Abstract
The primary role of autophagy is adaption to starvation. However, increasing evidence suggests that autophagy inhibition also plays an important role in tumorigenesis. Upregulation of X-linked inhibitor of apoptosis (XIAP) has been associated to a variety of human cancers, yet the underlying mechanisms remain obscure. Here, we report that XIAP suppresses autophagy by exerting a previously unidentified ubiquitin E3 ligase activity towards Mdm2, which is a negative regulator of p53. XIAP controls serum starvation-induced autophagy downstream of the PI3K/Akt pathway. In mouse models, inhibition of autophagy by XIAP promotes tumorigenecity of HCT116 cells. XIAP-mediated autophagy inhibition is also largely validated in clinical tumour samples. These findings reveal a novel XIAP-Mdm2-p53 pathway that mediates the inhibition of autophagy, by which XIAP may contribute to tumorigenesis.
Keywords: autophagy, Mdm2, XIAP
Introduction
Autophagy is an intracellular bulk degradation process that mediates the clearance of most long-lived proteins and damaged organelles (Levine and Klionsky, 2004; Mizushima, 2007). Upon induction of autophagy, cytosolic proteins and organelles are first sequestered within multimembrane-bound autophagosomes and subsequently delivered to lysosomes, where the autophagic cargo undergoes protease-dependent degradation (Klionsky, 2007). The basal level of autophagy is important for maintaining normal cellular homeostasis. Inhibition of basal autophagy in the brain leads to neurodegeneration in mouse models (Hara et al, 2006; Komatsu et al, 2006). In response to nutrient starvation, autophagy is induced as a pro-survival mechanism, which generates new metabolic substrates to meet the bioenergetic needs of cells and thus promotes cell survival (Lum et al, 2005; Mizushima and Komatsu, 2011). However, autophagy is also considered as a self-destructive process under certain circumstances (Levine and Kroemer, 2008).
Recent studies suggest that autophagy dysfunction plays an important role in the development of cancer (Mathew et al, 2007; Mizushima et al, 2008; Mah and Ryan, 2012). For example, allelic loss of the essential autophagy gene Beclin-1 is associated with high frequency of human ovarian, breast, and prostate cancers (Aita et al, 1999; Liang et al, 1999). Beclin-1 heterozygous mice with impaired autophagy are more prone to the development of spontaneous tumours, including lymphomas, lung carcinomas, hepatocellular carcinomas, and mammary precancerous lesions (Qu et al, 2003; Yue et al, 2003). Moreover, restoration of Beclin-1 expression in MCF-7 cells decreases their cellular proliferation, in vitro clonigenicity and tumour formation in nude mice (Liang et al, 1999). These genetic evidence strongly suggest that autophagy is a tumour suppressor pathway. However, the detailed molecular mechanisms by which autophagy suppresses tumour development remain largely undefined.
Inhibitor of apoptosis proteins (IAPs) were initially identified in baculoviruses, where they prevent defensive apoptosis of host cells (Crook et al, 1993; Birnbaum et al, 1994). Structurally, IAPs are characterized by the presence of one to three copies of BIR (baculovirus IAP repeat) motif (Salvesen and Duckett, 2002). In addition to BIR domains, many members of the IAPs also contain the RING (really interesting new gene) zinc finger domain at the very C-terminus of these proteins and possess ubiquitin E3 ligase activity that regulates auto- or trans-ubiquitination and protein degradation (Yang et al, 2000; Vaux and Silke, 2005; Hanson et al, 2012). Among the mammalian IAPs, X-linked inhibitor of apoptosis (XIAP) is most extensively studied and well characterized, containing three BIR domains and c-terminal RING domain. XIAP has the most potent anti-apoptotic ability (Eckelman et al, 2006), which is believed to be primarily related to direct binding and inhibiting of caspases, the apoptotic proteases that are responsible for the initiation and execution of apoptosis (Mizushima et al, 2008). XIAP strongly binds to both an initiator caspase (caspase-9) and effector caspases (caspase-3 and -7) via different parts of the BIR domains (Deveraux et al, 1997; Shiozaki and Shi, 2004). In addition, a recent study suggests that the ubiquitin E3 ligase activity of XIAP, which mediates proteasome-dependent degradation of caspases, is needed for its anti-apoptotic function (Schile et al, 2008).
Although XIAP has been well known for its anti-apoptotic function in regulating cell survival and cell proliferation (Liston et al, 1996; Schile et al, 2008; Gyrd-Hansen and Meier, 2010), it remains unclear whether XIAP also participates in the regulation of autophagy. Here, we provide evidence that XIAP functions as an endogenous repressor of autophagy. XIAP exerts its anti-autophagic function via acting as a ubiquitin E3 ligase for Mdm2 and thereby preventing Mdm2-mediated p53 degradation in the cytoplasm. We also show that XIAP-mediated autophagy inhibition indeed occurs in clinical tumour samples. Thus, our results suggest that XIAP-mediated autophagy inhibition may contribute to tumorigenesis.
Results
XIAP is a physiological inhibitor of autophagy
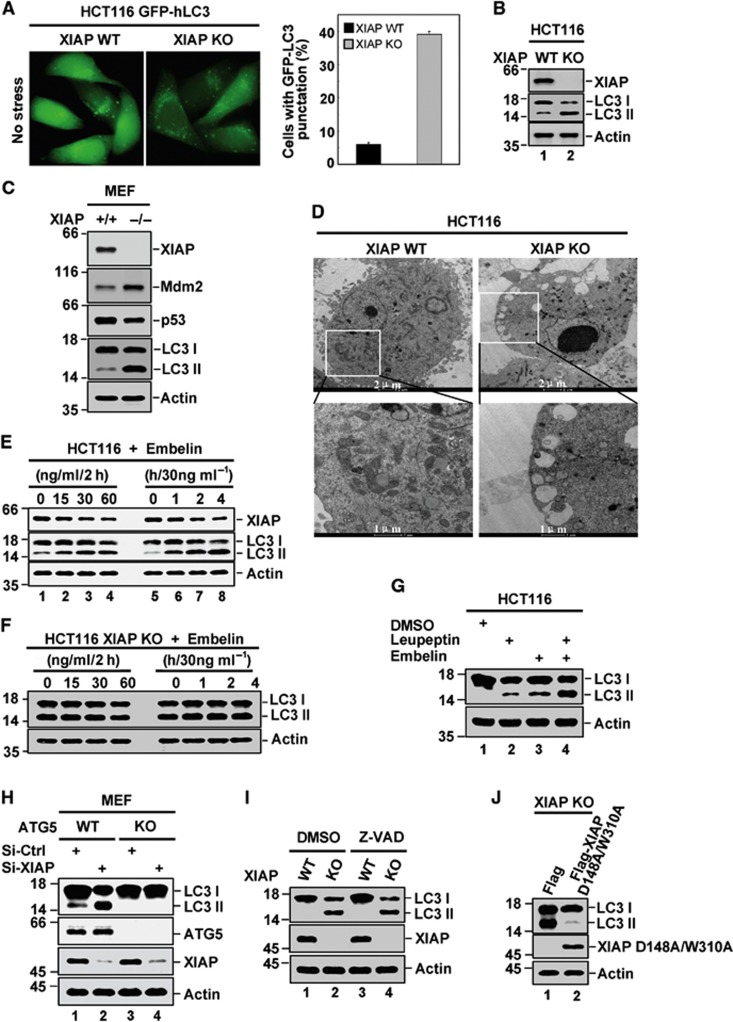
To determine whether XIAP involves in the regulation of autophagy, we first assessed the levels of basal autophagy in HCT116 XIAP+/+ (WT) and HCT116 XIAP−/− (KO) cells by tracking the conversion of LC3-I to LC3-II and evaluating autophagic vesicle formation using electron microscopy. We found that the percentage of cells with GFP-LC3 puncta was much higher in HCT116 XIAP KO cells than in HCT116 XIAP WT cells (Figure 1A). The subsequent western blot analysis also showed that conversion of LC3-I to LC3-II was greatly increased when XIAP was depleted in HCT116 cells (Figure 1B; Supplementary Figure S1A). Similar result was obtained using XIAP KO and the matching XIAP wild-type mouse embryonic fibroblasts (MEFs) (Figure 1C). In addition, XIAP deficiency led to a dramatic increase in autophagic vesicle formation in HCT116 cells (Figure 1D). Consistent with these results, treatment of HCT116 cells with Embelin, a specific inhibitor of XIAP (Nikolovska-Coleska et al, 2004), enhanced both conversion of LC3-I to LC3-II and formation of GFP-LC3 puncta in a dose- and time-dependent manner (Figure 1E; Supplementary Figure S1B and C). By contrast, Embelin failed to enhance autophagy in HCT116 XIAP KO cells (Figure 1F). In addition, the levels of LC3-II were further increased in the presence of leupeptin, a lysosomal protease inhibitor (Figure 1G, lane 3 versus 4; Supplementary Figure S1D), indicating that XIAP-mediated autophagy inhibition is through autophagic/lysosomal pathway. Knocking down XIAP expression in ATG5+/+ (WT) MEF cells also dramatically increased LC3-I to LC3-II conversion (Figure 1H, lane 1 versus 2; Supplementary Figure S1E). In contrast, ATG5−/− (KO) MEF cells, which are unable to undergo autophagy, showed no enhanced conversion of LC3-I to LC3-II by XIAP knockdown (Figure 1H, lane 3 versus 4; Supplementary Figure S1E). These results strongly suggest that XIAP is a physiological inhibitor of autophagy.
