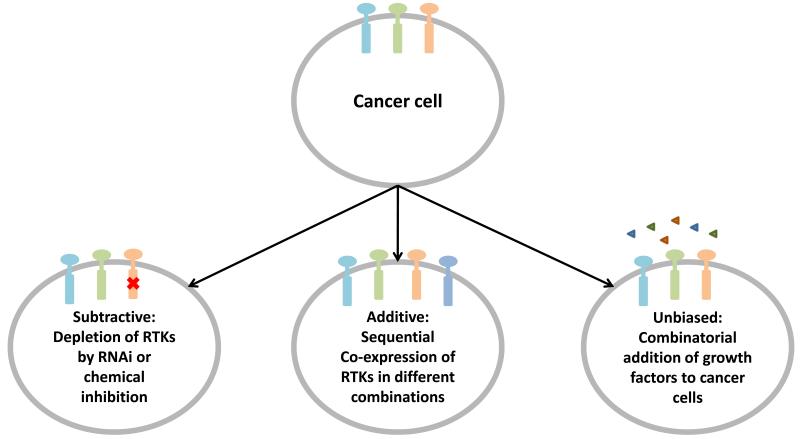
One approach to explore this hypothesis is to employ a subtractive strategy where coactivated RTKs are systematically eliminated either genetically with RNA interference or chemically using small molecule inhibitors, prior to phosphoproteomic profiling of downstream signalling. For instance, in the case of the A431 cells, RTKs such as PDGFR and ErbB2 can be depleted either singly or in combination and upon stimulation with EGF, phosphoproteomics can be used establish if these RTKs are essential for propagating EGFR signal transduction networks. Additionally, this approach will complement the study by Ciaccio et al. by validating the directionality of interactions between coactivated RTKs and the proposed order of the RTK activation cascade that was predicted using the Bayesian modelling approach 55. Alternatively, one can also use the additive strategy, where RTKs are sequentially added to cells by exogenous co-expression to establish the effect of RTK coactivation on signalling networks. The attractiveness of the additive approach is that RTKs can be titrated in specific concentrations and in different combinations in cells to investigate the cooperative nature of RTK coactivation. Finally, the unbiased approach previously employed by Ranganathan and co-workers can also be exploited for profiling RTK coactivation. Many cancer cell lines have been profiled for their endogenous RTK expression and activation levels 60. Stimulation of cancer cell lines with a panel of growth factors in a combinatorial fashion should identify the non-additive effects downstream of RTK coactivation and potentially elucidate candidate integrators for cellular signal processing. Since most cells express of a number of different endogenous RTKs, many of which are required for survival, great care must be taken to account for ‘confounding signals’ that may arise from simultaneous activation and crosstalk of multiple endogenous RTKs. A combination of the additive, subtractive and non-biased strategies will provide a valuable experimental handle for dissecting the functional relevance of RTK coactivation in cancer (Figure 4).
Figure 4.
Analysis of RTK coactivation. Three strategies can be used in a complementary fashion to elucidate RTK coactivation networks. A subtractive strategy can be employed to systematically deplete endogenous RTK levels in cancer cells. Alternatively, an additive approach of co-expressing different combinations of RTKs in cancer cells can be used to investigate the cooperative nature of RTK coactivation. A third approach is to administer growth factors to cancer cells in an unbiased fashion to identify non-additive effects of RTK coactivation on signal transduction.
CONCLUSION
Promising early studies have highlighted the potential of mass spectrometry-based phosphoproteomics for illuminating the signalling mechanisms that underlie many of the recent advances in RTK biology. More than 50% of known RTKs are poorly characterised and it remains necessary to continue the systematic profiling of these RTK signalling networks in a temporal/dose-dependent manner so as to expand our knowledge of these understudied RTKs 1. It is also critical that novel developments in phosphoproteomics are continually applied to new areas of RTK signalling such as those described in this article. It is important to note that these areas of RTK biology are not mutually exclusive and that a combination of different experimental strategies will be required to generate novel insights into RTK signalling. In this manner, phosphoproteomics can be used effectively to make further discoveries in RTK biology and continue contributing richly to this exciting area of signal transduction.
