Abstract
The role of water molecules in the selective transport of potassium ions across cell membranes is important. Experimental investigations of valinomycin–water interactions remain huge challenge due to the poor solubility of valinomycin in water. Herein, we removed this experimental obstacle by introducing gaseous water and valinomycin onto a Cu(111) surface to investigate the hydration of valinomycin. By combining scanning tunneling microscopy (STM) with density functional theory (DFT) calculations, we revealed that water could affect the adsorption structure of valinomycin. Hydrogen bond interactions occurred primarily at the carbonyl oxygen of valinomycin and resulted in the formation of valinomycin hydrates. The single-molecule perspective revealed in our investigation could provide new insight into the role of water on the conformation transition of valinomycin, which might provide a new molecular basis for the ion transport mechanism at the water/membrane interface.
Ion channels and ion carriers are essential for numerous important physiological processes, such as signal transduction, membrane potential generation and muscle contraction1,2,3,4. Valinomycin is a cyclic peptide and an important ion carrier for the specific transport of potassium ions (K+ ions) responsible for maintaining K+ ions balance between the two sides of the cell membrane5. Many studies, including theoretical simulations and experimental measurements, have been performed to elucidate the conformations, ion-transport dynamics and ion-transport mechanism of valinomycin6,7,8,9,10,11. The conformations under various environments have been widely studied because ion transport is always accompanied by a conformational transition6. X-ray crystallography has revealed that valinomycin can be crystallized into a number of conformations, including a “twisted propeller” conformation from dimethyl sulfoxide7 and a “twisted bracelet” conformation from octane8. Molecular dynamics simulations and spectroscopic measurements (i.e., nuclear magnetic resonance, circular dichroism and infrared spectroscopy) have indicated that the structure of valinomycin in solution is diverse and sensitive to the polarity of the solvent9,10. Some investigations have also been executed at the solvent/membrane interfaces taking the importance of interfaces account11; however, the central role of water was not considered in most of these studies.
Evaluating the importance of water as an integral part of biomolecular structural organization and its role in a variety of biological processes is a worthwhile endeavor12,13,14. Water, which exists on both sides of a cell membrane, is potentially an integral part of K+ transport due to its strong abilities to solvate K+ ions and to form hydrogen bonds with the peptide backbone13. Investigating the role of water in K+ transport is important. Molecular dynamics simulations have revealed that uncomplexed valinomycin acts as a surfactant with hydrophobic groups embedded in the membrane, whereas the hydrophilic carbonyl groups form hydrogen bonds with water15,16. The effective hydration of the carbonyl groups accompanied by a conformational transition causes the transfer of ions from the valinomycin carrier to the water/membrane interface16. Theoretical simulations have also indicated that the release of K+ ions involves the formation of intermediate water adducts and the gradual replacement of ester carbonyl groups by water molecules in the first coordination sphere of the K+ ions15. Water might play a role in all of the ion transport steps because peptide–water interactions might govern valinomycin's folding, conformation and biological function13,17.
Elucidating valinomycin–water interactions and the effects of water on the structure of valinomycin are important for understanding the mechanism of ion transport at the water/membrane interface under physiological conditions. Until now, the role of water in a selective transport mechanism has not been clear. The determination of the structure that valinomycin adopts at the water/membrane interface and valinomycin-water interactions using modern experimental techniques remains huge challenge because valinomycin is practically insoluble in water and the K+-valinomycin complexes are unstable in water18,19. Herein, we remove these experimental obstacles by introducing gaseous water and valinomycin onto a Cu(111) surface under ultra-high vacuum (UHV) conditions. In combination with density functional theory (DFT) calculations, the hydration of valinomycin was studied using scanning probe microscopy, which has the potential to be a powerful tool for revealing the fine structures and molecular mechanisms of biomolecules20,21,22,23,24,25,26,27,28. Two sample preparation methods for the valinomycin hydrates were employed to investigate the valinomycin–water interactions.