Figure 1.
XIAP inhibits autophagy. (A) Representative images of GFP-LC3 staining in HCT116 XIAP WT and XIAP KO cells with stable expression of GFP-LC3. Quantification of LC3 punctate cells was shown on the right. The data are represented as means±s.d. of three independent experiments. (B) Endogenous LC3 expression in HCT116 XIAP WT and XIAP KO cells was analysed by western blotting with anti-LC3 antibody. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S1A. (C) Lysates from XIAP+/+ and XIAP−/− mouse embryonic fibroblasts were analysed by western blotting with the indicated antibodies. The data are representative of two biological replicates. (D) Autophagic vesicles in HCT116 XIAP WT and XIAP KO cells were evaluated by electron microscopy. The images are representative of three biological replicates. (E) HCT116 cells were treated with Embelin as indicated. LC3-II accumulation was determined by western blot analysis with anti-LC3 antibody. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S1B. (F) HCT116 XIAP KO cells were treated with Embelin as indicated. LC3-II accumulation was determined by western blot analysis with anti-LC3 antibody. The data are representative of three biological replicates. (G) HCT116 cells were treated with Leupeptin and Embelin in the indicated combinations. Cell lysates were analysed by western blotting. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S1D. (H) MEF ATG5 WT and ATG5 KO cells were individually transfected with XIAP-specific or control siRNAs. Forty-eight hours after transfection, cell lysates were analysed by western blotting with the indicated antibodies. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S1E. (I) HCT116 XIAP WT and XIAP KO cells were treated with 20 μM Z-VAD-FMK or DMSO for 2 h. Cell lysates were then analysed by western blotting. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S1F. (J) HCT116 XIAP KO cells were transfected with either Flag-XIAP (D148A/W310A) or control plasmid. Twenty-four hours after transfection, cell lysates were subjected to western blot analysis with the indicated antibodies. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S1G.
We next determined whether XIAP-mediated autophagy inhibition is related to apoptosis inhibition. HCT116 XIAP WT and XIAP KO cells were treated with caspase inhibitor Z-VAD-FMK, followed by detecting LC3-I to LC3-II conversion. The enhancement of autophagy in HCT116 XIAP KO cells was not affected by Z-VAD-FMK treatment (Figure 1I, lane 2 versus 4; Supplementary Figure 1F). The reintroduction of a mutated form of XIAP (XIAP D148A/W310A) that cannot inhibit caspases into these XIAP KO cells was still able to protect the cells from autophagy (Figure 1J; Supplementary Figure 1G). These indicate that XIAP inhibits autophagy independent of caspase-mediated apoptosis.
XIAP inhibits autophagy via the Mdm2-p53 pathway
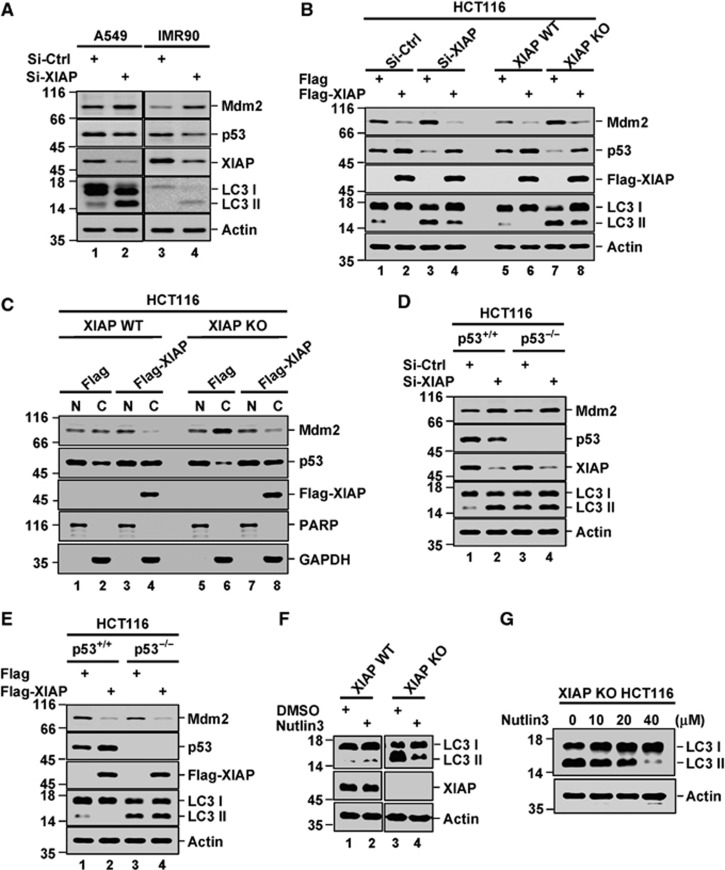
p53 has been recently recognized as a negative regulator of autophagy (Tasdemir et al, 2008). We sought to determine whether XIAP could regulate autophagy via a p53-dependent mechanism. We first examined whether XIAP affects the steady-state levels of p53 and its main negative regulator Mdm2. Intriguingly, in several tested human cell lines, small-interfering RNA (siRNA)- or small-hairpin RNA (shRNA)-mediated knockdown of XIAP led to a marked increase in Mdm2 levels and a dramatic decrease in p53 levels, which was accompanied with the enhanced conversion of LC3-I to LC3-II (Figure 2A; Supplementary Figure S2A–C). Similarly, compared to XIAP wild-type MEFs, XIAP KO MEFs expressed increased levels of Mdm2 and decreased levels of p53 (Figure 1C). Furthermore, ectopic expression of Flag-XIAP greatly reversed XIAP knockdown or knockout effects on the levels of Mdm2 and p53 and the conversion of LC3-I to LC3-II (Figure 2B, lane 3 versus 4 and lane 7 versus 8; Supplementary Figure 2D).
Figure 2.
XIAP inhibits autophagy via the Mdm2-p53 pathway. (A) A549 and IMR90 cells were transfected with XIAP-specific or control siRNAs. Cell lysates were subjected to western blot analysis with the indicated antibodies. The data are representative of two biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S2A. (B) HCT116 cells expressing XIAP-specific or control siRNAs, HCT116 XIAP WT and XIAP KO cells were individually transfected with Flag-XIAP or control vector. Cell lysates were analysed by western blotting. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S2D. (C) HCT116 XIAP WT and XIAP KO cells were individually transfected with Flag-XIAP or control vector. Cytoplasmic/nuclear fractionation was performed to analyse cellular localization of Mdm2 and p53. PARP and GAPDH were used as nuclear (N) and cytoplasmic (C) fraction markers, respectively. The data are representative of three biological replicates. (D) HCT116 p53+/+ and p53−/− cells were individually transfected with XIAP-specific or control siRNAs. LC3 conversion was detected by western blot analysis. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S2E. (E) HCT116 p53+/+ and p53−/− cells were individually transfected with Flag-XIAP or control vector. LC3 conversion was detected by western blot analysis. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S2F. (F) HCT116 XIAP WT and XIAP KO cells treated with 10 μM Nutlin3 for 6 h. LC3 conversion was evaluated by western blot analysis. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S2H. (G) HCT116 XIAP KO cells were treated with the indicated amounts of Nutlin for 8 h. Cell lysates were analysed by western blotting with anti-LC3 antibody. The data are representative of three biological replicates.