ACKNOWLEDGEMENTS
PHH is supported by the Wellcome Trust and The Institute of Cancer Research.
Abbreviations
- RTK
Receptor Tyrosine Kinases
- MS
mass spectrometry
- EGFR
Epidermal Growth Factor Receptor
REFERENCES
- 1.Lemmon MA, Schlessinger J. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gnad F, Gunawardena J, Mann M. Nucleic Acids Res. 2011;39:D253–260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 4.Dengjel J, Kratchmarova I, Blagoev B. Mol Biosyst. 2009;5:1112–1121. doi: 10.1039/b909534a. [DOI] [PubMed] [Google Scholar]
- 5.Kolch W, Pitt A. Nat Rev Cancer. 2010;10:618–629. doi: 10.1038/nrc2900. [DOI] [PubMed] [Google Scholar]
- 6.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 7.Marshall CJ. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Engholm-Keller K, Hansen TA, Palmisano G, Larsen MR. J Proteome Res. 2011 doi: 10.1021/pr200641x. [DOI] [PubMed] [Google Scholar]
- 11.Dengjel J, Akimov V, Olsen JV, Bunkenborg J, Mann M, Blagoev B, Andersen JS. Nat Biotechnol. 2007;25:566–568. doi: 10.1038/nbt1301. [DOI] [PubMed] [Google Scholar]
- 12.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Proc Natl Acad Sci U S A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar N, Hendriks BS, Janes KA, de Graaf D, Lauffenburger DA. Drug Discov Today. 2006;11:806–811. doi: 10.1016/j.drudis.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Janes KA, Lauffenburger DA. Curr Opin Chem Biol. 2006;10:73–80. doi: 10.1016/j.cbpa.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Nat Methods. 2010;7:383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 17.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 18.Monetti M, Nagaraj N, Sharma K, Mann M. Nat Methods. 2011;8:655–658. doi: 10.1038/nmeth.1647. [DOI] [PubMed] [Google Scholar]
- 19.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stratton MR. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 21.Pines G, Kostler WJ, Yarden Y. FEBS Lett. 2010;584:2699–2706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Garay M. L. Gonzalez, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radivojac P, Baenziger PH, Kann MG, Mort ME, Hahn MW, Mooney SD. Bioinformatics. 2008;24:i241–247. doi: 10.1093/bioinformatics/btn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youn A, Simon R. Bioinformatics. 2011;27:175–181. doi: 10.1093/bioinformatics/btq630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chumbalkar V, Latha K, Hwang Y, Maywald R, Hawley L, Sawaya R, Diao L, Baggerly K, Cavenee WK, Furnari FB, Bogler O. J Proteome Res. 2011;10:1343–1352. doi: 10.1021/pr101075e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Proc Natl Acad Sci U S A. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Proc Natl Acad Sci U S A. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, Comb MJ. Sci Signal. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guha U, Chaerkady R, Marimuthu A, Patterson AS, Kashyap MK, Harsha HC, Sato M, Bader JS, Lash AE, Minna JD, Pandey A, Varmus HE. Proc Natl Acad Sci U S A. 2008;105:14112–14117. doi: 10.1073/pnas.0806158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pines G, Huang PH, Zwang Y, White FM, Yarden Y. Oncogene. 2010;29:5850–5860. doi: 10.1038/onc.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang PH, Cavenee WK, Furnari FB, White FM. Cell Cycle. 2007;6:2750–2754. doi: 10.4161/cc.6.22.4922. [DOI] [PubMed] [Google Scholar]
- 32.Streuli CH, Akhtar N. Biochem J. 2009;418:491–506. doi: 10.1042/BJ20081948. [DOI] [PubMed] [Google Scholar]
- 33.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 34.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. Nat Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 35.Peschard P, Park M. Oncogene. 2007;26:1276–1285. doi: 10.1038/sj.onc.1210201. [DOI] [PubMed] [Google Scholar]