Results
“One-step dosing” method
Valinomycin is a cyclic dodecadesipeptide composed of 12 α-amino acid residues that appear as three repeated tetrapeptide structural sequences (l-Val – d-Hyv – d-Val – l-Lac), as shown in Figure 1a. Valinomycin is important in the selective transport of K+ ions across the cell membrane due to its chemical composition. An abundance of hydrophobic alkyl side chains in the amino acid residues allow valinomycin to enter the core of the lipid phospholipid bilayer, whereas the 12 carbonyl and six hydrophilic ether oxygens prefer the hydrophilic environment. A typical STM image (Figure 1b) shows that valinomycin monomers and tetramers coexisted on the Cu(111) surface when the valinomycin molecules were dosed onto a 78 K surface and subsequently flash-annealed at a room-temperature (RT) carousel for 2 min to obtain their stable adsorption configurations. Valinomycin appears as two types of monomers with different adsorption configurations when it is adsorbed onto the perfect Cu(111) terrace. The L-type monomer, which is denoted ML, appears as three lobes with a central protrusion in the STM image. The R-type monomer, which is denoted MR, appears as three lobes with a central cavity in the STM image. Both types of monomers exhibit nearly three-fold symmetry in the STM images. In addition, the valinomycin tetramer (marked as “T” in Figure 1b) also exhibits a nearly three-fold symmetrical STM pattern.
Figure 1. Comparison between valinomycin and the valinomycin–water complexes prepared using the “one-step dosing” method.
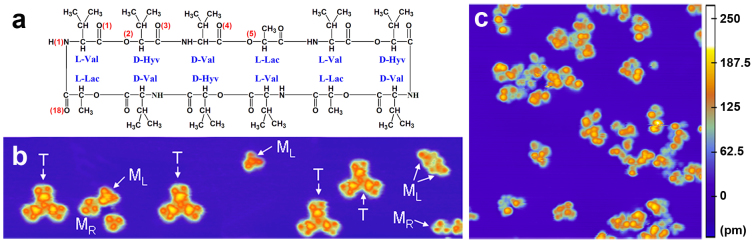
(a) Structural formula of valinomycin. Some of oxygen and nitrogen atoms are marked by the numbers in the brackets. (b) STM image (55 nm × 12 nm) of valinomycin monomers and tetramers coexisting on the Cu(111) surface. The L-type monomer, R-type monomer and tetramer are denoted ML, MR and T, respectively. (c) STM image (25 nm × 25 nm) of the valinomycin–water complexes. Both of the STM images were obtained at −2 V and 0.03 nA.
After determining the adsorption structure of the uncomplexed valinomycin molecules, we also investigated valinomycin–water complexes. In the “one-step dosing” method, we obtained valinomycin–water complexes by co-evaporating water and valinomycin molecules onto the 78 K Cu(111) surface, which was followed by flash annealing at the RT carousel for 2 min. In contrast to the identical three-fold symmetrical STM patterns for the uncomplexed valinomycin molecule (Figure 1b), the valinomycin–water complexes were scattered randomly and disorderly on the surface (Figure 1c). The intervention of water molecules disturbed the formation of the identical adsorbed structure of valinomycin on the surface. This result indicates that significant interactions occurred between the gaseous water and valinomycin molecules when valinomycin and water were simultaneously dosed. We believe that the initial contact between water and valinomycin occurs in the UHV environment and that the dynamic adjustment of the valinomycin–water interactions occurs primarily on the Cu(111) surface in the subsequent annealing process. The “one-step dosing” method suggests that valinomycin–water interactions strongly affect the adsorption conformation of valinomycin, which is very critical for K+ transport at the water/membrane interface.
“Two-step dosing” method
Valinomycin molecules have well-defined, nearly three-fold symmetrical adsorption configurations (both the monomer and tetramer) when anchored on a perfect Cu(111) terrace (Figure 1b), which enables an investigation of the effect of water on the adsorbed valinomycin at the single-molecule level. In addition to the previously mentioned “one-step dosing” method, a “two-step dosing” method has also been used to prepare valinomycin hydrates. In the first step, valinomycin molecules were evaporated onto the 78 K Cu(111) surface followed by flash annealing at the RT carousel for 2 min to obtain their stable adsorption configurations. In the second step, gaseous water molecules were dosed onto the 78 K Cu(111) surface, where valinomycin molecules had been previously fixed in the first step. The dynamic adjustment of the valinomycin–water binding occurred primarily during the subsequent annealing process. The interactions between the water and valinomycin molecules, including the individual L-type valinomycin monomer (ML), the R-type monomer (MR) and the tetramer (T) are individually discussed in the subsequent sections.