The subsequent fractionation assays revealed that lack of XIAP in HCT116 cells led to the elevated levels of Mdm2 and the reduced levels of p53 in cytosol, while the nuclear levels of Mdm2 and p53 remained relatively unaffected (Figure 2C, lanes 1 and 2 versus 5 and 6). Consistently, ectopic expression of XIAP specifically altered the cytosolic levels of Mdm2 and p53 (Figure 2C, lanes 1 and 2 versus 3 and 4, and lanes 5 and 6 versus 7 and 8). Previous study has shown that cytosolic but not nuclear p53 is able to antagonize autophagy (Tasdemir et al, 2008). Therefore, our results suggest that XIAP inhibits autophagy through upregulating cytosolic p53 levels. To further confirm this, we first determined the effect of XIAP expression on autophagy in HCT116 p53+/+ and HCT116 p53−/− cells by evaluating the conversion of LC3-I to LC3-II. In HCT116 p53+/+ cells, XIAP knockdown resulted in a dramatic increase in autophagy (Figure 2D, lane 1 versus 2; Supplementary Figure 2E), whereas XIAP overexpression showed the opposite effect (Figure 2E, lane 1 versus 2; Supplementary Figure 2F). However, when p53 was ablated, neither of these effects occurred (Figure 2D, lane 3 versus 4; Figure 2E, lane 3 versus 4; Supplementary Figure S2E and F). Moreover, ectopic expression of wild-type p53 or its tumour-derived mutants, including p53 R175H, p53 R273H, p53 G279E, and p53 K386R, effectively prevented autophagy occurred in HCT116 XIAP KO cells (Supplementary Figure S2G). We also evaluated the levels of autophagy in the presence and absence of Nutlin, a pharmacological inhibitor of Mdm2 that disrupts the Mdm2-p53 association. Nutlin was shown to greatly inhibit autophagy induction by XIAP depletion (Figure 2F and G; Supplementary Figure S2H). Together, these results indicate that XIAP inhibits autophagy via regulating the Mdm2-p53 pathway.
XIAP is a novel ubiquitin E3 ligase for Mdm2
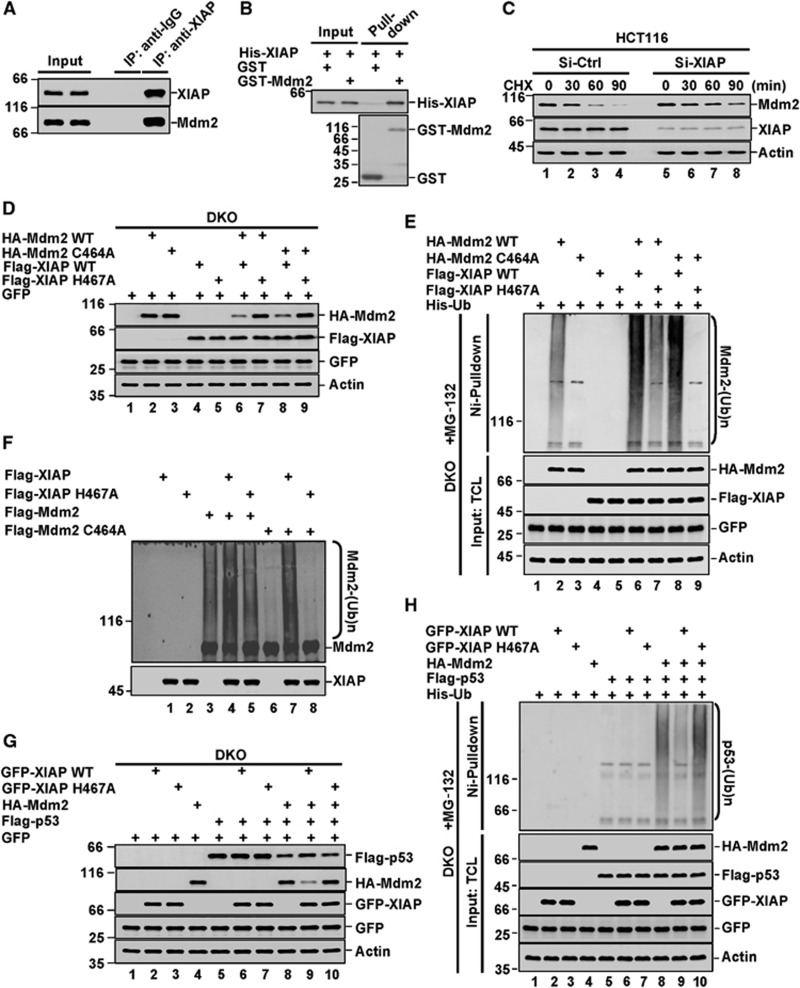
We next investigated the molecular mechanism underlying the regulation of Mdm2 and p53 by XIAP. Using reciprocal co-immunoprecipitation assays with anti-XIAP or anti-Mdm2 antibodies, the interaction between endogenous XIAP and Mdm2 was readily detected (Figure 3A; Supplementary Figure S3A). This XIAP-Mdm2 interaction appears to be direct, as shown by an in vitro GST-pulldown assay with recombinant proteins (Figure 3B). Furthermore, the half-life of endogenous Mdm2 was increased in cycloheximide-treated XIAP knockdown cells compared to control cells (Figure 3C; Supplementary Figure S3B). Similar result was also obtained when XIAP was depleted (Supplementary Figure S3C and D). These results indicate that increased Mdm2 levels by XIAP depletion occur through its stabilization.
Figure 3.
XIAP is a novel E3 ligase for Mdm2. (A) Lysates from HCT116 cells were immunoprecipitated separately with anti-XIAP antibody and an isotype-matched control IgG. Immunoprecipitates and input were analysed by western blotting. The data are representative of two biological replicates. (B) Recombinant His-XIAP protein was incubated separately with purified GST and GST-Mdm2 fusion proteins on glutathione beads for 4 h followed by western blot analysis using anti-His antibody. GST and GST-Mdm2 were analysed by Coomassie blue staining. The data are representative of two biological replicates. (C) HCT116 cells were treated with XIAP-specific or control siRNAs. Twenty-four hours later, cells were cultured in the presence of 50 μg ml−1 cycloheximide for the indicated periods of time, and subsequently analysed by western blotting. We should mention that amounts of cell lysates were adjusted to achieve similar expression levels of Mdm2 at time 0, while the same amounts of cell lysates were used to examine levels of XIAP and actin. The data are representative of three biological replicates. The ratio of Mdm2 to actin is presented in Supplementary Figure S3B. (D) p53−/−Mdm2−/− MEF cells were co-transfected with the indicated Mdm2, XIAP, and p53 constructs. Cell lysates were analysed by western blotting with the indicated antibodies. We should mention that XIAP H467A expressing plasmid was always used less than XIAP expressing plasmid to ensure expression levels of XIAP and XIAP H467A were similar. The data are representative of three biological replicates. (E) p53−/−Mdm2−/− MEF cells were transfected with the indicated plasmids. Twenty-four hours after transfection, cells were treated with 20 μM MG-132 for additional 4 h. Cell lysates were denatured before proteins conjugated to His-ubiquitin were pulled down by Ni2+-NTA beads. The bead-bound proteins and total cell lysates (TCLs) were analysed by western blot with anti-Mdm2 antibody. The data are representative of three biological replicates. (F) In all, 2 μM Mdm2 or its C464A mutant and 1 μM XIAP or its H467A mutant proteins were incubated with 100 nM E1, 2 μM E2 and 200 μM Ub in a total 20 μl in vitro ubiquitination reaction buffer at 37°C for 1 h. The reaction mixtures were analysed by western blotting with anti-Mdm2 antibody. The data are representative of three biological replicates. (G) p53−/−Mdm2−/− MEF cells were transfected with the indicated plasmids. Cell lysates were analysed by western blotting. The data are representative of three biological replicates. (H) p53−/−Mdm2−/− MEF cells were transfected with the indicated plasmids. Twenty-four hours after transfection, cells were treated with 20 μM MG-132 for additional 4 h. Cell lysates were denatured before proteins conjugated to His-ubiquitin were pulled down by Ni2+-NTA beads. The bead-bound proteins were analysed by western blotting with anti-p53 antibody. The data are representative of three biological replicates.
