Abstract
Squamous cell carcinomas (SCCs) are highly heterogeneous tumours, resulting from deranged expression of genes involved in squamous cell differentiation. Here we report that microRNA-34a (miR-34a) functions as a novel node in the squamous cell differentiation network, with SIRT6 as a critical target. miR-34a expression increases with keratinocyte differentiation, while it is suppressed in skin and oral SCCs, SCC cell lines, and aberrantly differentiating primary human keratinocytes (HKCs). Expression of this miRNA is restored in SCC cells, in parallel with differentiation, by reversion of genomic DNA methylation or wild-type p53 expression. In normal HKCs, the pro-differentiation effects of increased p53 activity or UVB exposure are miR-34a-dependent, and increased miR-34a levels are sufficient to induce differentiation of these cells both in vitro and in vivo. SIRT6, a sirtuin family member not previously connected with miR-34a function, is a direct target of this miRNA in HKCs, and SIRT6 down-modulation is sufficient to reproduce the miR-34a pro-differentiation effects. The findings are of likely biological significance, as SIRT6 is oppositely expressed to miR-34a in normal keratinocytes and keratinocyte-derived tumours.
Keywords: actinic keratosis, miR-34a, p53, SIRT6, squamous cell carcinoma
Introduction
Squamous cell carcinoma (SCC) is the most frequent type of solid human tumours and a main cause of cancer-related death, occurring in many internal organs as well as skin. A network of squamous cell differentiation connected genes has emerged that is either deregulated or mutated in SCC, and plays a demonstrated or likely driver role (Agrawal et al, 2011; Stransky et al, 2011; Wang et al, 2011b). The skin provides an excellent model system to understand integrated control of squamous cell differentiation under normal conditions, in response to external insults and in tumourigenesis (Lefort and Dotto, 2004). Complex biological systems such as the skin appear to be intrinsically ‘robust’, that is, organized as scale-free networks, with most connections being made to critical ‘hubs’ (or signalling centres). Several ‘hubs’ that integrate and maintain skin homeostasis have been identified, including the transcriptional regulators p53 and Notch1 (Dotto, 2009).
A main role of p53 is that of a sensor of acute or chronic alterations in normal cellular physiology and, more specifically, DNA and chromosomal integrity (Riley et al, 2008; Olivier et al, 2009). However, while initial analysis of p53 knockout mice suggested that this gene is dispensable for development (Donehower et al, 1992), subsequent studies pointed to its involvement in differentiation of several tissues (Dotto, 2009; Olivier et al, 2009). Importantly, p53 has also been implicated as a negative regulator of stem cell potential with clear implications for cancer development (Jerry et al, 2008; Zhang et al, 2008; Zheng et al, 2008).
In keratinocytes of the proliferative compartment, increased p53 activity caused by UVB exposure or inhibition of EGFR signalling triggers a pro-differentiation transcriptional response connected with increased Notch1 expression and activity (Kolev et al, 2008; Mandinova et al, 2008; Guinea-Viniegra et al, 2012). Down-regulation of Notch1 transcription in keratinocyte-derived tumours can also be explained, at least in part, by compromised p53 function: in primary human keratinocytes, endogenous p53 binds to the Notch1 promoter and p53 knockdown results in down-modulation of Notch1 expression, whereas increased p53 levels lead to Notch1 upregulation (Lefort et al, 2007; Yugawa et al, 2007; Mandinova et al, 2008).
In concert with protein-encoding genes, microRNAs (miRNAs) play a key role in integrating multiple signalling inputs and coordinating the overall gene expression response of cells and tissues. miRNAs are often expressed in a lineage- and time-specific fashion and can control cell fate decisions as well as tumour development (reviewed in Lee and Dutta, 2009). In the skin, miRNAs appear to play an important role in control of epidermal and hair follicle development and function (Andl et al, 2006; Yi et al, 2006; Lena et al, 2008; Zhang et al, 2011). Keratinocyte-specific deletion of the Dicer, DGCR8, or Ago 1 and 2 genes, encoding essential miRNA processing enzymes, results in severely altered hair follicle morphogenesis and in their degeneration (Yi et al, 2006, 2009; Teta et al, 2012; Wang et al, 2012). miRNAs have also been implicated in epidermal homeostasis (Botchkareva, 2012; Rivetti di Val Cervo et al, 2012), wound healing (Banerjee et al, 2011; Pastar et al, 2012), various skin disorders (Bostjancic and Glavac, 2008; Schneider, 2012), epidermal cell cycle control and carcinogenesis (Antonini et al, 2010; Dziunycz et al, 2010; Darido et al, 2011; Sand et al, 2012; Xu et al, 2012).
miR-34 family members (a, b, and c) are among the most highly studied miRNAs (Hermeking, 2010). They are best understood as mediators of p53 action on growth arrest, senescence, and apoptosis (Hermeking, 2007; Raver-Shapira et al, 2007), as well as inhibition of epithelial–mesenchymal transition (EMT) (Siemens et al, 2011). miR-34a is the most prevalent form, except in lung and testis where miR-34b and miR-34c are more abundant (Bouhallier et al, 2010; Hermeking, 2010). Expression of miR-34a is down-modulated in a variety of cancers, including melanoma, prostate, pancreatic, colorectal, and non-small-cell lung cancers and neuroblastoma (Hermeking, 2010). miR-34a maps to the 1p36 genomic region that is frequently deleted in human cancers. In cancers where this region is intact, increased methylation of the miR-34a promoter region has been found (Hermeking, 2010).
Besides its role as downstream mediator of p53, miR-34a can enhance p53 activity through a mechanism involving decreased deacetylation via down-modulation of SIRT1 expression (Yamakuchi and Lowenstein, 2009). The sirtuin family comprises seven members, SIRT1 to −7. They are NAD+-dependent protein deacetylases and/or mono-[ADP-ribosyl] transferases. These proteins diverge in localization and functions, with SIRT1, −2, −6, and −7 acting as critical modulators of epigenetic modifications, while others, SIRT3, −4 and −5, functioning mostly in the mitochondria. Through one or more of these mechanisms, sirtuins are emerging as key players in development, cell differentiation, and ageing (Bosch-Presegue and Vaquero, 2011; Carafa et al, 2012).
Here, as part of a study of miRNAs that are aberrantly deregulated in keratinocyte-derived cancer, we have uncovered miR-34a as a novel node in the squamous cell differentiation network, with SIRT6 as critical target.
Results
miR-34a expression is suppressed in skin and oral SCCs and in keratinocytes with a compromised differentiation programme
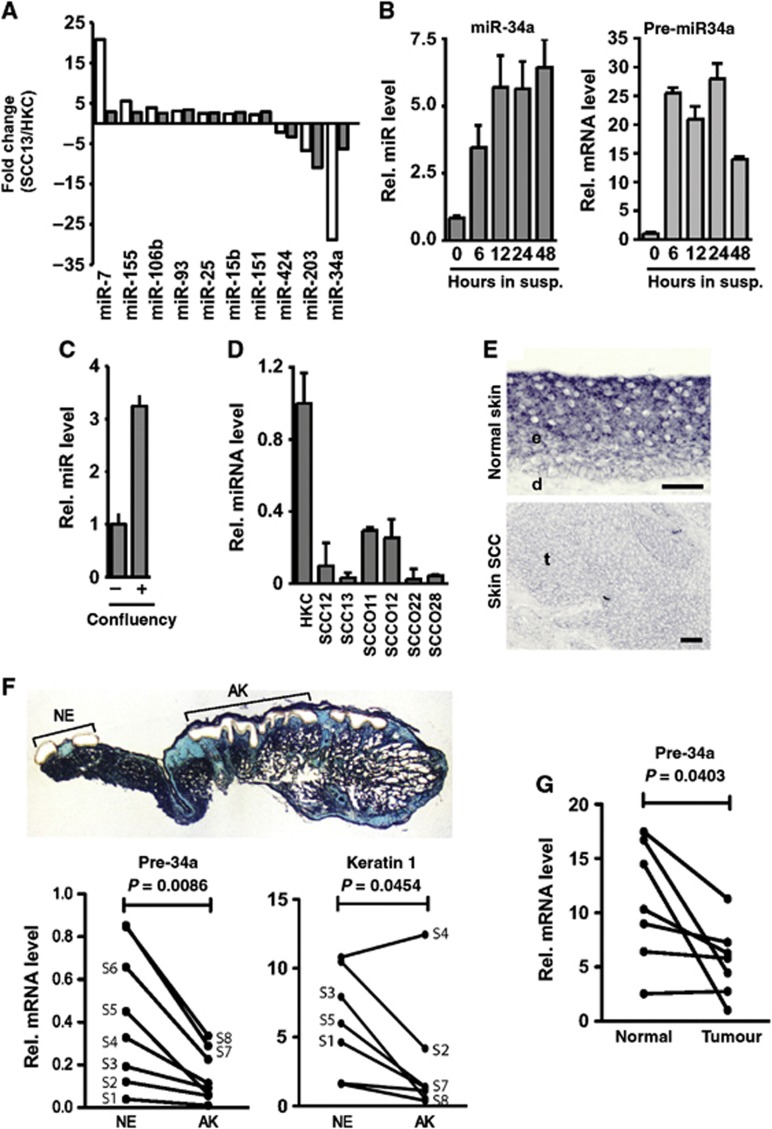
Keratinocyte differentiation and tumour development are inversely related events. We hypothesized that specific miRNAs participate in keratinocyte tumour suppression through a mechanism linked with differentiation. As an initial test of this possibility, the total pattern of microRNA expression, as assessed by microarray hybridization, was compared in exponentially growing primary human keratinocytes (HKCs) versus the keratinocyte-derived SCC13 cell line (Rheinwald and Beckett, 1980) using two different platforms (LC Sciences and Agilent Technologies). A number of miRNAs showed opposite expression in the two cell types, with miR-34a and miR-203 being the most significantly downregulated ones in the SCC cells (Figure 1A). While the role of miR-203 in keratinocyte differentiation is well established (Lena et al, 2008; Yi et al, 2008), a role of miR-34a has been only studied in the context of the cell cycle (Antonini et al, 2010). Of the three isoforms (miR-34a, b, and c), miR-34a is the one mainly expressed in HKCs, while miR-34b and c are present only at low levels (Supplementary Figure S1A). qRT–PCR analysis showed strong upregulation of miR-34a but not miR-34b and c in differentiating primary human keratinocytes (HKCs) (Figure 1B and C and Supplementary Figure S1B), and significant down-modulation in several keratinocyte-derived SCC cell lines (Figure 1D). Consistent with these results, in situ hybridization revealed more pronounced expression of miR-34a in the suprabasal than basal layers of normal epidermis and drastic suppression in a skin SCC of the same patient (Figure 1E). Several human skin samples of in situ squamous cell carcinoma lesions (actinic keratoses) excised together with flanking normal epidermis were utilized for laser capture microdissection (LCM) followed by qRT–PCR analysis. As shown in Figure 1F, miR-34a expression was found to be significantly downregulated in the neoplastic versus normal epidermis areas (P-value=0.0086), in parallel with down-modulation of the keratin 1 differentiation marker in all samples except one (Figure 1F). MiR-34a expression was also reduced in a set of oral SCCs versus normal mucosa from the same patients (Figure 1G).