The hydration of the L-type valinomycin monomer
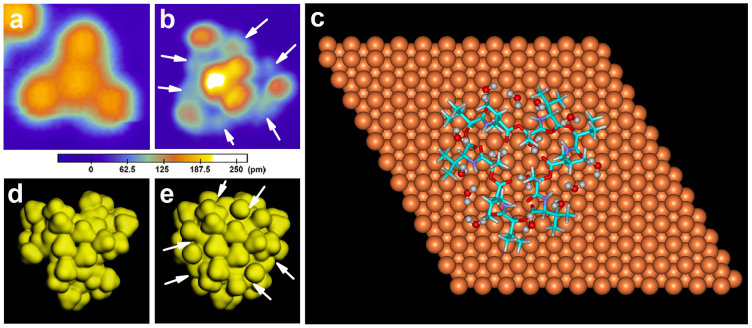
Figure 2a shows a typical STM image of an individual L-type valinomycin monomer (ML), which can be viewed as three lobes with a central protrusion. The high-resolution STM image (Fig. S1 in the supplementary information) reveals that the central protrusion becomes three smaller protrusions if the resolution of the STM tip is sufficiently high. Figure 2b is a typical STM image of an individual L-type valinomycin monomer hydrate (ML'). By comparing Figures 2b and 2a, we determined that the three-fold symmetry is retained in the STM pattern of ML and ML'; however, the morphologies, sizes and numbers of protrusions are completely different. The central protrusion becomes small protrusions and the three external protrusions become smaller after gaseous water molecules are introduced, exhibiting characteristics similar to those of the high-resolution STM image of ML shown in Fig. S1. These changes might be due to the improvement in the imaging resolution induced by the water-modified STM tip, similar to the improvements observed with a carbon-monoxide-functionalized tip29,30. We also noticed that six additional protrusions emerged after the sample was dosed with water. These protrusions are marked by the arrows in Figure 2b. The STM image of ML' is different from that of ML, which suggests the possibility of valinomycin hydration. The possible binding of water may account for the increase of electron density at the binding sites because STM signals reflect the electron density of a sample.
Figure 2. STM images and DFT calculation results for the L-type valinomycin monomer and its hydrate prepared using the “two-step dosing” method.
(a, b) Typical STM images of an individual L-type valinomycin monomer (ML) and its hydrate (ML') adsorbed onto the Cu(111) surface at 78 K. Both of the STM image sizes are 3 nm × 3 nm. Both of the STM images were collected at −2 V and 0.03 nA. (c) Calculated adsorption configuration of ML' on the Cu(111) surface. (d, e) Calculated electron density of ML and ML'.
To elucidate the binding details of water molecules and the valinomycin–water interactions, we performed density functional theory (DFT) calculations. All of the possible sites for the binding of water molecules in ML were evaluated. Detailed adsorption configurations of the water molecules binding to ML are shown in Figure 2c, revealing that there are 15 possible binding sites for water molecules. These possible sites can be classified into three groups because of the nearly three-fold symmetry of the ML adsorption structure. For simplification, we focus on one group in the following discussion of valinomycin–water interactions. Five representative binding modes for an individual water molecule are illustrated in Figs. S2a–e, and the binding sites are marked by arrows (see the supplementary information). The calculated interaction energies between water and ML and the respective binding sites are summarized in Table 1. The theoretical results show that water molecules primarily bind to valinomycin via the formation of one or two hydrogen bonds to the carbonyl oxygen, ester oxygen or amino nitrogen of valinomycin. In addition, water molecules preferentially bind to the carbonyl oxygen. For example, mode “e” in which an individual water molecule interacts with two adjacent carbonyl oxygens, is the most energetically favorable binding mode. For mode “a”, the binding of a water molecule with a nitrogen atom is the most unstable mode.
Table 1. Calculated binding sites and interaction energies between water and the L-type valinomycin monomers (ML) on the Cu(111) surface. A more negative energy indicates that the calculated system is more stable.
| ML | Mode a | Mode b | Mode c | Mode d | Mode e |
|---|---|---|---|---|---|
| Binding sites | N(1)H | O = C − O(2); C = O(18) | C = O(3) | O = C − O(5) | C = O(1); C = O(4) |
| Interaction energy (kcal/mol) | −6.00 | −12.57 | −9.07 | −7.44 | −13.25 |
Note: The code number of the nitrogen or oxygen atoms is consistent with the structural formula of valinomycin in Figure 1a.