Since Mdm2 stability is largely regulated by its autoubiquitination, we then examined the effect of XIAP on the RING finger-domain mutant MDM2 C464A which is inactive in autoubiquitination. Strikingly, similar to the effect of XIAP on wild-type Mdm2, induction of XIAP also led to the reduced levels of Mdm2 C464A (Figure 3D, lane 2 versus 6 and lane 3 versus 8), and the accompanying enhancement of its polyubiquitination (Figure 3E, lane 2 versus 6 and lane 3 versus 8) in MEF cells deficient in both p53 and Mdm2 (DKO). Consistently, when XIAP was co-expressed, the half-life of both wild-type Mdm2 and mutant Mdm2 C464A was markedly decreased (Supplementary Figure S3E). Of note, the caspase-binding mutant XIAP D148A/W310A still can bind to Mdm2 and mildly decreased the half-life of Mdm2 (Supplementary Figure S4A and B). Collectively, these results demonstrate that XIAP regulates Mdm2 stability is not through promoting its autoubiquitination activity. We also found that XIAP H467A, a mutant that lost intrinsic E3 ligase activity in RING domain (Yang et al, 2000), failed to suppress the levels of wild-type Mdm2 or mutant Mdm2 C464A (Figure 3D, lane 2 versus 7 and lane 3 versus 9; Supplementary Figure S3E), although XIAP H467A still retained the Mdm2-binding ability (Supplementary Figure S4C). In addition, XIAP H467A neither promoted the polyubiquitination of wt-Mdm2 or mutant Mdm2 C464A (Figure 3E, lane 2 versus 7 and lane 3 versus 9) nor decreased their half-life (Supplementary Figure S3E), suggesting that XIAP could be a ubiquitin E3 ligase for Mdm2.
To confirm this, an in vitro ubiquitination assay was performed with purified recombinant proteins. Consistent with previous study (Fang et al, 2000), Mdm2 underwent autoubiquitination, which was diminished by its C464A mutation (Figure 3F, lane 3 versus 6). Interestingly, when recombinant XIAP protein was included in the reaction mixture, ubiquitination of both wild-type Mdm2 and mutant Mdm2 C464A was strongly enhanced (Figure 3F, lane 3 versus 4 and lane 6 versus 7). However, compared with wild-type XIAP, the RING-inactive mutant XIAP H467A failed to show any effect on the ubiquitination of Mdm2 and its C464A mutant (Figure 3F, lane 3 versus 5 and lane 6 versus 8). The specificity of XIAP as a ubiquitin E3 ligase towards Mdm2 was further concluded by the observation that XIAP promoted Mdm2 C464A ubiquitination in a dose-dependent fashion in vitro (Supplementary Figure S5A).
To further test whether XIAP regulates p53 degradation through Mdm2, a co-transfection assay was performed in DKO MEF cells. When wild-type XIAP or mutant XIAP H467A was co-expressed with p53 in these cells, levels of p53 remained unaltered (Figure 3G, lanes 5–7). However, by additional expression of Mdm2, wild-type XIAP, but not mutant XIAP H467A, was shown to increase p53 level (Figure 3G, lanes 8–10). This correlated well with a decline in Mdm2 levels by expression of wild-type XIAP but not mutant XIAP H467A (Figure 3G, lanes 8–10). The subsequent in vivo ubiquitination assay further revealed that Mdm2-mediated ubiquitination of p53 was inhibited by wild-type XIAP, but not mutant XIAP H467A (Figure 3H, lanes 8–10). Knockdown of XIAP decreased ubiquitination of Mdm2, but increased ubiquitination of p53 (Supplementary Figure S5B). Additionally, XIAP did not affect the Mdm2-p53 interaction (Supplementary Figure S5C). Therefore, these data demonstrate that XIAP possesses an intrinsic ubiquitin E3 ligase activity towards Mdm2, by which XIAP regulates Mdm2-p53 axis.
XIAP regulates serum deprivation-induced autophagy
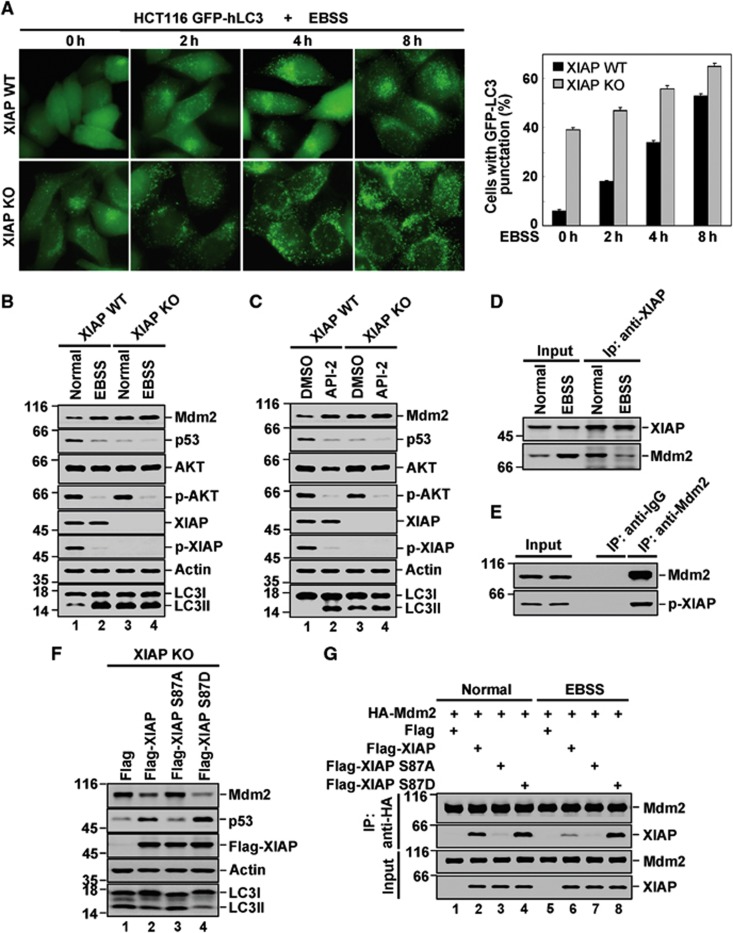
The inhibitory effect of XIAP on the basal autophagy prompted us to investigate whether XIAP regulates serum deprivation-induced autophagy. In HCT116 cells with wild-type XIAP expression, EBSS (Earle’s balanced salt solution) treatment increased autophagy in a time-dependent manner as manifested by enhanced GFP-LC3 puncta formation and increased conversion of endogenous LC3-I to LC3-II. However, the EBSS-induced autophagic effect was substantially minimized by XIAP depletion (Figure 4A; Supplementary Figure S6A). The subsequent analysis showed that, in wild-type XIAP expressing cells, EBSS treatment resulted in increased levels of Mdm2 and decreased levels of p53, which was accompanied by elevated conversion of LC3-I to LC3-II (Figure 4B, lane 1 versus 2; Supplementary Figure 6B). However, when XIAP was ablated, these EBSS-mediated effects were inhibited (Figure 4B, lane 3 versus 4; Supplementary Figure 6B). These results strongly suggest the critical role of XIAP in regulating serum deprivation-induced autophagy. In agreement with previous report that XIAP is phosphorylated and stabilized by Akt at Ser87 (Dan et al, 2004), the levels of phosphorylated XIAP (p-XIAP) were found to be significantly reduced when the endogenous PI3K/Akt pathway was inhibited by serum starvation, although total levels of XIAP were not affected (Figure 4B, lane 1 versus 2). This indicates that dephosphorylation of XIAP by Akt inhibition might set up a switch for autophagy induction in response to serum deprivation. In supporting of this, treatment of API-2, a specific inhibitor of Akt, dramatically increased autophagy in the cells with wild-type XIAP expression, but not in the XIAP-deficient cells (Figure 4C, lane 1 versus 2 and lane 3 versus 4; Supplementary Figure 6C).