Figure 1.
miR-34a expression is downregulated in keratinocyte-derived SCC cell lines and tumours, while it is induced with differentiation. (A) Differential miRNA expression in human skin SCC13 cells versus normal human primary keratinocytes (HKCs) was analysed using both the LC Sciences (white bars) and Agilent Technologies miRNA microarray platforms (grey bars). Results are expressed as fold change of miRNA levels in SCC13 cells versus HKCs. Shown are only miRNAs with fold change >2 or <−2 and P-values<0.01 in both platforms. (B, C) HKCs were induced to differentiate either by culture in suspension (susp.) for the indicated time periods (B) or at confluency for 7 days (C). Levels of mature (miR-34a) were measured by qRT–PCR using specific Taqman probes using the snRNA Z30 for normalization and levels of precursor miR-34a (pre-miR-34a) were measured by conventional qRT–PCR using 36β4 for normalization. (D) Mature miR-34a expression was assessed in HKCs and in different skin- (SCC12 and SCC13) and oral mucosa- (SCCO11, SCCO12, SCCO22, and SCCO28) derived SCC cell lines by qRT–PCR as previously described (Raymond et al, 2005) with 5S RNA for normalization. (E) Human skin (upper panels) as well as skin SCC (lower panels) from the same patient were probed with double digoxigenin (DIG)-miR-34a probes for in situ hybridization. Bars, 50 μm. (e refers to epidermis, d to dermis, and t to tumour). (F) Excised skin samples (S1-S8), containing, in each case, a field of normal epidermis (NE) well separated from one with actinic keratosis (AK) lesions were utilized for laser capture microdissection (LCM) of the AK epithelium versus epidermis further away (as shown for one of the cases—S7, by the histological image after LCM), followed by qRT–PCR analysis of precursor miR-34a and keratin 1 marker expression, using β-actin for normalization. Values are shown as matched pairs of AK versus normal epidermis for each of the samples. Statistical significance of the differences between the AK versus normal epidermis groups was calculated by Student’s paired t-test. (G) Levels of precursor miR-34a (pre-miR-34a) were measured by qRT–PCR in oral SCC versus matched normal oral mucosa from the same patient with 36β4 for normalization. Statistical significance of the differences between tumour and oral mucosa values was calculated by Student’s paired t-test.
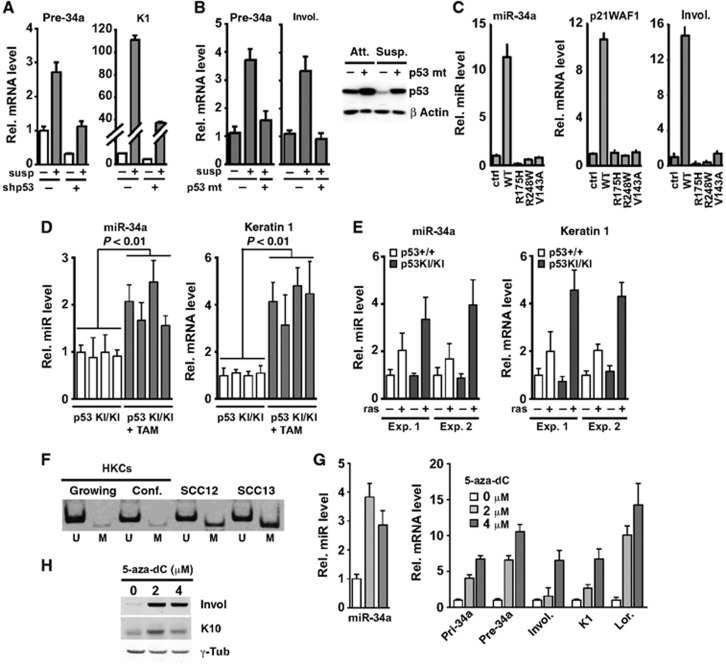
Loss or mutations of p53 is an important mechanism underlying altered differentiation of keratinocyte-derived cancer cells (Lefort et al, 2007; Kolev et al, 2008). In HKCs with either p53 knockdown or expression of a mutant p53 protein, differentiation-induced levels of miR-34a and differentiation markers keratin 1 (K1) and involucrin were concomitantly reduced (Figure 2A and B). Even in the absence of any exogenous manipulations, there is a basal level of p53 activity as well as spontaneously occurring differentiation in keratinocyte cultures (Mandinova et al, 2008), and, even under these conditions, knockdown of p53 resulted in a parallel down-modulation of differentiation markers and miR-34a expression (Figure 2A and Supplementary Figure S2A). Conversely, miR-34a and involucrin as well as p21WAF1/Cip1 were induced in SCC cells upon expression of wild-type p53, while no such effects were elicited by expression of different p53 mutant forms (Figure 2C). Similar results were obtained by analysis of a mouse genetic model for p53 function in skin carcinogenesis. The model is based on mice with a homozygous knock-in replacement of the p53 gene with a gene encoding a p53ER-TAM fusion protein, whose activity can be induced by treatment with tamoxifen (Christophorou et al, 2005; Guinea-Viniegra et al, 2012). In chemically-induced papillomas of these mice with silent p53, restoration of p53 activity by tamoxifen treatment resulted in a concomitant induction of K1 differentiation marker expression and miR-34a levels (Figure 2D). Parallel induction of K1 and miR-34a was also observed in mouse keratinocytes with wild-type p53 or a tamoxifen-induced p53ER-TAM knock-in gene upon oncogenic H-rasV12 expression (Figure 2E).
Figure 2.
Control of miR-34a expression in keratinocyte differentiation and SCC cells. (A) HKCs stably transduced with a shRNA retrovirus against p53 (+) or empty vector control (−) were kept under proliferating conditions (−) or induced to differentiate in suspension (+) for 24 h. Levels of the indicated transcripts were measured by qRT–PCR with 36β4 for normalization. (B) HKCs stably transduced with a lentiviral vector for the doxycycline-inducible expression of p53 mutant R248W (p53 mt) were either untreated (−) or treated with doxycycline (+) for 5 days. Cells were kept under proliferating conditions (−) or induced to differentiate in suspension (+) for 6 h. Left panel: Levels of the indicated transcripts were measured by qRT–PCR with 36β4 for normalization. Right panel: Parallel cultures were examined by immunoblotting for p53 protein expression. (C) SCC13 cells were transduced for 5 days with retroviruses expressing either p53 wild type (WT), three different p53 mutants harbouring the indicated single amino-acid substitutions (R175H, R248W, and V143A) or empty vector control (ctrl). Levels of mature miR-34a were measured by qRT–PCR as previously described in (Raymond et al, 2005) with 5S RNA for normalization. Levels of p21WAF1/CIP1 and involucrin were measured as in A. (D) p53KI/KI mice with chemically-induced papillomas were treated for 15 days with tamoxifen or vehicle (black and white bars respectively) as reported (Guinea-Viniegra et al, 2012). Levels of mature miR-34a were measured in papillomas from four mice per group by qRT–PCR with Taqman probes and U6 RNA for normalization, with parallel assessment of keratin 1 mRNA levels. (E) Cultured primary keratinocytes from wild-type (p53+/+) and p53KI/KI (p53KI/KI) mice were transduced for 6 days with a H-RasV12 expressing retrovirus (+) or empty vector control (−), followed by treatment with 1 μM 4-OH tamoxifen for 48 h as previously described (Guinea-Viniegra et al, 2012). Levels of mature miR-34a were measured by qRT–PCR, with 5S RNA for normalization, in parallel with keratin 1 mRNA. (F) DNA was extracted from the indicated cells and subjected to bisulphite conversion. Methylation of miR-34a was assayed by PCR using primers specific for unmethylated (U) and methylated (M) miR-34a promoter sequences (primer sequences are given in Supplementary Table 3). (G, H) SCC13 cells were treated with the indicated concentrations (μM) of 5-aza-2′deoxycytidine (5-aza-dC) for 4 days. Levels of mature (miR-34a), primary (pri-34a), and precursor (pre-34a) miR-34a as well as indicated gene expression were determined by qRT–PCR (G) and immunoblotting for Keratin 10 and involucrin (with γ-tubulin for equal loading control) (H).
Source data for this figure is available on the online supplementary information page.
It has been previously shown that, in mouse keratinocytes, under proliferative conditions, miR-34a expression is under negative control of p63 (Antonini et al, 2010). Given its frequent deregulation in SCCs (Perez and Pietenpol, 2007), we tested whether p63 was also involved in control of miR-34a expression in HKC differentiation and/or SCC cells. Consistent with the previous report (Antonini et al, 2010), sustained ΔNp63α expression via retroviral vector infection reduced miR-34a levels in HKCs under basal conditions, but did not prevent its induction with differentiation, and even slightly enhanced it (Supplementary Figure S2B–D). As a control, FGF21, a known p63 target (Vigano et al, 2006), was upregulated in p63 overexpressing HKCs under both growing and differentiating conditions (Supplementary Figure S2C). In converse experiments, p63 knockdown in human keratinocytes caused only a slight upregulation of miR-34a expression (in one experiment, with no upregulation in another) that could also be explained by a concomitant upregulation of p53 (Supplementary Figure S2E). This is at variance with the strong upregulation of miR-34a reported for mouse keratinocytes upon ΔNp63α knockdown (Antonini et al, 2010), possibly due to species-specific differences in regulation of this miRNA and/or different culture conditions. p63 knockdown had also no consistent effects on miR-34a expression in SCC cells (Supplementary Figure S2F).