- 36.Kermorgant S, Parker PJ. J Cell Biol. 2008;182:855–863. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joffre C, Barrow R, Menard L, Calleja V, Hart IR, Kermorgant S. Nat Cell Biol. 2011;13:827–837. doi: 10.1038/ncb2257. [DOI] [PubMed] [Google Scholar]
- 38.Haugh JM, Huang AC, Wiley HS, Wells A, Lauffenburger DA. J Biol Chem. 1999;274:34350–34360. doi: 10.1074/jbc.274.48.34350. [DOI] [PubMed] [Google Scholar]
- 39.Haugh JM, Schooler K, Wells A, Wiley HS, Lauffenburger DA. J Biol Chem. 1999;274:8958–8965. doi: 10.1074/jbc.274.13.8958. [DOI] [PubMed] [Google Scholar]
- 40.Demory ML, Boerner JL, Davidson R, Faust W, Miyake T, Lee I, Huttemann M, Douglas R, Haddad G, Parsons SJ. J Biol Chem. 2009;284:36592–36604. doi: 10.1074/jbc.M109.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang YN, Yamaguchi H, Hsu JM, Hung MC. Oncogene. 2010;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cvrljevic AN, Akhavan D, Wu M, Martinello P, Furnari FB, Johnston AJ, Guo D, Pike L, Cavenee WK, Scott AM, Mischel PS, Hoogenraad NJ, Johns TG. J Cell Sci. 2011;124:2938–2950. doi: 10.1242/jcs.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Bohmer FD, Gerke V, Schmidt-Arras DE, Berdel WE, Muller-Tidow C, Mann M, Serve H. Mol Cell. 2009;36:326–339. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 44.Lu Z, Hunter T. Annu Rev Biochem. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunter T. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Wiley HS, Burke PM. Traffic. 2001;2:12–18. doi: 10.1034/j.1600-0854.2001.020103.x. [DOI] [PubMed] [Google Scholar]
- 47.Huang F, Goh LK, Sorkin A. Proc Natl Acad Sci U S A. 2007;104:16904–16909. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sorkin A, Goh LK. Exp Cell Res. 2008;314:3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Argenzio E, Bange T, Oldrini B, Bianchi F, Peesari R, Mari S, Di Fiore PP, Mann M, Polo S. Mol Syst Biol. 2011;7:462. doi: 10.1038/msb.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akimov V, Rigbolt KT, Nielsen MM, Blagoev B. Mol Biosyst. 2011 doi: 10.1039/c1mb05185g. [DOI] [PubMed] [Google Scholar]
- 51.Bielen A, Perryman L, Box GM, Valenti M, de Haven Brandon A, Martins V, Jury A, Popov S, Gowan S, Jeay S, Raynaud FI, Hofmann F, Hargrave D, Eccles SA, Jones C. Mol Cancer Ther. 2011 doi: 10.1158/1535-7163.MCT-11-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brevet M, Shimizu S, Bott MJ, Shukla N, Zhou Q, Olshen AB, Rusch V, Ladanyi M. J Thorac Oncol. 2011;6:864–874. doi: 10.1097/jto.0b013e318215a07d. [DOI] [PubMed] [Google Scholar]
- 53.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, Chin L, DePinho RA. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 54.Xu AM, Huang PH. Cancer Res. 2010;70:3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciaccio MF, Wagner JP, Chuu CP, Lauffenburger DA, Jones RB. Nat Methods. 2010;7:148–155. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.King I, Sartorelli AC. Cancer Res. 1989;49:5677–5681. [PubMed] [Google Scholar]
- 57.Konger RL, Chan TC. J Cell Physiol. 1993;156:515–521. doi: 10.1002/jcp.1041560310. [DOI] [PubMed] [Google Scholar]
- 58.Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. Nat Cell Biol. 2006;8:571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 59.Gordus A, Krall JA, Beyer EM, Kaushansky A, Wolf-Yadlin A, Sevecka M, Chang BH, Rush J, MacBeath G. Mol Syst Biol. 2009;5:235. doi: 10.1038/msb.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, Burns M, Julian B, Peng XP, Hieronymus H, Maglathlin RL, Lewis TA, Liau LM, Nghiemphu P, Mellinghoff IK, Louis DN, Loda M, Carr SA, Kung AL, Golub TR. Nat Biotechnol. 2009;27:77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]