Figure 4.
XIAP regulates serum starvation-induced autophagy. (A) HCT116 XIAP WT and XIAP KO cells were treated with EBSS for the indicated periods of time. GFP-LC3 puncta formation was observed by a microscope. Quantification of LC3 punctate cells was shown on the right. The data are represented as means±s.d. of three independent experiments. (B) HCT116 XIAP WT and XIAP KO cells were treated with EBSS for 4 h followed by western blot analysis with the indicated antibodies. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S6B. (C) HCT116 XIAP WT and XIAP KO cells were treated with 1 μM API-2 for 6 h. Cell lysates were analysed by western blotting. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S6C. (D) HCT116 cells were treated with or without EBSS for 4 h. Cell lysates were subjected to immunoprecipitation with anti-XIAP antibody followed by western blot analysis. The data are representative of two biological replicates. (E) Lysates from HCT116 cells were immunoprecipitated with anti-Mdm2 antibody. Immunoprecipitates and cell lysates were analysed by western blotting with anti-p-XIAP antibody. The data are representative of two biological replicates. (F) HCT116 XIAP KO cells were transfected with Flag-XIAP, Flag-XIAP S87A, Flag-XIAP S87D or control vector. Twenty-four hours after transfection, cells were harvested. LC3 conversion was evaluated by western blot analysis. The data are representative of three biological replicates. The ratio of LCII/LC3I to actin is presented in Supplementary Figure S6D. (G) p53−/−Mdm2−/− MEF cells were transfected with HA-Mdm2 plus Flag-XIAP, Flag-XIAP S87A or Flag-XIAP S87D. Twenty-four hours later, cells were treated with 20 μM MG-132 for another 4 h. Cell lysates were subjected to immunoprecipitation with anti-HA antibody. Immunoprecipitates were analysed by western blotting. The data are representative of two biological replicates.
We next examined whether EBSS treatment regulates the interaction of XIAP and Mdm2. Treatment of HCT116 cells with EBSS greatly abolished the XIAP-Mdm2 interaction (Figure 4D). The strong correlation between decreased levels of p-XIAP and increased levels of Mdm2 upon EBSS or API-2 treatment (Figure 4B, lane 1 versus 2; Figure 4C, lane 1 versus 2) led us to explore the possibility that it is the p-XIAP that preferably binds to Mdm2 and controls its stability. The interaction between endogenous Mdm2 and p-XIAP was confirmed by a co-immunoprecipitation assay (Figure 4E). The effects of wild-type XIAP, phosphorylation defective mutant XIAP S87A, and phosphorylation mimics XIAP S87D on expression levels of Mdm2 and p53 were also assessed. Ectopic expression of XIAP consistently led to a decrease in Mdm2 and an increase in p53 (Figure 4F, lane 1 versus 2). Differing from XIAP S87A, which failed to show any effect on the steady-state levels of Mdm2 or p53, XIAP S87D exhibited an even stronger ability to regulate levels of Mdm2 and p53 than wild-type XIAP (Figure 4F, lanes 2–4). Correlating with the effect on Mdm2 and p53, XIAP S87D showed the strongest ability to inhibit autophagy (Figure 4F, lanes 2–4; Supplementary Figure 6D). These results were in good agreement with the differential binding preferences of Mdm2 towards wt-XIAP, XIAP S87A, and XIAP S87D. Under unstressed condition, unlike wild-type XIAP, XIAP S87A had lost its ability to interact with Mdm2, and XIAP S87D showed even stronger binding affinity with Mdm2 than wild-type XIAP (Figure 4G, lanes 2–4). Interestingly, under stressed condition of EBSS treatment, the interaction of wild-type XIAP and Mdm2 was strongly inhibited (Figure 4G, lane 2 versus 6), which further confirmed our endogenous co-immunoprecipitation result (Figure 4D). However, the interaction between XIAP S87D and Mdm2 was little, if any affected by EBSS treatment (Figure 4G, lane 4 versus 8). Taken together, these results indicate that dephosphorylation of XIAP by Akt inhibition is essential for the dissociation of XIAP from Mdm2, thereby enhancing Mdm2 ubiquitin E3 ligase activity towards p53 and facilitating EBSS-induced autophagy.
Biological significance of XIAP-mediated autophagy inhibition
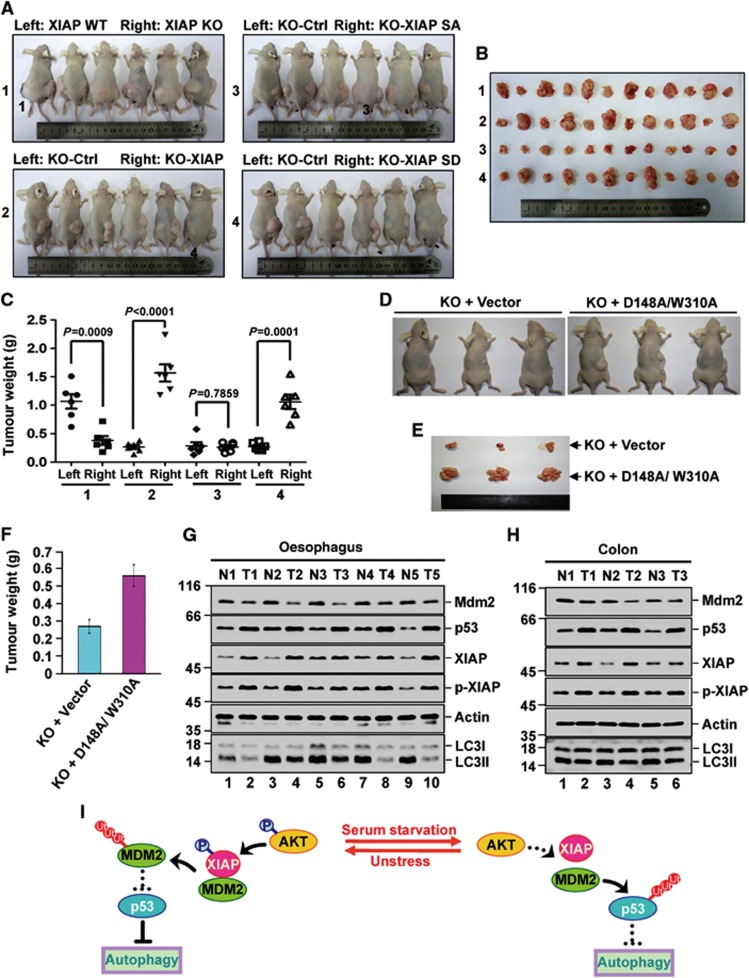
Autophagy malfunction has been recently linked to promotion of tumorigenesis (Mathew et al, 2007). The effect of p-XIAP on autophagy inhibition prompted us to examine whether phosphorylation of XIAP is essential for its ability to promote tumorigenecity of human cancer cells. We used a xenograft mouse model. XIAP ablation significantly suppressed tumorigenecity of HCT116 cells as revealed by measuring tumour mass (Figure 5A1, B1, and C1). When wild-type XIAP was reconstituted into HCT116 XIAP KO cells, their tumorigenecity was greatly improved (Figure 5A2, B2, and C2). However, phosphorylation defective mutant XIAP S87A was unable to recover tumorigenecity of HCT116 XIAP KO cells (Figure 5A3, B3, and C3), whereas phosphorylation mimics mutant XIAP S87D showed the comparable activity to wild-type XIAP in promoting tumorigenecity of HCT116 XIAP KO cells (Figure 5A4, B4, and C4).
Figure 5.