Increased promoter DNA methylation could contribute to the low miR-34a expression in SCC cells, as it has been reported for other cancer types (Lodygin et al, 2008; Chim et al, 2010). Bisulphite DNA conversion followed by PCR analysis with methylated- versus unmethylated-specific primers revealed much greater methylation at the miR-34a promoter in SCC cells than either growing or differentiating HKCs (Figure 2F). Treatment of SCC cells with 5-aza-2′-deoxycytidine (5-aza-dC), a DNA methyltransferase inhibitor, resulted in the concomitant induction of miR-34a and differentiation marker expression (Figure 2G and H).
miR-34a has pro-differentiation functions
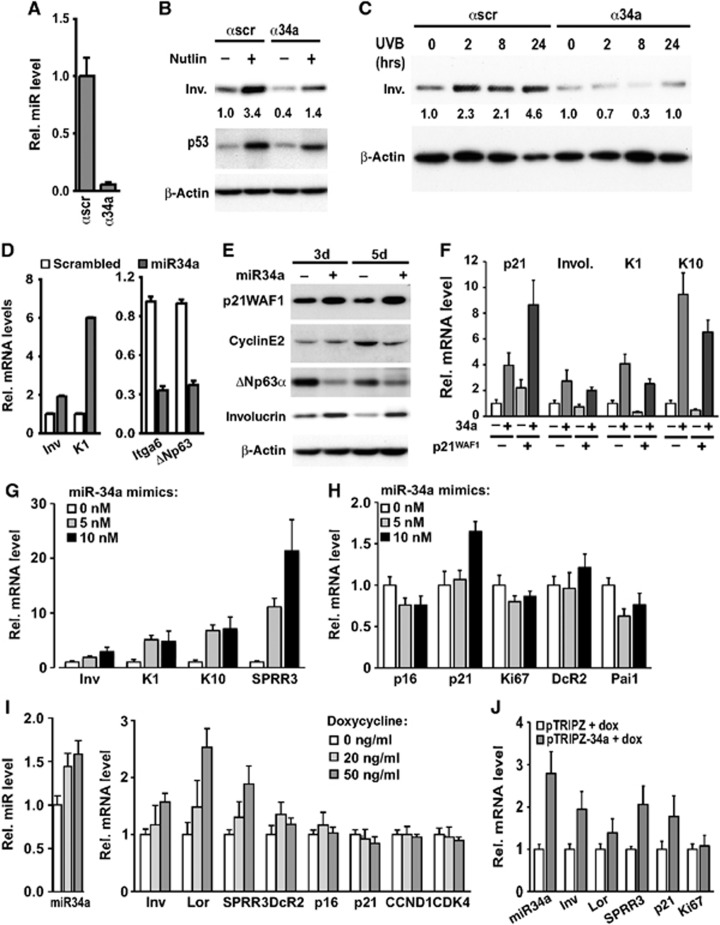
To assess whether the pro-differentiation effects of increased p53 activity are linked to miR-34a, HKCs were transfected with specific antagomiRs, causing >10-fold down-modulation of this miRNA (Figure 3A). As previously reported, for increased p53 activity in proliferating keratinocytes (Yugawa et al, 2007; Kolev et al, 2008; Guinea-Viniegra et al, 2012), nutlin-3a treatment resulted in increased involucrin differentiation marker expression, under conditions that resulted in little or no change in apoptosis (Supplementary Figure S2G) in accordance with a previous report on nutlin-3a-treated keratinocytes (Kranz and Dobbelstein, 2006). The nutlin-3a effects on differentiation were paralleled by increased miR-34a expression (Supplementary Figure S2H) and counteracted by antagomiR-mediated silencing of this miRNA (Figure 3B). Basal levels of differentiation marker expression were also suppressed by the miR-34a antagomiRs (Figure 3B). Similar results were observed upon UVB exposure of HKCs plus/minus miR-34a silencing (Figure 3C and Supplementary Figure S2I and J).
Figure 3.
miR-34a is a positive determinant of keratinocyte differentiation. (A) Proliferating HKCs were transfected for 3 days with miR-34a-specific antagomiRs (α34a) (50 nM) or control scrambled antagomiR (αscr) and levels of mature miR-34a were measured by qRT–PCR using specific Taqman probes. (B) HKCs were transfected as in A and, 24 h later, treated or not with nutlin-3a (10 μM) for additional 48 h. Involucrin and p53 protein levels were analysed by immunoblotting with β-actin as equal loading control. Numbers correspond to folds of induction of involucrin expression relative to untreated control, calculated after densitometric scanning of the autoradiographs and normalization for β-actin expression. (C) HKCs transfected with antagomiR against miR-34a (α34a) or control (αscr) as in A for a total of 3 days were UVB irradiated (50 mJ/cm2) at the indicated times from the end of the experiment, followed by immunoblot analysis for involucrin expression and data quantification by densitometric analysis as in the previous panel. Pattern of p53 protein expression is shown in Supplementary Figure S2J. (D) HKCs transfected for 3 days with miR-34a precursor (+) (25 nM) or scrambled control oligonucleotides (−) were examined for involucrin (inv), keratin 1 (K1), integrin α6 (itga6) and ΔNp63α expression by qRT–PCR with 36β4 for normalization. (E) HKCs at 3 and 5 days after transfection as in the previous panels were analysed by immunoblotting for expression of the indicated proteins. p21WAF1, cyclin E2 and β-actin blots were performed by sequential blotting of the same membrane without stripping, while ΔNp63α and involucrin expression was assessed by a parallel gel/blot with β-actin giving the same pattern of expression. (F) HKCs were stably infected with a lentivirus for doxycycline-inducible expression of p21WAF1/Cip1. Cells were either untreated (−) or treated with doxycycline (500 ng/ml) for 5 days (+) for sustained p21WAF1/Cip1 expression and associated cell cycle arrest. Cells were transfected with miR-34a mimics (25 nM) or scrambled controls for the last 3 days of the experiment. Expression of the indicated genes was assessed by qRT–PCR with 36β4 for normalization. (G, H) HKCs were transfected for 2 days with miR-34a precursor at low concentrations (5 and 10 nM) with scrambled controls used to keep equal total amounts of transfected oligonucleotides. Cells were examined for expression of the indicated differentiation marker (G) and cell cycle and senescence genes (H), by qRT–PCR with 36β4 for normalization. (I) HKCs were stably infected with a lentivirus for doxycycline-inducible expression of miR-34a. Cells were either untreated or treated with doxycycline at the indicated concentrations for 2 days followed by determination of mature miR-34a levels in parallel with expression of the indicated genes by qRT–PCR with 36β4 for normalization. (J) HKCs stably infected with a lentivirus for doxycycline-inducible expression of miR-34a (pTRIPZ-34a) or empty vector control (pTRIPZ) were treated with doxycycline (50 ng/ml) for 4 days, followed by measurement of mature miR-34a levels in parallel with expression of the indicated genes by qRT–PCR as in I.
Source data for this figure is available on the online supplementary information page.
To assess the more direct consequences of increased miR-34a expression, HKCs were transfected with miR-34a precursor oligonucleotides in parallel with scrambled controls. Transfection of miR-34a mimics at commonly used doses (25 nM) resulted in significant morphological changes associated with growth arrest and senescence rather than apoptosis (Supplementary Figure S3 A–D). miR-34a is part of a positive feedback mechanism that reinforces p53 activity through down-modulation of SIRT1 deacetylation (Yamakuchi and Lowenstein, 2009). Consistent with this, p53 protein levels were found to oscillate in keratinocytes with miR-34a over-expression (Supplementary Figure S3D), as expected from increased p53 activity causing destabilization of the p53 protein through induction of MDM2 (Wu et al, 1993).
In parallel, miR-34a overexpression caused increased expression of terminal differentiation markers like involucrin and keratin 1, and down-modulation of genes associated with the proliferative compartment, like the genes encoding integrin α6 and ΔNp63α (Figure 3D and E). To assess whether the induction of keratinocyte differentiation was a secondary consequence of cell cycle arrest and/or senescence, HKCs were infected with a lentiviral vector for inducible expression of p21WAF1/Cip1, another potent blocker of the cell cycle and inducer of cell senescence (Dotto, 2000). Consistent with previous studies (Di Cunto et al, 1998), increased p21WAF1/Cip1 expression did not induce but repressed keratinocyte differentiation markers, while expression of these genes was induced by increased miR-34a levels even in the growth-arrested p21WAF1/Cip1 overexpressing cells (Figure 3F and Supplementary Figure S4A). Similarly, treatment of HKCs with TGF-β1, a potent cell cycle inhibitor and inducer of p15INK4b and p21WAF1/Cip1 expression (Massague et al, 2000), did not induce but suppressed differentiation markers, which were induced by miR-34a even in TGF-β1-treated cells (Supplementary Figure S4B and C).
While comparable to those used in most other studies, the levels of exogenous miR-34a mimics transfected into HKCs may be excessive and have effects of little physiological significance. This is hard to assess by direct measurements of miR34a levels, as the vast majority of exogenously delivered miRNA mimics detected by qRT–PCR are not functional and sequestered in vesicles, while only a small free active pool exists (Thomson et al, 2013). Therefore, two alternative functional approaches were adopted. In the first, HKCs were transfected with substantially lower amounts of miR-34a mimics (5–10 nM, i.e., 1/10–1/4 of what is usually used in the literature). This was sufficient to induce differentiation marker expression while there were little or no effects on cell cycle- or senescence-related genes (Figure 3G and H), indicating that differentiation control is particularly sensitive to miR-34a modulation. In a second, more conclusive approach, HKCs were stably transduced with a lentiviral vector for doxycycline-inducible expression of the miR-34a DNA spanning region (Yang et al, 2012). In a dose–response experiment, minimal doxycycline concentrations (20–50 ng/ml) resulting in less than two-fold increase of mature miR-34a expression (comparable or even lower than that occurring in differentiation) were sufficient to induce expression of several differentiation markers while, as after transfection of low amounts of miR-34a mimics, expression of several cell cycle- or senescence-related genes was mostly unaffected (Figure 3I). Doxycycline treatment has the potential of exerting some significant biological effects on its own. Accordingly, similar experiments were repeated with keratinocytes infected with the miR-34a expressing lentivirus versus empty vector control, with doxycycline treatment causing selective induction of differentiation markers only in cells transduced with the miR-34a delivering lentivirus (Figure 3J).