Additional detailed electron densities of the L-type valinomycin monomer hydrate, which contains one water molecule in five binding modes, are presented in Figs. S2a'–e'. The local changes of electron density caused by the binding of the water molecules are clearly marked. An increase in the local electron densities at the binding sites was observed for all of the binding modes except mode “e” (Fig. S2e'). Notably, not all binding of the water molecules resulted in the appearance of new protrusions. If the 15 total possible binding sites in ML are all occupied by water molecules, the electron densities of valinomycin hydrate (ML') will be different from those of valinomycin (ML). Figures 2d and 2e show the calculated electron densities of ML and ML', respectively. The distinct differences in the calculated electron densities between ML and ML' are marked by the arrows in Figure 2e. These theoretical results agree well with the experimental results. When comparing Figure 2e with Figure 2b, we noticed that the binding of water molecules accounts for the appearance of the six additional protrusions marked by the arrows in the STM images. We also noticed that the number of newly observed protrusions in Figure 2b is not equal to the number of binding water molecules. The appearance of new protrusions in the STM image depends on the actual contribution of adsorbed water molecules to the total electron density. Numerous factors, such as the number of binding water molecules, the binding sites and the interaction strength, can affect the final STM image of the valinomycin hydrate.
The hydration of the R-type valinomycin monomer
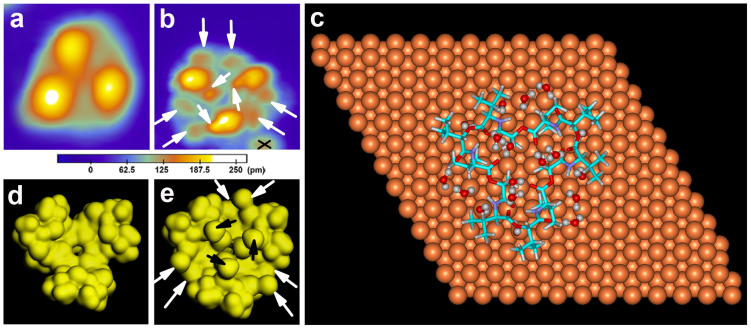
We also investigated the interactions between the R-type valinomycin monomer (MR) and water using STM and DFT calculations. The high-resolution STM images of the R-type valinomycin monomer (MR) and its hydrate (MR') are displayed in Figures 3a and 3b, respectively. The STM patterns of MR' and MR are obviously different. In MR', six additional protrusions appear at the edge, and an additional three protrusions emerge at the center, which are marked by the arrows in Figure 3b. We speculated that the adsorption of water is responsible for these changes in the electronic structure. The calculated adsorption configuration of MR' indicates that 18 possible binding sites for water molecules exist in MR (Figure 3c). These 18 possible binding sites were also divided into three groups because MR is nearly three-fold symmetrical. Six different binding modes displaying the sites binding with an individual water molecule are shown in Fig. S3 in the supplementary information. The calculated interaction energy between water and MR and the respective binding sites are summarized in Table 2. Water molecules bind with valinomycin by forming one or two hydrogen bonds to the carbonyl oxygen, ester oxygen or amino nitrogen. The energetically favorable and unfavorable adsorptions still occur in modes “e” and “a” respectively.
Figure 3. STM images and DFT calculation results of the R-type valinomycin monomer and its hydrate prepared using the “two-step dosing” method.
(a, b) Typical STM images of an individual R-type valinomycin monomer (MR) and its hydrate (MR') adsorbed onto the Cu(111) surface at 78 K. The crossed black lines in Figure 3b mark a protrusion belonging to another adsorbate that is not shown here. Both of the STM image sizes are 3 nm × 3 nm. Both of the STM images were collected at −2 V and 0.03 nA. (c) Calculated adsorption configuration of MR' on the Cu(111) surface. (d, e) Calculated electron density of MR and MR'.