XIAP-modulated autophagy is associated with the tumorigenecity. (A–C) 1 × 106 HCT116 XIAP WT and 1 × 106 XIAP KO cells (group 1), 1 × 106 HCT116 XIAP KO cells (KO-Ctrl) and 1 × 106 HCT116 XIAP KO cells stably expressing wild-type XIAP (KO-XIAP) (group 2), 1 × 106 HCT116 XIAP KO cells (KO-Ctrl) and 1 × 106 HCT116 XIAP KO cells stably expressing XIAP S87A mutant (KO-XIAP SA) (group 3), or 1 × 106 HCT116 XIAP KO cells (KO-Ctrl) and 1 × 106 HCT116 XIAP KO cells stably expressing XIAP S87D mutant (KO-XIAP SD) (group 4) were individually injected to the left flank (Left) and right flank (Right) of nude mice as indicated. Five weeks after injection, the mice were sacrificed and photographed (A). The volumes of all flank tumours excised from six mice in each group were compared (n=6) (B). Tumour weights were represented as means±s.d. from six mice in each group (C). (D–F) 1 × 106 HCT116 XIAP KO cells or 1 × 106 HCT116 XIAP KO cells stably expressing XIAP D148A/310A were injected to the right flank of nude mice as indicated (n=3). Five weeks after injection, the mice were sacrificed and photographed (D). The volumes of excised tumours were shown (E). Tumour weights were represented as means±s.d. from three mice in each group (F). (G, H) The human tumour tissues (T) from Oesophagus (G) and Colon (H) and their adjacent normal tissues (N) were homogenized for protein extraction. Protein extracts were analysed by western blotting with the indicated antibodies. The data are representative of three biological replicates. The blots in (G, H) were qualified and the ratio of LC3II/LC3I to actin was then calculated and shown in Supplementary Figure 9A and B. (I) The schematic model of autophagic regulation by XIAP is shown. Under non-stressed condition, phosphorylated XIAP interacts with Mdm2 and mediates its rapid degradation, thus maintaining the relative high levels of p53 and inhibiting basal levels of autophagy. In response to serum starvation, XIAP undergoes dephosphorylation due to AKT inhibition, thereby promoting the dissociation of XIAP from Mdm2. This in turn accelerates the degradation of p53 and facilitates serum starvation-induced autophagy.
To verify whether the effect of wild-type XIAP or XIAP mutants on tumorigenecity is indeed an autophagy-related process, the extent of autophagy in respective excised tumours was evaluated by tracking the conversion of LC3-I to LC3-II using western blot analysis. As was expected, increased autophagy levels were observed in XIAP KO tumours compared with those in the XIAP WT samples (Supplementary Figure S7, upper left panels). However, this XIAP depletion-induced autophagy was greatly inhibited by re-expressed wild-type XIAP (Supplementary Figure S7, lower left panels) or XIAP S87D (Supplementary Figure S7, lower right panels), but not by re-expressed XIAP S87A (Supplementary Figure S7, upper right panels). Overall, the extent of autophagy was correlated well with the expression levels of Mdm2 and p53 in their respective tumours (Supplementary Figure S7). These were further confirmed by the immunohistochemical analyses (Supplementary Figure S8). More importantly, we found that when XIAP caspase-binding mutant (XIAP D148A/310A) was reconstituted into HCT116 XIAP KO cells, the tumorigenecity of these transfected cells was markedly improved (Figure 5D–F), indicating that the inhibition of apoptosis contributes little if any to the tumour-promoting effect of XIAP. Together, these results suggest that phosphorylation of XIAP is essential in regulating tumorigenecity and indicate the critical role of autophagy inhibition mediated by the XIAP-Mdm2-p53 signalling pathway in facilitating tumorigenesis.
To further validate the biological significance of XIAP-mediated autophagy inhibition in cancer, we analysed the expression levels of XIAP, p-XIAP, Mdm2, and p53 and conversion of LC3-I to LC3-II in several different types of human primary tumours, including oesophagus, colon, breast, lung, and stomach, along with their matched adjacent normal tissues. We found that 5 of 16 oesophagus cancers, 3 of 5 colon cancers, 1 of 3 breast cancers, 2 of 8 lung cancers, and 3 of 10 stomach cancers showed great autophagy inhibition along with the elevated expression of p-XIAP, XIAP, and p53 and the decreased expression of Mdm2 compared to those from their matched adjacent normal tissues (Figure 5G and H; Supplementary Figures S9 and S10). The XIAP-mediated autophagy inhibition in these tumours was also confirmed by the subsequent immunohistochemical analyses (Supplementary Figure S11). Since p53 status could affect p53 expression levels in the different tumors, we currently cannot totally exclude the possibility that increased levels of p53 are due, at least in part, to mutant p53 in human clinical tumour samples. However, based on the data presented here, autophagy inhibition mediated by the XIAP-Mdm2-p53 signalling pathway will likely play a significant role in promoting human tumour formation.
Discussion
The biological effects of XIAP have been largely attributed to its effects on the apoptotic pathway, although it has been recognized that XIAP is involved in the regulation of the NFkB pathway, inflammatory signalling, and cell migration (Lu et al, 2007; Gyrd-Hansen and Meier, 2010; Wu et al, 2010; Damgaard et al, 2012). Here, we demonstrate a novel and unexpected function of XIAP in negatively regulating the autophagy pathway.
In unstressed cells, p-XIAP interacts with Mdm2 and mediates its rapid degradation, thereby maintaining the relative high levels of cytosolic p53 (Figure 5I). Mdm2 is the major ubiquitin E3 ligase of p53, both of them are labile proteins. Although the trans-E3 activity of Mdm2 towards p53 has been well established, how Mdm2 itself is regulated still remains largely unknown. Previous studies suggest that cis-E3 ligase activity of Mdm2 is not responsible for its short-lived nature (Itahana et al, 2007). Recently, PCAF and SCFβ-TRCP were reported to be involved in the rapid degradation of Mdm2 (Linares et al, 2007; Inuzuka et al, 2010). Our finding that XIAP is an intrinsic ubiquitin E3 ligase for Mdm2 uncovers a novel, unexpected molecular mechanism that controls Mdm2 stability and indicates the complexity of the regulation of Mdm2. p53 has been suggested to play an two-faceted function in the regulation of autophagy (Maiuri et al, 2010). Within the nucleus, p53 induces autophagy through transcriptional effects (Crighton et al, 2006). Within the cytosol, p53 acts as a master repressor of autophagy (Tasdemir et al, 2008). Thus, XIAP may inhibit autophagy through regulating the levels of cytosolic p53. Our data also show that in response to serum starvation, XIAP undergoes dephosphorylation due to the inhibition of Akt, thus promoting the dissociation of XIAP from Mdm2. This is likely to make Mdm2 stable and subsequently lead to the accelerated degradation of cytosolic p53, thereafter facilitating the induction of autophagy (Figure 5I). Overall, XIAP appears to be an important molecular switch that controls initiation of serum starvation-induced autophagy.
It has been recently shown that Mdm2 induces XIAP expression via enhancing its translation following irradiation (Gu et al, 2009). Interestingly, our data show that XIAP negatively regulates Mdm2 stability through its ubiquitin E3 ligase activity. These combined findings suggest the existence of a feedback loop between XIAP and Mdm2 under certain situation, but this hypothesis needs to be further investigated.
Autophagy is well considered as a pro-survival mechanism for supporting the long-term survival of mammalian cells. Recent studies suggest that autophagy dysfunction plays an important role in various human diseases, such as cancer, neurodegenerative diseases, and ageing (Mizushima et al, 2008; Kimmelman, 2011; Rubinsztein et al, 2011). A causal effect of autophagy inhibition on tumorigenesis has been initially revealed by the finding that Beclin-1 heterozygous knockout mice with decreased autophagy levels are more prone to spontaneous tumour development. The subsequent studies with mice lacking Atg4C and BIF1 also show that deficiency in these autophagic factors leads to an increased frequency of tumour formation (Marino et al, 2007; Takahashi et al, 2007). Furthermore, mutations in other autophagy-related genes such as Atg2B, Atg5, Atg12, and UVRAG have been associated with the development of gastric and colorectal cancers (Kim et al, 2008; Kang et al, 2009). These findings uncover the importance of autophagy inhibition in promoting tumorigenesis. In this study, we show that XIAP is a physiological inhibitor of autophagy. XIAP-mediated autophagy inhibition appears to be important for tumour promoting effect, as revealed by the finding that reintroduction of XIAP S87A, which has lost its anti-autophagy activity, into HCT116 XIAP KO cells failed to recover the tumorigenecity of these cells in a xenograft mouse model. More importantly, the autophagy inhibition mediated by the XIAP-Mdm2-p53 pathway was also largely confirmed in clinical tumour samples. These data strongly suggest that XIAP-mediated autophagy inhibition may play an important role in tumour formation.