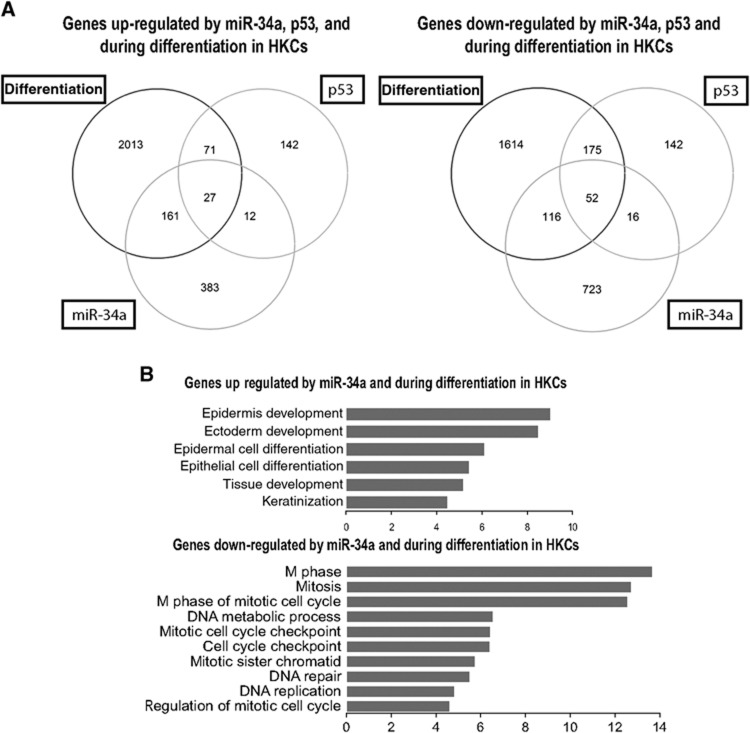
Further insights were sought by global gene expression profiling. Several hundred genes were significantly modulated by increased miR-34a expression in HKCs. Of these, 356 transcripts (188 up- and 168 downregulated) were similarly regulated in HKCs induced to differentiate (Figure 4A and Supplementary Tables 1 and 2). The genes upregulated in the two cases aggregated into six main gene ontology (GO) categories, with the most represented ones being epidermis/ectoderm development and epidermal/epithelial cell differentiation (Figure 4B, upper panel). The genes downregulated in the two conditions were classified into 10 categories with an over-representation of processes involved in M phase and mitosis (Figure 4B, lower panel). Further insights were provided by comparative analysis of HKCs with increased p53 activity by treatment with the MDM2 inhibitor nutlin-3a. This revealed a substantial set of genes commonly modulated in the three conditions. Importantly, however, the overlap was only partial. Many transcripts commonly regulated by miR-34a and differentiation, were not regulated by p53, pointing to a possible p53-independent role of miR-34a in differentiation (Figure 4A and Supplementary Tables 1 and 2).
Figure 4.
Distinct impact of p53 and miR-34a on keratinocyte gene expression. (A) Venn diagrams of up- and down-modulated genes (fold change >2 or <−2, P-value<0.01) in HKCs transfected for 3 days with miR-34a mimics (25 nM) versus scrambled control (miR-34a) or HKCs induced to differentiate by suspension culture for 24 h (differentiation) or HKCs treated with nutlin-3a for 30 h (p53) versus corresponding controls. Analysis was based on cDNA microarray hybridization using the Affymetrix HG-U133A 2.0 platform as described in Supplementary Materials and methods. Total number of exclusively up- and down-regulated genes (left and right panels, respectively) is indicated for each condition, as well as, in the intersections, number of similarly modulated genes. The list of each group of genes is given in Supplementary Tables 1 and 2. (B) Gene ontology analysis of genes similarly up- or down-regulated (upper and lower panels, respectively) in HKCs induced to differentiate and overexpress miR-34a. Biological processes with—log10 (P-value)≥4 are shown.
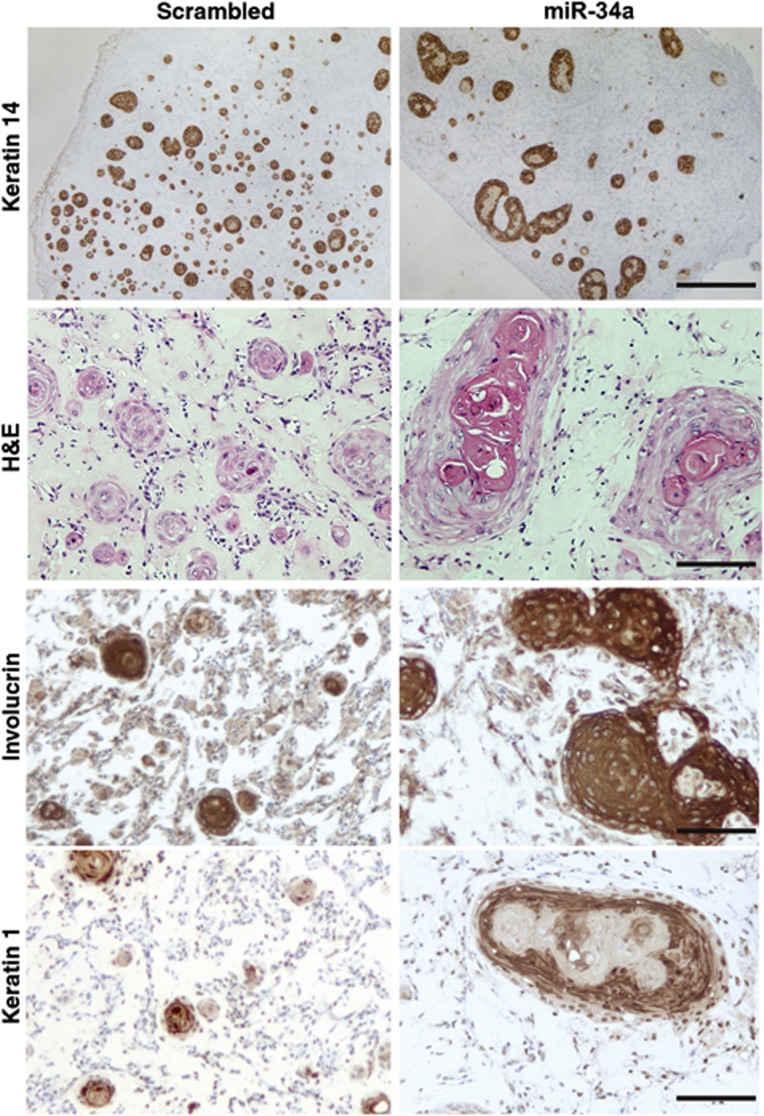
To assess whether miR-34a can play a pro-differentiation function in vivo, HKCs were transfected with precursor miR-34a oligonucleotides or scrambled controls followed by intradermal injections into mice. Relative to grafting onto the back of mice, intradermal injection assays provide a reliable method to assess in vivo growth/differentiation of keratinocytes in the absence of any inflammatory/wound healing reaction that can substantially modify behaviour of cells (Restivo et al, 2011). Intradermal injections of highly proliferating HKCs (maintained by low confluence culture conditions), resulted, within a week, in formation of a large number of small nests of cells with basal cell characteristics and little differentiation, as revealed by immunohistochemical analysis of the corresponding markers (Figure 5). By contrast, HKCs with elevated miR-34a expression formed a much lesser number of ‘basal cell nests’, giving rise preferentially to larger keratinized cysts intensely positive for suprabasal markers (Figure 5 and Supplementary Figure S5).
Figure 5.
Impact of miR-34a on keratinocyte differentiation in vivo. HKCs transfected with miR-34a precursor or scrambled oligonucleotides and kept under low density proliferating conditions were injected, 48 h later, at the dermal–epidermal junction of immunocompromised NOD/SCID mice. Nodules formed 8 days after injection were processed for immunohistochemistry with antibodies against the basal maker K14 for comprehensive detection of small islands of keratinocytes with little or no terminal differentiation markers, as well as more differentiated larger cysts. The latter were also detected by haematoxylin and eosin (H&E) staining and immunostaining with antibodies against suprabasal differentiation markers (involucrin and keratin 1). Bars, 900 μm for top panel and 180 μm for the others. Similar results are shown in Supplementary Figure S5.
miR-34a is a regulator of p53-induced Notch1 expression and activity
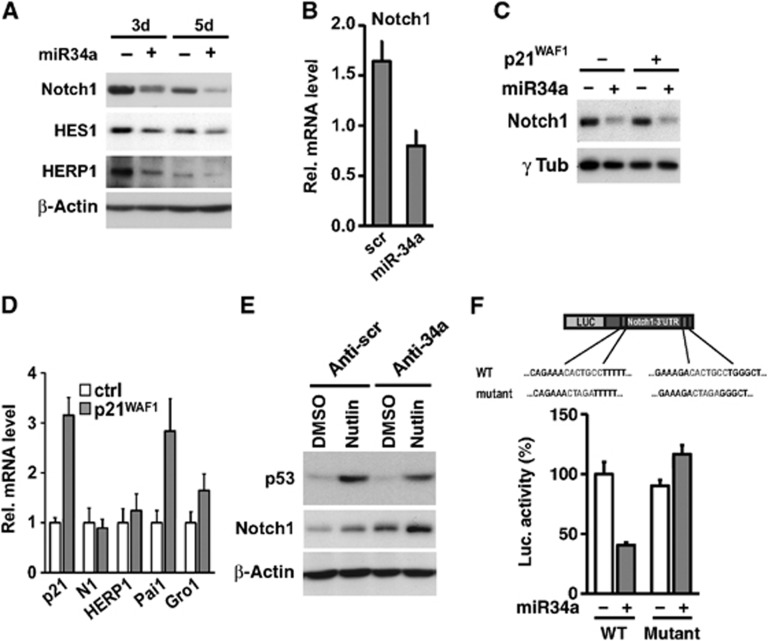
We previously showed that the Notch1 gene is a p53 target and mediator of its effects on differentiation (Lefort et al, 2007). Given that miR-34a is a p53 effector (Hermeking, 2010), we assessed whether Notch1 expression could be also regulated by miR-34a. To this end, we measured Notch1 mRNA and protein levels in HKC overexpressing miR-34a or control. Increased levels of miR-34a led to a sustained reduction of Notch1 expression, rather than an increase, as it would have been expected if the pro-differentiation function of miR-34a were linked to that of Notch1 (Figure 6A and B). No effect on Notch1 mRNA or protein levels was observed in growth-arrested p21WAF1/Cip1 overexpressing HKCs, while Notch1 expression was suppressed by miR-34a even in the p21WAF1/Cip1 overexpressing cells (Figure 6C and D). Interestingly, concomitant activation of p53 signalling (by nutlin-3a treatment) and miR-34a knockdown, led to an over accumulation of the Notch1 protein, suggesting that miR-34a is part of a negative feedback mechanism for control of Notch1 expression by p53 (Figure 6E).
Figure 6.
miR-34a is a modulator of Notch1 expression by p53. (A) HKCs were transfected with miR-34a precursor (+) or scrambled control oligonucleotides (−) for the indicated times followed by immunoblot analysis of the indicated proteins. HES1, HERP1, and β-actin blots were performed by sequential blotting of the same membrane without stripping, while Notch1 expression was assessed by a parallel gel/blot with β-actin giving the same pattern of expression. (B) HKCs transfected with the miR-34a precursor or scrambled control for 3 days were analysed for levels of Notch1 mRNA expression by qRT–PCR with 36β4 for normalization. (C, D) HKCs stably infected with a lentivirus for doxycycline-inducible expression of p21WAF1/Cip1 were either untreated (−) or treated with doxycycline (500 ng/ml) for 5 days (+). In (C), cells were transfected with miR-34a mimics (25 nM) or scrambled controls for the last 3 days of the experiment followed by immunoblot analysis of Notch1 expression. In (D), cells were analysed by qRT–PCR for the indicated genes. (E) HKCs were transfected for 3 days with antagomiR-34a (anti-34a) or negative (anti-scr) control plus/minus treatment with nutlin-3a (10 μM) for the last 30 h. Protein extracts were analysed by immunoblotting for expression of p53 and Notch1 with β-actin as equal loading control. Notch1 and β-actin blots were performed by sequential blotting of the same membrane without stripping, while p53 expression was assessed by a parallel gel/blot with β-actin giving the same pattern of expression. (F) HeLa cells were co-transfected in triplicate wells with a luciferase reporter construct with the wild type (WT) Notch1-3′UTR sequence or one containing the indicated nucleotide substitutions abrogating the two miR-34a recognition sequences (mutant) together with miR-34a precursor or scrambled control oligonucleotides. Luciferase activity was measured 48 h later, with total protein normalization.
Source data for this figure is available on the online supplementary information page.
Studies in other cellular systems have shown that Notch1 can be a direct miR-34a target (Chen and Hu, 2012; Bu et al, 2013). In agreement with these reports, we found that, in HKCs transfected with a luciferase reporter with the 3′-UTR of the Notch1 transcript, activity of the reporter was significantly reduced by co-transfection with miR-34a mimics. No such effects were observed with a luciferase reporter with the 3′-UTR of the Notch1 transcript devoid of its two miR-34a target sequences (Figure 6F).
SIRT6 as a miR-34a target and determinant of its impact on differentiation
The above findings indicated that the pro-differentiation function of miR-34a cannot be explained by modulation of Notch1 expression. To identify other miR-34a targets that may be involved in differentiation, we undertook a combined bioinformatics and functional approach. Gene transcripts commonly down-modulated in HKCs by increased miR-34a expression and differentiation (Supplementary Table 2), were examined for the presence, in their 3′UTR, of predicted miR-34a target sequences. Among the 10 genes identified by this approach (Supplementary Figure S6A), there were two well-known tyrosine kinase receptors, Axl and c-Met, as well as a transcription factor, FosL1, with a known or likely role in keratinocyte growth/differentiation control (Cortesina et al, 2000; Murai et al, 2004; Mangone et al, 2005; Green et al, 2006). qRT–PCR and immunoblot analysis showed that these genes were all effectively down-modulated in HKCs with increased miR-34a expression (Supplementary Figure S6B and C). To assess the functional significance of the findings, we tested whether down-modulation of one or more of these genes could reproduce, at least in part, the impact of miR-34a on differentiation. However, siRNA-mediated knockdown of the genes did not induce but, if anything, decreased differentiation marker expression (Supplementary Figure S6D).
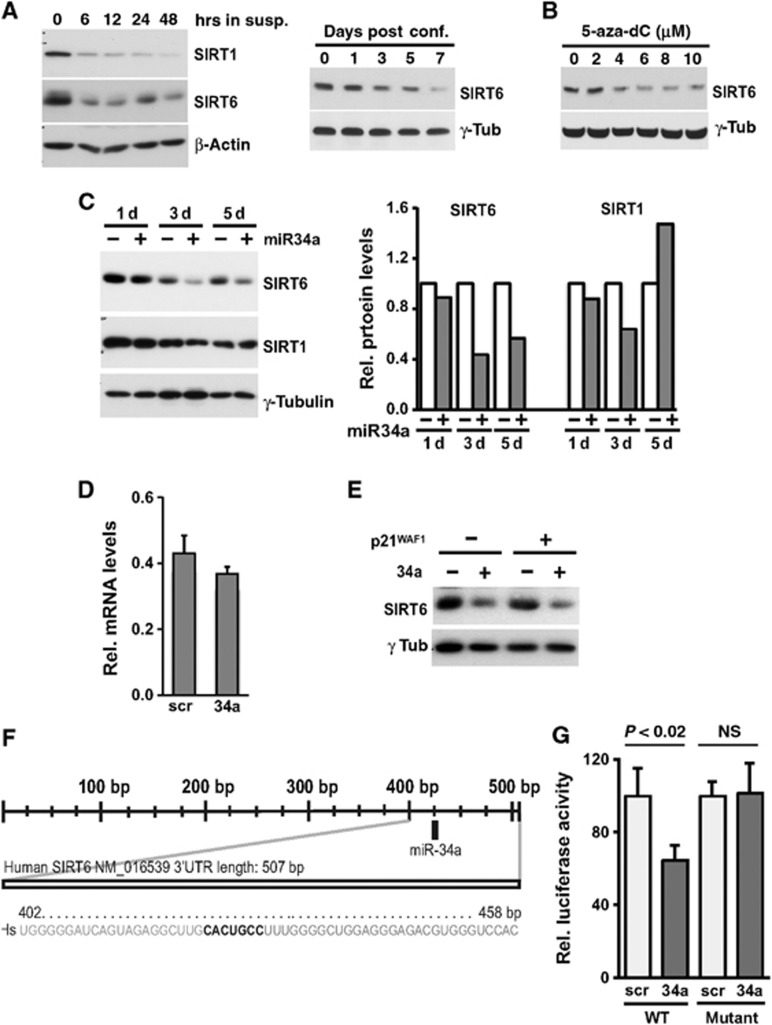
Epigenetic modifications play a key role in keratinocyte differentiation (Pickard et al, 2010; Sen et al, 2010). Sirtuins are NAD+-dependent protein deacetylases and/or ADP-ribosyl transferases, involved in a variety of cellular functions like chromatin regulation, development/differentiation, metabolism, and cell survival (Rajendran et al, 2011). Sirtuin 1 (SIRT1) is a well-known miR-34a target (Yamakuchi et al, 2008). Of the other Sirtuin family members, by Targetscan 5.2 and MiRanda sequence analysis we found that only SIRT6 harbours a miR-34a seed sequence in its 3′UTR. In differentiating HKCs, in which miR-34a is upregulated, SIRT1 and SIRT6 protein expressions were both suppressed (Figure 7A). Similar suppression of SIRT6 was observed in 5-aza-dC-treated SCC13 cells in which miR-34a expression was induced in parallel with differentiation (Figure 7B). More directly, in HKCs with increased miR-34a expression, there was little change of SIRT1 expression, while SIRT6 was significantly suppressed, at the protein but not RNA level (Figure 7C and D). No changes in SIRT6 expression were caused by increased expression of p21WAF1/Cip1, as an unrelated inducer of cell cycle arrest and senescence, while miR-34a caused down-modulation of SIRT6 even in the p21WAF1/Cip1 overexpressing cells (Figure 7E). To assess whether SIRT6 is a direct miR-34a target in these cells, we performed a heterologous reporter assay with a luciferase reporter gene inserted upstream of wild-type SIRT6 3′UTR or mutant UTR lacking the miR-34a recognition sequence. As shown in Figure 7F and G, luciferase activity of the wild-type SIRT6 3′UTR construct was reduced by concomitant miR-34a overexpression, while activity of the mutant 3′UTR construct was unaffected.
Figure 7.
SIRT6 is a miR-34a target downregulated during differentiation. (A) HKCs were induced to differentiate either by culture in suspension or high confluency conditions for the indicated times (left and right panels, respectively) and levels of SIRT1 and SIRT6 expression were determined by immunoblot analysis with β-actin and γ-tubulin, respectively, as equal loading controls. (B) SCC13 cells were treated with the indicated concentrations (μM) of 5-aza-2′-deoxycytidine for 4 days and levels of SIRT6 expression were determined by immunoblot analysis with γ-tubulin as equal loading control. (C) HKCs were transfected with miR-34a precursor (+) or scrambled control oligonucleotides (−) for the indicated times (days) followed by immunoblot analysis of the indicated proteins. SIRT1 and SIRT6 blots were performed on parallel gel/blot by sequential blotting of the same membrane without stripping with γ-tubulin giving the same pattern of expression (left panel). Relative protein levels were quantified after densitometric scanning of the autoradiographs and normalization for γ-tubulin expression (right panel). (D) HKCs were transfected for 3 days with miR-34a precursor (+) or scrambled control oligonucleotides (−) and analysed for levels of SIRT6 mRNA by qRT–PCR. (E) HKCs stably infected with a lentivirus for doxycycline-inducible expression of p21WAF1/Cip1 were either untreated (−) or treated with doxycycline (500 ng/ml) for 5 days (+). Cells were transfected with miR-34a mimics (25 nM) or scrambled controls for the last 3 days of the experiment followed by immunoblot analysis of SIRT6 expression with γ-tubulin as equal loading control. (F) The human SIRT6 3′-UTR was analysed for the presence of miR-34a recognition sites by Targetscan 5.2 and Miranda softwares. Shown in bold is the position of the miR-34a binding site in the human SIRT6 3′-UTR. Numbers refer to the nucleotide position starting from the stop codon of the SIRT6 coding region. (G) HKCs were co-transfected in triplicate wells with a luciferase reporter construct with the WT SIRT6-3′UTR sequence or one lacking the miR-34a binding site (mutant) together with miR-34a precursor or scrambled control oligonucleotides. Luciferase activity was measured 48 h later, with total protein levels for normalization.
Source data for this figure is available on the online supplementary information page.
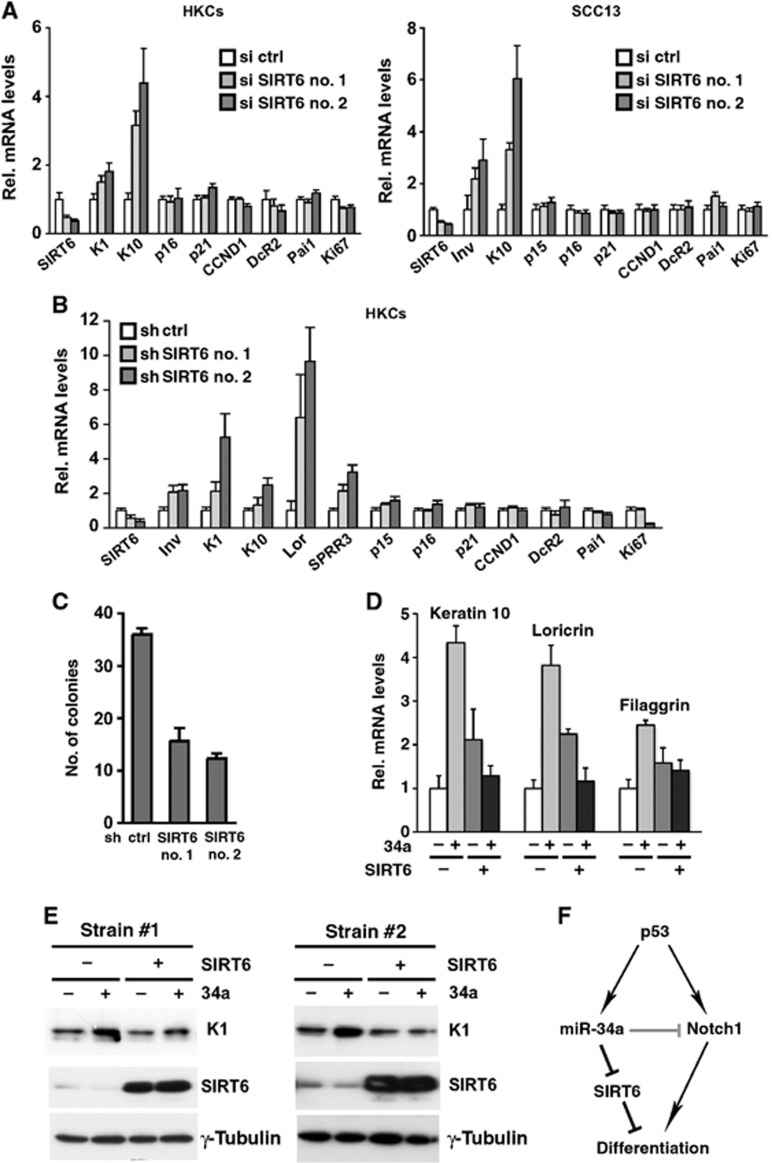
Functionally, si- or shRNA-mediated knockdown of SIRT6 was sufficient to reproduce the effects of increased miR-34a on differentiation in HKCs as well as in SCC cells, causing upregulation of a number of suprabasal markers, while having little or no effect on a number of cell cycle regulatory genes (Figure 8A and B and Supplementary Figure S7A). Interestingly, in parallel with enhanced differentiation, SIRT6 knockdown resulted in reduced proliferation potential, as assessed by clonogenicity assays (Figure 8C). Conversely, infection of two different strains of HKCs with a lentivirus expressing a SIRT6 cDNA devoid of the 3′ miR-34a recognition sequence rendered cells resistant to the pro-differentiation effects of miR-34a (Figure 8D and E).