Overexpression of XIAP has been reported in a variety of human cancers (Tamm et al, 2000). Studies in human cancer cell culture models have demonstrated that ectopic expression of XIAP inhibits, whereas downregulation of XIAP enhances, caspase activation and apoptosis triggered by various apoptotic stimuli (Gyrd-Hansen and Meier, 2010). Moreover, studies in xenograft tumour models also reveal that loss of XIAP abrogates tumour growth (Ravi et al, 2006). These evidence suggest that XIAP may facilitate cancer development by exerting its anti-apoptotic function. However, XIAP knockout mice exhibit normal development and have no obvious apoptosis-related phenotype, indicating that the anti-apoptotic function of XIAP may not be essential for its tumour-promoting effect (Harlin et al, 2001). Based on our findings, it is conceivable that inhibition of autophagy by XIAP may represent an important mechanism by which XIAP functions as an oncogenic molecule to promote tumorigenesis.
Materials and methods
Antibodies and reagents
The following reagents were purchased from the designated suppliers: Embelin (Santa Cruz), EBSS (Sigma), MG-132 (Sigma), Cycloheximide (Sigma), API-2 (Sigma), Etoposide (Sigma), Doxorubicin (Sigma), and Ubiquitination Kit (Boston Biochem). The antibodies against the following proteins/epitopes were purchased from the indicated sources: XIAP (E-2, Santa Cruz; H-202, Santa Cruz), p-XIAP (S87, Abcam), Mdm2 (Ab-1, Calbiochem; HDM2-323, Santa Cruz), p53 (DO-1 HRP, Santa Cruz; C-term, Abgent), LC3 (NB100, Novus; L7543, Sigma; #2775, Cell Signaling), Ub (U5379, Sigma), p-AKT (pS473, Epitomics), Actin (#4967, Cell Signaling), GAPDH (A-3, Santa Cruz), PARP (5A5, Santa Cruz), HA (HA-7, Sigma; Y-11, Santa Cruz), Flag (M2, Sigma), GFP (JL-8, Clontech), His (27E8, Cell Signaling), AP/ HRP-conjugated secondary antibodies (Promega).
Cell culture and transfection
HCT116, p53−/−Mdm2−/− MEF, HEK293T cell lines were cultivated in Dulbecco’s modified Eagle’s medium (GIBCO) supplemented with 10% FBS and antibiotics (GIBCO) at 37°C under an atmosphere of 5% CO2 in air. IMR90, A549, LO2, HepG2, MCF10A, and MCF7 cell lines were maintained using standard culture conditions specified by ATCC. HCT116 cells stably transfected with GFP-LC3 were pooled after culturing for 2–3 weeks in medium containing 40 μg ml−1 G418. Transfection of HCT116 XIAP WT and XIAP KO cells and Mdm2−/−p53−/− MEF cells by lipofectamine 2000 (Invitrogen, USA) was performed according to manufacturer’s instruction, which gives ∼85% transfection efficiency.
RNAi
Transfection of cells with RNA oligos by Oligofectamine or Lipofectamine 2000 (Invitrogen, USA) was carried out according to manufacturer’s specification. XIAP siRNA sequence: 5′-GCTGAAACAGGACTACCAC-3′ (GenePharma), XIAP shRNA sequences: (1) 5′-CCGGAGCTGTAGATAGATGGCAATACTCGAGTATTGCCATCTATCTACAGCTTTTTT-3′; (2) 5′-CCGGGCACTCCAACTTCTAATCAAACTCGAGTTTGATTAGAAGTTGGAGTGCTTTTT-3′; (3) 5′-CCGGCAGAATGGTCAGTACAAAGTTCTCGAGAACTTTGTACTGACCATTCTGTTTTT-3′ (Sigma). Lentivirus-mediated knockdown system (pLKO.1, VSV-G, Gag and Rev) and overexpression system (PWPI, psPAX2, and pMD2.G) were performed as described elsewhere. Stable knockdown or overexpression transfectants was selected in medium containing puromycin (1 μg ml−1) and pooled after culturing for 1–2 weeks. Cells were then cultured in growth medium for further analysis.
Autophagy tests
Autophagy was measured by light microscopic quantification of cells transfected with GFP-LC3 as Furuya described or by western blot analysis of the LC3II level. For most experiments, the percentage of cells with GFP-LC3 punctuation was determined under fluorescence microscopy (Olympus DP71X). Cells presenting a mostly diffuse distribution of GFP-LC3 in the cytoplasm and nucleus were considered as non-autophagic, whereas cells representing intense GFP-LC3 aggregates in the cytosol but not in the nucleus were classified as autophagic. After serum starvation in EBSS for 2–8 h, cells were fixed in methanol and the GFP-LC3-positive cells were counted in four random areas. Levels of LC3II, which directly correlate with autophagosome numbers, were detected by western blotting and densitometric analysis relative to actin.
Electron microscopy
For ultrastructural analysis by EM, HCT116 XIAP WT or HCT116 XIAP KO cells were harvested and washed twice with PBS (pH 7.4). Cells were then fixed in 3% glutaraldehyde in 0.1 M MOPS buffer (pH 7.0) for 8 h at RT, in 3% glutaraldehyde/1% paraformaldehyde in 0.1 M MOPS buffer (pH 7.0) for 16 h at 4°C, and in 1% osmium tetroxide for 1 h at RT. The cells were embedded in Spurr’s resin at RT for 4–6 h and then polymerized at 60°C for 2 days. The blocks were cut into micrometre sections with a diamond knife, picked up on 200 mesh grids, stained and observed according to the standard electron microscopy procedures. At least 100 cells from randomly chosen electron microscopy fields were analysed for quantification of morphological features.
Cell fractionation
After treatment, cells were harvested and the cytoplasmic and nuclear protein fractions were separated using Beyotime Nuclear and Cytoplasmic Protein Extraction Kit according to manufacturer’s recommended procedures.
Immunoprecipitation
Cells were resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 0.5% NP-40, 0.5% deoxycholate, 100 mM NaF, 1 mM PMSF, 1 mM DTT, 10% glycerol and complete protease inhibitors Cocktail; Roche) on ice for 1 h, and precleared with protein A/G-coupled Sepharose beads for 2 h, then centrifugated at 12 000 g for 15 min. The supernatants were incubated with beads coated with the indicated antibodies for at least 4 h or overnight at 4 °C. Beads were washed three times with lysis buffer and boiled in 2 × loading buffer.
Western blot analysis
Protein samples were boiled in 2 × loading buffer, resolved by SDS–PAGE and transferred onto nitrocellulose membrane. Membranes were then blocked in 5% skim milk and incubated with the indicated antibodies in TBST. Immunolabelling was developed with Lumi-Phos WB Chemiluminescent Substrate (Thermo Pierce) or ECL Western Blotting Detection Reagent (GE Amersham). Visualized images were obtained using ImageQuant LAS-4000 mini (GE Fujifilm). During capture of the images, the nearly identical standard setting for brightness and contrast was used.
Ubiquitylation assay
In vivo ubiquitination assay was performed in the denaturing conditions as previous described (Tang et al, 2006). Briefly, cells were treated with MG-132 for 4–6 h before harvesting to allow for the accumulation of protein that are degraded in the cytoplasm and were then lysed in SDS lysis buffer (50 mM Tris pH 6.8, 1% SDS, 10% glycerol, 1 mM Na3VO4, 1 mM NaF and protease inhibitors). After boiling for 5 min, whole-cell lysates were diluted 10 times with cold lysis buffer supplemented with 1 × complete inhibitor and 10 mM NEM (Sigma). The diluted lysates were immunoprecipitated with the indicated antibodies and probed with anti-ubiquitin antibody. Alternatively, the diluted lysates were incubated with N2+-NTA beads to pulldown proteins conjugated to His-ubiquitin. Beads were then analysed by western blotting with anti-Mdm2 or anti-p53 antibody.