Figure 8.
SIRT6 silencing promotes differentiation of HKCs and SCC13 cells and its persistent expression counteracts miR34 effects. (A) HKCs (left panel) or SCC13 (right panel) transfected for 3 and 2 days, respectively with siRNAs against SIRT6 (si SIRT6 no. 1 and no. 2) or scrambled control (si ctrl) were analysed for expression of the indicated genes by qRT–PCR using 36β4 for normalization. (B) HKCs stably transduced with shRNA retroviruses against SIRT6 (sh SIRT6 no. 1 and no. 2) or empty vector control (sh ctrl) were analysed as in A. (C) The same cells as in B were plated at low density on triplicate dishes for clonogenicity assay. Shown are numbers of colonies counted after crystal violet staining. (D, E) HKCs stably infected with a lentivirus for doxycycline-inducible expression of a SIRT6 cDNA devoid of the 3′ UTR miR-34a recognition sequence were either untreated (−) or treated with doxycycline for either 4 or 5 days (+). Cells were transfected with miR-34a mimics (25 nM) or scrambled controls for the last 3 days of the experiment followed by qRT–PCR (D) or immunoblot (E) analysis of expression of the indicated genes. In (E) results are shown for two different strains of HKCs. For strain 1, the blot was sequentially probed with antibodies against the indicated proteins without stripping. For strain 2, the blot was sequentially probed with keratin 1 and γ tubulin antibodies without stripping while SIRT6 was detected on a parallel blot. (F) Diagram showing the proposed dual function of miR-34a as an inducer of keratinocyte differentiation through negative control of SIRT6 expression and, at the same time, fine tune modulator of Notch1 expression downstream of p53.
Source data for this figure is available on the online supplementary information page.
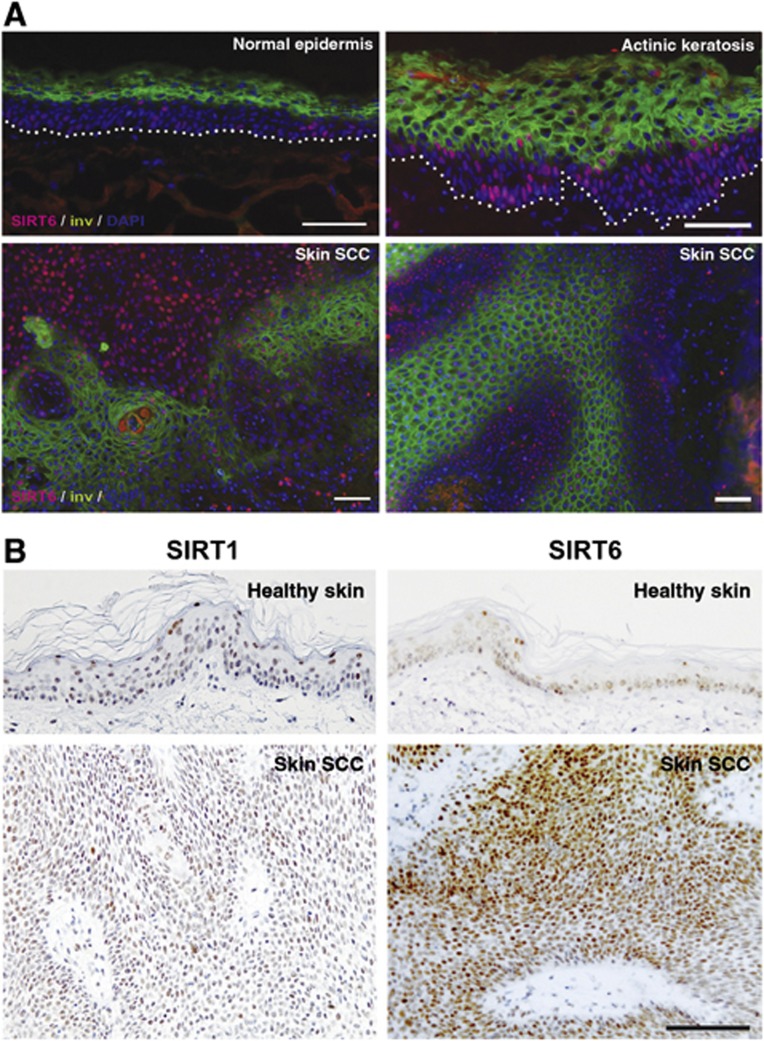
The inverse relation between SIRT6 expression and miR-34a and differentiation is of likely in vivo significance. Immunofluorescence analysis of intact human skin showed only few SIRT6 positive cells mostly located in the basal layer of the epidermis, while a large number of these cells was found in premalignant actinic keratoses (Figure 9A and Supplementary Figure S7B), in which, as shown in Figure 1F, miR-34a is down-modulated. Importantly, the pattern of SIRT6 expression in these lesions was mutually exclusive with that of the involucrin differentiation marker (Figure 9A and Supplementary Figure S7B). Massive increase of SIRT6 positive cells (and to a lesser extent, SIRT1) was also observed in cutaneous SCCs (Figure 9B) in which differentiation, as well as miR-34a expression, were compromised (Figure 1F).
Figure 9.
Upregulated SIRT6 expression in premalignant and malignant keratinocyte lesions inversely to differentiation. (A) Double immunofluorescence analysis of SIRT6 (red) and involucrin (green) expression in actinic keratosis lesions, surrounding normal skin and skin SCCs, with DAPI for counterstaining. Immunofluorescence analysis of SIRT6 expression in an additional actinic keratosis sample (same as S4 used for LCM analysis in Figure 1F) and skin SCCs is shown in Supplementary Figure S7B. (B) Immunohistochemical analysis of SIRT1 versus SIRT6 expression in another human skin SCC sample versus surrounding healthy skin. Bars, 200 μm.
Discussion
p53 plays a key role in the stress response of cells, controlling the decision between growth arrest and repair versus apoptosis (Dotto, 2009; Olivier et al, 2009). In addition, in keratinocytes of the proliferative compartment, increased p53 activity, as a consequence of UVB exposure (Lefort et al, 2007; Yugawa et al, 2007; Mandinova et al, 2008) or inhibition of mitogenic EGFR/ERK signalling (Kolev et al, 2008; Guinea-Viniegra et al, 2012), promotes differentiation-related changes in gene expression. Conversely, compromised p53 function accounts, in part, for the deranged differentiation programme of keratinocyte-derived cancer cells (Lefort et al, 2007; Kolev et al, 2008). Sustained DNA methylation, which is required for maintenance of normal proliferating keratinocyte populations (Sen et al, 2010), is another mechanism that can contribute to the block of differentiation of cancer cells (Momparler, 2005).
miR-34a is a well established target and mediator of p53 function with a similar context-dependent involvement in growth arrest, senescence, or apoptosis (Hermeking, 2010). Like p53, we have found that increased miR-34a levels in proliferating keratinocytes are sufficient to trigger a differentiation response and that this microRNA is required for the p53 and UVB pro-differentiation effects. miR-34a impact on differentiation is only partially overlapping with p53 and depends on down-modulation of SIRT6, a novel miR-34a target that is strongly upregulated in keratinocyte-derived SCC tumours, in which this miRNA is suppressed (Figure 8F). The gene encoding miR-34a, the form prevalently expressed in keratinocytes, resides on chromosome 1p36, a region of frequent deletion in neuroblastomas and melanomas (Hermeking, 2010). Loss of miR-34a expression, by gene deletion, compromised p53 function or increased promoter DNA methylation, has been implicated in resistance of tumour cells to chemotherapeutic pro-apoptotic agents (Bader, 2012 and references therein). The fact that ectopic expression of miR-34 genes has substantial growth-inhibitory or pro-apoptotic effects has prompted attempts to deliver this miRNA for gene therapy of tumours (Bader, 2012 and references therein). Besides the suppression of proliferation and apoptosis, miR-34a has been implicated as a determinant of embryonic stem (ES) cell/induced pluripotent stem cell (iPS) fate determination, colon stem cell commitment, neuroblast differentiation, and organismal ageing, with a number of possible targets involved in one or more of these processes (Agostini et al, 2011; Aranha et al, 2011; Choi et al, 2011; Yang et al, 2011; Jain et al, 2012; Liu et al, 2012; Bu et al, 2013).
SCC of the skin, oral cavity, lung, and other internal organs, are the most common type of solid human tumours (Ratushny et al, 2012). They are characterized by highly heterogeneous cell populations at various stages of differentiation, which can explain their resilience to various forms of therapy. Squamous cell differentiation is the combined result of complex transcription and cell structural/adhesion events (Lefort and Dotto, 2004). A number of studies have indicated that miRNAs are involved in control of squamous cell differentiation, as it occurs in normal epidermis and is perturbed in keratinocyte-derived neoplasia and other hyperplastic conditions like psoriasis (Bostjancic and Glavac, 2008; Dziunycz et al, 2010; Banerjee et al, 2011; Darido et al, 2011; Botchkareva, 2012; Pastar et al, 2012; Rivetti di Val Cervo et al, 2012; Sand et al, 2012; Schneider, 2012; Xu et al, 2012). MiR-203 has drawn special attention as a positive determinant of epidermal differentiation and, possibly, tumour suppression, through direct negative control of p63 expression. Consistent with previous reports (Dziunycz et al, 2010), we found that miR-203 is significantly less expressed in SCC cells than normal HKCs.