In vitro ubiquitination assay: Flag-tagged Mdm2 and XIAP were generated in 293T cells and purified using M2 beads as previously described (Tang et al, 2006). The non-specific binding proteins were removed by sequential washes with lysis buffers containing 0.25, 0.5, and 1 M KCl. Recombinant proteins were eluted using Flag peptide, followed by mixing with 100 nM E1, 2 μM E2 and 200 μM Ub (Boston Biochem) in a final volume of 20 μl reaction buffer (50 mM Hepes pH 8.0, 100 mM NaCl, 10 mM Mg2+-ATP, 0.5 mM DTT). The reaction was carried out at 37°C for 1 h and products were analysed by immunoblotting with anti-Mdm2 antibody.
Tumorigenecity assays
HCT116 XIAP−/− cells (KO), XIAP+/+ cells (WT) or HCT116 XIAP−/− cells stably expressing XIAP WT, XIAP S87A, XIAP S87D or XIAP 148A/D310A (1 × 106) were subcutaneously injected into the dorsal flanks of 4-week-old male athymic nude mice (Shanghai SLAC Laboratory Animal Co. Ltd.). After 5 weeks, mice were photographed and tumours were excised and weighed. The tumours were homogenized and proteins were extracted for western blot analysis. A total of 24 athymic nude mice were used and all animal experiments were conducted strictly in accordance with the Guide for the local Animal Care and Use Committee.
Human specimens
Total samples consisted of 42 cases of primary cancer tissues (oesophagus, colon, breast, lung, and stomach) together with the matched adjacent non-cancerous tissues were collected from patients who underwent surgery at the Affiliated Hospital of Anhui Medical University (Hefei, China) between 2010 and 2012. The pathohistological diagnosis of the specimens at the Department of Pathology was based on the WHO guidelines. Proteins extracted from these tissues were used for immunoblotting. The protocol for the use of tissue samples from patients and follow-up study was approved by our Institutional Review Board, and every patient had signed a consent form.
Immunohistochemistry
Immunohistochemical analysis of LC3, XIAP, p53, and Mdm2 protein expression was performed on formalin-fixed paraffin-embedded tissue sections (3 μm thick) with polyclonal antibodies against LC3 (1:100), XIAP (1:25), p53 (1:100), and Mdm2 (1:100) by the two-step method (Maixin Biotechnologies, Fuzhou, China) as previously described (Wu et al, 2011).
Statistical analyses
Statistical analysis was carried out using Microsoft Excel software and GraphPad Prism to assess the differences between experimental groups. Statistical significance was analysed by Student’s t-test and expressed as a P-value. P-values lower than 0.05 was considered to indicate statistical significance.
Supplementary Material
Acknowledgments
We thank Professor David Vaux for XIAP wild-type and XIAP KO mouse embryonic fibroblasts, Prof. Bert Vogelstein for HCT116 XIAP WT/KO cell lines, Professor Li Yu for GFP-LC3 (Human) construct, and Professor Tao Zhu for breast cancer cell lines. This work was supported by grants from the Ministry of Science and Technology of China (2010CB912804 and 2011CB966302); National Natural Science Foundation of China 31030046 (to MW) and 31171316 (to YM); and Chinese Academy of Sciences (XDA01020104).
Author contributions: XH planned and performed most of the experiments and helped to write the manuscript. ZW provided the human samples. YM and MW coordinated the study, oversaw the results and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B (1999) Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 59: 59–65 [DOI] [PubMed] [Google Scholar]
- Birnbaum MJ, Clem RJ, Miller LK (1994) An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol 68: 2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM (2006) DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126: 121–134 [DOI] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK (1993) An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol 67: 2168–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, Rieser E, Rickard JA, Bankovacki A, Peschel C, Ruland J, Bekker-Jensen S, Mailand N, Kaufmann T, Strasser A, Walczak H, Silke J, Jost PJ, Gyrd-Hansen M (2012) The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell 46: 746–758 [DOI] [PubMed] [Google Scholar]
- Dan HC, Sun M, Kaneko S, Feldman RI, Nicosia SV, Wang HG, Tsang BK, Cheng JQ (2004) Akt phosphorylation and stabilization of X-linked inhibitor of apoptosis protein (XIAP). J Biol Chem 279: 5405–5412 [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC (1997) X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388: 300–304 [DOI] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL (2006) Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep 7: 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M (2009) Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell 15: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P (2010) IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer 10: 561–574 [DOI] [PubMed] [Google Scholar]
- Hanson AJ, Wallace HA, Freeman TJ, Beauchamp RD, Lee LA, Lee E (2012) XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol Cell 45: 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB (2001) Characterization of XIAP-deficient mice. Mol Cell Biol 21: 3604–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, Stommel JM, Gygi S, Lahav G, Asara J, Xiao ZX, Kaelin WG Jr, Harper JW, Wei W (2010) Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell 18: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y (2007) Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12: 355–366 [DOI] [PubMed] [Google Scholar]
- Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, Ahn CH, Yoo NJ, Lee SH (2009) Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol 217: 702–706 [DOI] [PubMed] [Google Scholar]
- Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ (2008) Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol 39: 1059–1063 [DOI] [PubMed] [Google Scholar]
- Kimmelman AC (2011) The dynamic nature of autophagy in cancer. Genes Dev 25: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477 [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, Lacroix M, Le Cam L, Coux O, Benkirane M (2007) Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol 9: 331–338 [DOI] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda JE, MacKenzie A, Korneluk RG (1996) Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature 379: 349–353 [DOI] [PubMed] [Google Scholar]
- Lu M, Lin SC, Huang Y, Kang YJ, Rich R, Lo YC, Myszka D, Han J, Wu H (2007) XIAP induces NF-kappaB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell 26: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB (2005) Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol 6: 439–448 [DOI] [PubMed] [Google Scholar]
- Mah LY, Ryan KM (2012) Autophagy and cancer. Cold Spring Harb Perspect Biol 4: a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G (2010) Autophagy regulation by p53. Curr Opin Cell Biol 22: 181–185 [DOI] [PubMed] [Google Scholar]
- Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C (2007) Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem 282: 18573–18583 [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E (2007) Role of autophagy in cancer. Nat Rev Cancer 7: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovska-Coleska Z, Xu L, Hu Z, Tomita Y, Li P, Roller PP, Wang R, Fang X, Guo R, Zhang M, Lippman ME, Yang D, Wang S (2004) Discovery of embelin as a cell-permeable, small-molecular weight inhibitor of XIAP through structure-based computational screening of a traditional herbal medicine three-dimensional structure database. J Med Chem 47: 2430–2440 [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112: 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R, Fuchs EJ, Jain A, Pham V, Yoshimura K, Prouser T, Jalla S, Zhou X, Garrett-Mayer E, Kaufmann SH, Schulick RD, Pardoll DM, Bedi A (2006) Resistance of cancers to immunologic cytotoxicity and adoptive immunotherapy via X-linked inhibitor of apoptosis protein expression and coexisting defects in mitochondrial death signaling. Cancer Res 66: 1730–1739 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G (2011) Autophagy and aging. Cell 146: 682–695 [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS (2002) IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol 3: 401–410 [DOI] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H (2008) Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev 22: 2256–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Shi Y (2004) Caspases, IAPs and Smac/DIABLO: mechanisms from structural biology. Trends Biochem Sci 29: 486–494 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG (2007) Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 9: 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I, Kornblau SM, Segall H, Krajewski S, Welsh K, Kitada S, Scudiero DA, Tudor G, Qui YH, Monks A, Andreeff M, Reed JC (2000) Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res 6: 1796–1803 [PubMed] [Google Scholar]
- Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, Yang X (2006) Critical role for Daxx in regulating Mdm2. Nat Cell Biol 8: 855–862 [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G et al. (2008) Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Silke J (2005) IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol 6: 287–297 [DOI] [PubMed] [Google Scholar]
- Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, Tergaonkar V (2010) ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell 40: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZS, Yang K, Wan Y, Qian PX, Perry JK, Chiesa J, Mertani HC, Zhu T, Lobie PE (2011) Tumor expression of human growth hormone and human prolactin predict a worse survival outcome in patients with mammary or endometrial carcinoma. J Clin Endocrinol Metab 96: E1619–E1629 [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD (2000) Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288: 874–877 [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA 100: 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.