Another miRNA strongly downregulated in SCC cells was miR-34a. Antonini et al (2010) have previously shown that in mouse keratinocytes, miR-34a expression is under negative p63 control and that increased expression of this miRNA mediates the growth-inhibitory effects of loss of p63 through negative regulation of cell cycle promoting genes. The impact of miR-34a expression on keratinocyte differentiation was not assessed. In the context of the present findings, an attractive possibility was that the low expression of miR-34a in differentiation-defective SCC cells was due to the deregulated p63 function that occurs is these cells (Perez and Pietenpol, 2007; Ferone et al, 2013). However, at least in human keratinocytes, persistently elevated p63 expression was not sufficient to prevent differentiation-dependent induction of miR-34a expression. Instead, compromised p53 function, by either knocked down or mutant p53 expression, led to a concomitant suppression of differentiation and miR-34a expression.
A second mechanism that can contribute to decreased miR-34a expression in SCC cells is DNA methylation. Our finding of strong induction of miR-34a as well as differentiation marker expression in SCC cells by treatment with a DNA methyltransferase inhibitor, is of potential clinical significance in the context of novel therapy attempts based on the recruitment of a self renewing cancer cell population into differentiation (Momparler, 2005). No obvious skin phenotype has been reported for mice with homozygous miR-34a gene deletion (Choi et al, 2011), suggesting that lack of miR-34a can be compensated by multiple parallel pathways that concur in the normal keratinocyte differentiation programme (Lefort and Dotto, 2004). Consistent with previous reports with other cells (Li et al, 2009; Du et al, 2012; Bu et al, 2013) even in keratinocytes miR-34a functions as a direct negative regulator, rather than inducer of Notch1 expression, therefore being part of a negative feed forward loop linking p53 and Notch and fine-tuning skin homeostasis (Figure 8F).
Like p53, miR-34a may be more important for differentiation of basal proliferating keratinocytes in response to exogenous insults, like UVB exposure, and its down-modulation may contribute to the defective differentiation programme of keratinocyte-derived SCC cells. Interestingly, the pattern of differentiation-related genes modulated by miR-34a and p53 is only partially overlapping, indicating that miR-34a plays a function in differentiation that is distinct from p53. Consistent with this possibility is the finding that SIRT6, a novel miR-34a target as discussed further below, is modulated like miR-34a by changes in DNA methylation but is not affected by changes in p53. Downstream of miR-34a, a large number of targets have been identified that impinge on cell cycle control, apoptosis, and stem cell potential (Hermeking, 2010). Searching for direct targets whose down-modulation mediates its impact on differentiation, we identified a few genes that are down-modulated in HKCs as a consequence of increased miR-34a expression. Among these were the genes for two tyrosine kinases, c-Met and Axl, and a transcription factor, FosL1, with a known or likely role in keratinocyte growth/differentiation control and/or transformation (Cortesina et al, 2000; Murai et al, 2004; Green et al, 2006; Papadakis et al, 2011). However, knockdown of these genes could not recapitulate the miR-34a pro-differentiation effects in keratinocytes. SIRT1 is another known miR-34a target that has been recently linked to keratinocyte senescence or differentiation (Blander et al, 2009; Pickard et al, 2010; Rivetti di Val Cervo et al, 2012). In HKCs, we did not find any significant regulation of this gene by miR-34a, while we observed significant down-modulation of SIRT6, the only other sirtuin family member with a miR-34a recognition sequence in its 3′UTR.
Our finding, that down-modulation of SIRT6 is sufficient to reproduce the miR-34a impact on differentiation, is of likely biological significance, as expression of this gene is inversely related to miR-34a in differentiating keratinocytes and keratinocyte-derived tumours. SIRT6 is a highly specific NAD+−dependent histone deacetylase and ADP-ribosyl transferase that has been implicated in DNA repair, genomic stability, glucose and fatty acid metabolism (Davalos et al, 2011; Jia et al, 2012). As such, SIRT6 has been proposed to be a potential tumour suppressor gene (Bosch-Presegue and Vaquero, 2011). While this conclusion is consistent with down-modulation of SIRT6 in precancerous lesions of hepatocellular carcinomas and colon adenocarcinomas (Min et al, 2012; Sebastian et al, 2012), this gene has been reported to be over-expressed in B cells of patients with chronic lymphocytic leukaemia (CLL) and in pancreatic cancer cell lines (Wang et al, 2011a; Bauer et al, 2012). Thus, depending on tissue type and condition, SIRT6 may behave as either an oncogene or tumour suppressor gene (like it is postulated for SIRT1 (Rajendran et al, 2011)), and additional studies will be required to define its precise role in skin SCCs. Given the degenerative ageing-related phenotypes developed by the SIRT6-deficient mice (Mostoslavsky et al, 2006), an interesting possibility for future studies is that the miR-34a-SIRT6 connection that we have uncovered applies also to senescence/ageing processes in which miR-34a is implicated (Li et al, 2011; Yang et al, 2011; Chen and Hu, 2012; Liu et al, 2012).
Materials and methods
Conditions for retrovirus/lentivirus production and infection were as previously reported (Nguyen et al, 2006; Lefort et al, 2007). The pBABE-puro-p53 wild-type and 3 mutants vectors were from Dr A Rustgi (University of Pennsylvania, PA) (Okawa et al, 2007). The pRS-p53 (sh-p53) vector was from Dr R Agami (The Netherlands Cancer Institute, Amsterdam, The Netherlands). The pINDUCER20-p21 was a gift from Dr SJ Elledge (Brigham and Women's Hospital, Boston, MA) (Meerbrey et al, 2011). The pLKO1-shp63 constructs were obtained from Dr L Ellisen (Mass Gen. Hospital., Boston, MA). The pTRIPZ and pTRIPZ-miR-34a plasmids were from Dr X-F Wang (Yang et al, 2012). The ΔNp63α expression plasmid was a gift of Dr C Missero (Naples, Italy). The pRS-SIRT6 (pRS-SIRT6 no. 1, 5′-AAGCTGGAGCCCAAGGAGGAA-3′; pRS-SIRT6 no. 2, 5′-AAGAATGTGCCAAGTGTAAGA-3′) were from Dr KF Chua (Stanford University, CA) (Michishita et al, 2008). The inducible pINDUCER20-p53R248W (p53 mt) was cloned using the pBABE-p53R248W (Okawa et al, 2007) as template into pENTR-TOPO (Invitrogen) using the following primers: 5′-CACCATGGAGGAGCCGCAGTCAGATCCTAGCGTCGA-3′ (forward) and 5′-TCAGTCTGAGTCAGGCCCTTCTGTCTTGAA-3′ (reverse). The inducible pINDUCER20-SIRT6 was cloned using cDNA from HKCs as template into pENTR-TOPO with the following primers: 5′-CACATGTCGGTGAATTACGCGGC-3′ (forward) and 5′-TCAGCTGGGGACCGCCTT-3′ (reverse). After sequence verification, the coding sequence of p53R248W or SIRT6 was transferred by recombination into the destination lentiviral vector pINDUCER-20 (Meerbrey et al, 2011) using the pENTR™ Directional TOPO® Cloning kit (Invitrogen). Cells stably infected with these lentiviruses (after G418 selection (500 μg/ml)) were induced to express p53R248W or SIRT6 by the presence of doxycycline in the culture medium.
Other detailed Materials and methods are provided as Supplementary materials and methods.
Supplementary Material
Acknowledgments
We thank Drs J Rheinwald, J Rocco, A Rustgi, R Agami, KF Chua, X-F Wang, C Missero, SE Elledge and L Ellisen for their gifts of cells or plasmids. We are also grateful to Elena Menietti in our group for cloning the pINDUCER20-p53R248W and to N Allioli (University of Lyon, France) for her technical assistance for ISH in paraffin embedded tissues. We thank Donald Singer, Einar Castillo, as well as Florine Favre for their technical assistance. This work was supported by grants from the Swiss National Foundation (grants CRSI33-130576/1; 3100A0-122281/1), Oncosuisse (grant 02361-02-2009) and NIH (grant AR39190) to GPD. KL is a recipient of the Swiss L’Oréal ‘For Women in Science’ award. PO was supported by a grant from Lauretana S.P.A.
Author contributions: The overall study was conceived and designed by GPD and KL with an initial help of IK. KL, YB, VC and MCA performed the experiments and analysed the data. PO performed the conventional microarray experiments and analysed the data relative to microarray experiments. JGV and EFW designed and analysed the data relative to the p53KI/KI mouse model. AAH and WH provided critical human oral and skin SCC samples respectively. SW gave conceptual input in interpretation of the experiments and technical suggestions. GPD and KL wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G (2011) microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA 108: 21099–21104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Treviño L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH et al. (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333: 1154–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE (2006) The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 16: 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C (2010) Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol 130: 1249–1257 [DOI] [PubMed] [Google Scholar]
- Aranha MM, Santos DM, Sola S, Steer CJ, Rodrigues CM (2011) miR-34a regulates mouse neural stem cell differentiation. PLoS One 6: e21396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG (2012) miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet 3: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee J, Chan YC, Sen CK (2011) MicroRNAs in skin and wound healing. Physiol Genomics 43: 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer I, Grozio A, Lasiglie D, Basile G, Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A, Zoppoli G, Cea M, Feldmann G, Mostoslavsky R, Ballestrero A, Patrone F, Bruzzone S, Nencioni A (2012) The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem 287: 40924–40937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Bhimavarapu A, Mammone T, Maes D, Elliston K, Reich C, Matsui MS, Guarente L, Loureiro JJ (2009) SIRT1 promotes differentiation of normal human keratinocytes. J Invest Dermatol 129: 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L, Vaquero A (2011) The dual role of sirtuins in cancer. Genes Cancer 2: 648–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostjancic E, Glavac D (2008) Importance of microRNAs in skin morphogenesis and diseases. Acta Dermatovenerol Alp Panonica Adriat 17: 95–102 [PubMed] [Google Scholar]
- Botchkareva NV (2012) MicroRNA/mRNA regulatory networks in the control of skin development and regeneration. Cell Cycle 11: 468–474 [DOI] [PubMed] [Google Scholar]
- Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP (2010) Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 16: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, Li J, Yang H, Milsom J, Lee S, Zipfel W, Jin MM, Gümüş ZH, Lipkin SM, Shen X (2013) A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell 12: 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafa V, Nebbioso A, Altucci L (2012) Sirtuins and disease: the road ahead. Front Pharmacol 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hu SJ (2012) Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: a review. J Biochem Mol Toxicol 26: 79–86 [DOI] [PubMed] [Google Scholar]
- Chim CS, Wong KY, Qi Y, Loong F, Lam WL, Wong LG, Jin DY, Costello JF, Liang R (2010) Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis 31: 745–750 [DOI] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, Ozturk A, Hicks GG, Hannon GJ, He L (2011) miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 13: 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI (2005) Temporal dissection of p53 function in vitro and in vivo. Nat Genet 37: 718–726 [DOI] [PubMed] [Google Scholar]
- Cortesina G, Martone T, Galeazzi E, Olivero M, De Stefani A, Bussi M, Valente G, Comoglio PM, Di Renzo MF (2000) Staging of head and neck squamous cell carcinoma using the MET oncogene product as marker of tumor cells in lymph node metastases. Int J Cancer 89: 286–292 [PubMed] [Google Scholar]
- Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q, Rank G, Srivastava S, Finlay MJ, Papenfuss AT, Pandolfi PP, Pearson RB, Jane SM (2011) Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 20: 635–648 [DOI] [PubMed] [Google Scholar]
- Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suárez Y, Lai EC, Fernández-Hernando C (2011) miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA 108: 9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto F, Topley G, Calautti E, Hsiao J, Ong L, Seth PK, Dotto GP (1998) Inhibitory function of p21Cip1/WAF1 in differentiation of primary mouse keratinocytes independent of cell cycle control. Science 280: 1069–1072 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr., Butel JS, Bradley A (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221 [DOI] [PubMed] [Google Scholar]
- Dotto GP (2000) p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta 1471: M43–M56 [DOI] [PubMed] [Google Scholar]
- Dotto GP (2009) Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer 9: 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, Wang H, Huang C, Sun S (2012) Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS One 7: e30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziunycz P, Iotzova-Weiss G, Eloranta JJ, Lauchli S, Hafner J, French LE, Hofbauer GF (2010) Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J Invest Dermatol 130: 2686–2689 [DOI] [PubMed] [Google Scholar]
- Ferone G, Mollo MR, Thomason HA, Antonini D, Zhou H, Ambrosio R, De Rosa L, Salvatore D, Getsios S, van Bokhoven H, Dixon J, Missero C (2013) p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum Mol Genet 22: 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Ikram M, Vyas J, Patel N, Proby CM, Ghali L, Leigh IM, O'Toole EA, Storey A (2006) Overexpression of the Axl tyrosine kinase receptor in cutaneous SCC-derived cell lines and tumours. Br J Cancer 94: 1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea-Viniegra J, Zenz R, Scheuch H, Jimenez M, Bakiri L, Petzelbauer P, Wagner EF (2012) Differentiation-induced skin cancer suppression by FOS, p53, and TACE/ADAM17. J Clin Invest 122: 2898–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H (2007) p53 enters the microRNA world. Cancer Cell 12: 414–418 [DOI] [PubMed] [Google Scholar]
- Hermeking H (2010) The miR-34 family in cancer and apoptosis. Cell Death Differ 17: 193–199 [DOI] [PubMed] [Google Scholar]
- Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC (2012) p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol 10: e1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerry DJ, Tao L, Yan H (2008) Regulation of cancer stem cells by p53. Breast Cancer Res 10: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Su L, Singhal S, Liu X (2012) Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Mol Cell Biochem 364: 345–350 [DOI] [PubMed] [Google Scholar]
- Kolev V, Mandinova A, Guinea-Viniegra J, Hu B, Lefort K, Lambertini C, Neel V, Dummer R, Wagner EF, Dotto GP (2008) EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol 10: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz D, Dobbelstein M (2006) Nongenotoxic p53 activation protects cells against S-phase-specific chemotherapy. Cancer Res 66: 10274–10280 [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol 4: 199–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort K, Dotto GP (2004) Notch signaling in the integrated control of keratinocyte growth/differentiation and tumor suppression. Semin Cancer Biol 14: 374–386 [DOI] [PubMed] [Google Scholar]
- Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, Neel V, Garlick J, Chiorino G, Dotto GP (2007) Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev 21: 562–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, di Val Cervo PR, Aberdam D, Knight RA, Melino G, Candi E (2008) miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ 15: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Li N, Muthusamy S, Liang R, Sarojini H, Wang E (2011) Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev 132: 75–85 [DOI] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R (2009) MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 69: 7569–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Zhu Y, Wang LS, Bonini NM (2012) The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H (2008) Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7: 2591–2600 [DOI] [PubMed] [Google Scholar]
- Mandinova A, Lefort K, Tommasi di Vignano A, Stonely W, Ostano P, Chiorino G, Iwaki H, Nakanishi J, Dotto GP (2008) The FoxO3a gene is a key negative target of canonical Notch signalling in the keratinocyte UVB response. Embo J 27: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone FR, Brentani MM, Nonogaki S, Begnami MD, Campos AH, Walder F, Carvalho MB, Soares FA, Torloni H, Kowalski LP, Federico MH (2005) Overexpression of Fos-related antigen-1 in head and neck squamous cell carcinoma. Int J Exp Pathol 86: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS (2000) TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103: 295–309 [DOI] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ, Schmitt EM et al. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA 108: 3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, Chen L, Scheuch H, Zheng H, Qin L, Zatloukal K, Hui L, Wagner EF (2012) Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol 14: 1203–1211 [DOI] [PubMed] [Google Scholar]
- Momparler RL (2005) Epigenetic therapy of cancer with 5-aza-2′-deoxycytidine (decitabine). Semin Oncol 32: 443–451 [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329 [DOI] [PubMed] [Google Scholar]
- Murai M, Shen X, Huang L, Carpenter WM, Lin CS, Silverman S, Regezi J, Kramer RH (2004) Overexpression of c-met in oral SCC promotes hepatocyte growth factor-induced disruption of cadherin junctions and invasion. Int J Oncol 25: 831–840 [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, di Vignano AT, Kitajewski J, Chiorino G, Roop DR, Missero C, Dotto GP (2006) Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev 20: 1028–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa T, Michaylira CZ, Kalabis J, Stairs DB, Nakagawa H, Andl CD, Johnstone CN, Klein-Szanto AJ, El-Deiry WS, Cukierman E, Herlyn M, Rustgi AK (2007) The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev 21: 2788–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Petitjean A, Marcel V, Petre A, Mounawar M, Plymoth A, De Fromentel CC, Hainaut P (2009) Recent advances in p53 research: an interdisciplinary perspective. Cancer Gene Ther 16: 1–12 [DOI] [PubMed] [Google Scholar]
- Papadakis ES, Cichon MA, Vyas JJ, Patel N, Ghali L, Cerio R, Storey A, O'Toole EA (2011) Axl promotes cutaneous squamous cell carcinoma survival through negative regulation of pro-apoptotic Bcl-2 family members. J Invest Dermatol 131: 509–517 [DOI] [PubMed] [Google Scholar]
- Pastar I, Khan AA, Stojadinovic O, Lebrun EA, Medina MC, Brem H, Kirsner RS, Jimenez JJ, Leslie C, Tomic-Canic M (2012) Induction of specific micro Ribonucleic acids (miRNAs) inhibits cutaneous wound healing. J Biol Chem 287: 29324–29335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CA, Pietenpol JA (2007) Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle 6: 246–254 [DOI] [PubMed] [Google Scholar]
- Pickard A, Wong PP, McCance DJ (2010) Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. J Cell Sci 123: 3718–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R, Garva R, Krstic-Demonacos M, Demonacos C (2011) Sirtuins: molecular traffic lights in the crossroad of oxidative stress, chromatin remodeling, and transcription. J Biomed Biotechnol 2011: 368276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT (2012) From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest 122: 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM (2005) Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. Rna 11: 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo G, Nguyen BC, Dziunycz P, Ristorcelli E, Ryan RJ, Ozuysal OY, Di Piazza M, Radtke F, Dixon MJ, Hofbauer GF, Lefort K, Dotto GP (2011) IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J 30: 4571–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Beckett MA (1980) Defective terminal differentiation in culture as a consistent and selectable character of malignant human keratinocytes. Cell 22: 629–632 [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A (2008) Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, Saintigny G, Dellambra E, Odorisio T, Mahe C, Calin GA, Candi E, Melino G (2012) p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci USA 109: 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand M, Skrygan M, Sand D, Georgas D, Hahn S, Gambichler T, Altmeyer P, Bechara FG (2012) Expression of microRNAs in basal cell carcinoma. Br J Dermatol 167: 847–855 [DOI] [PubMed] [Google Scholar]
- Schneider MR (2012) MicroRNAs as novel players in skin development, homeostasis and disease. Br J Dermatol 166: 22–28 [DOI] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BMM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE (2012) The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151: 1185–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA (2010) DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H (2011) miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 10: 4256–4271 [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta M, Choi YS, Okegbe T, Wong G, Tam OH, Chong MM, Seykora JT, Nagy A, Littman DR, Andl T, Millar SE (2012) Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Development 139: 1405–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, Bracken CP, Szubert JM, Goodall GJ (2013) On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One 8: e55214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, Robert A, Candi E, Melino G, Gidrol X, Mantovani R (2006) New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J 25: 5105–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O'Loughlin E, Lee T, Houel S, O'Carroll D, Tarakhovsky A, Ahn NG, Yi R (2012) Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 26: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Kafeel MI, Avezbakiyev B, Chen C, Sun Y, Rathnasabapathy C, Kalavar M, He Z, Burton J, Lichter S (2011a) Histone deacetylase in chronic lymphocytic leukemia. Oncology 81: 325–329 [DOI] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L, Pennypacker S, Zou Y, Sharma M, North JP, Vemula SS, Mauro TM, Neuhaus IM, Leboit PE, Hur JS, Park K (2011b) Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA 108: 17761–17766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ (1993) The p53-mdm-2 autoregulatory feedback loop. Genes Dev 7: 1126–1132 [DOI] [PubMed] [Google Scholar]
- Xu N, Zhang L, Meisgen F, Harada M, Heilborn J, Homey B, Grander D, Stahle M, Sonkoly E, Pivarcsi A (2012) MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration and invasion. J Biol Chem 287: 29899–29908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA 105: 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ (2009) MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle 8: 712–715 [DOI] [PubMed] [Google Scholar]
- Yang J, Chen D, He Y, Melendez A, Feng Z, Hong Q, Bai X, Li Q, Cai G, Wang J, Chen X (2011) MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr) 35: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, Wang HY, Cheng SQ, Xie D, Wang XF (2012) TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 22: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E (2006) Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38: 356–362 [DOI] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, Fuchs E (2009) DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA 106: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E (2008) A skin microRNA promotes differentiation by repressing 'stemness'. Nature 452: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T (2007) Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol 27: 3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Stokes N, Polak L, Fuchs E (2011) Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 8: 294–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Tsimelzon A, Hilsenbeck S, Green JE, Michalowska AM, Rosen JM (2008) Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res 68: 4674–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA (2008) p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.