Table 2. Calculated binding sites and interaction energies between water and the R-type valinomycin monomers (MR) on the Cu(111) surface. A more negative energy indicates that the calculated system is more stable.
| MR | Mode a | Mode b | Mode c | Mode d | Mode e | Mode f |
|---|---|---|---|---|---|---|
| Binding sites | N(1)H | O = C − O(2); C = O(18) | C = O(3) | O = C − O(5) | C = O(1); C = O(4) | C = O(1) |
| Interaction energy (kcal/mol) | −5.59 | −11.95 | −8.20 | −7.21 | −13.80 | −6.54 |
Note: The code number of the nitrogen or oxygen atoms is consistent with the structural formula of valinomycin in Figure 1a.
In addition, the effect of an individual water molecule on the local electron densities of MR has also been elucidated using DFT calculations and is shown in Fig. S3. Only four binding modes exhibit an obvious increase in the local electron density at the binding sites, with the exceptions of modes “a” (Fig. S3a') and “b” (Fig. S3b'). We determined the overall effect of all 18 possible binding waters by comparing the calculated electron densities of MR (Figure 3d) and MR' (Figure 3e). The differences in the electron density between MR and MR', which is marked by the arrows in Figure 3e, is attributed to the binding of water molecules, which is in agreement with the changes observed in the STM experiments. The appearance of the nine additional protrusions in Figure 3b confirms the formation of valinomycin hydrates.
This “two-step dosing” method provides a single-molecule perspective of the valinomycin–water interactions, which is valuable for understanding the role of water in the conformational transition of valinomycin under physiological conditions.
The hydration of the valinomycin tetramer
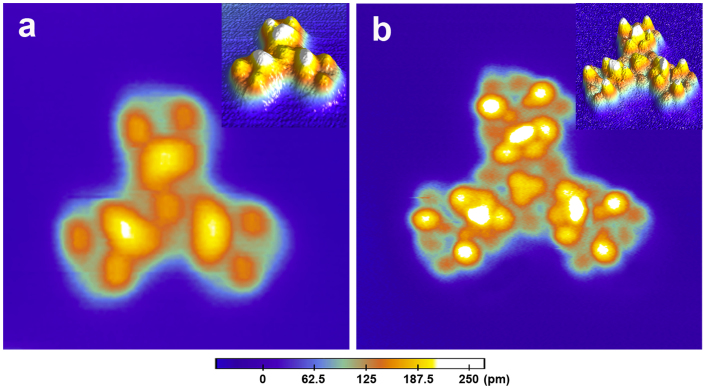
The effect of water on the adsorption structure of valinomycin was further confirmed in the case of valinomycin tetramer using the “two-step dosing” method. A typical STM image (Figure 4a) shows that the individual valinomycin tetramer (T) has highly symmetrical architectures with three “arms” on the Cu(111) surface. After obtaining discrete valinomycin tetramers in the first step, we dosed gaseous water molecules onto the Cu(111) surface in the second step. STM images reveal that the three-fold symmetry of the adsorbate is well maintained; however, additional bright protrusions appear after the dosing of gaseous water molecules onto the surface (Figure 4b). The original protrusion, which is composed of the joint of the central and surrounding subunits, becomes four smaller protrusions after dosing the water vapor. Like ML' and MR', the high resolution structure of the tetramer's hydrate (T') is more easily obtained, which is potentially due to the water-functionalized STM tip. In addition to the three joints, the other portions of the ML' and MR' subunits exhibit features similar to the monomer's hydrates (i.e., ML' in Figure 2b and MR' in Figure 3b). These changes could be intuitively discovered via comparison of their three-dimensional images, which are shown in Figure 4. The hydration of valinomycin was further verified by the results from the valinomycin tetramers in the “two-step dosing” experiment.
Figure 4. STM images of an individual valinomycin tetramer and its hydrate prepared using the “two-step dosing” method.
(a) Typical STM image of an individual valinomycin tetramer (T) adsorbed onto the Cu(111) surface at 78 K. (b) Typical STM image of an individual valinomycin tetramer hydrate (T') adsorbed onto the Cu(111) surface at 78 K. Both of the STM image sizes are 8 nm × 8 nm. Both of the STM images were collected at −2 V and 0.01 nA. Their corresponding three-dimensional STM images are inserted in the upper-right corners.
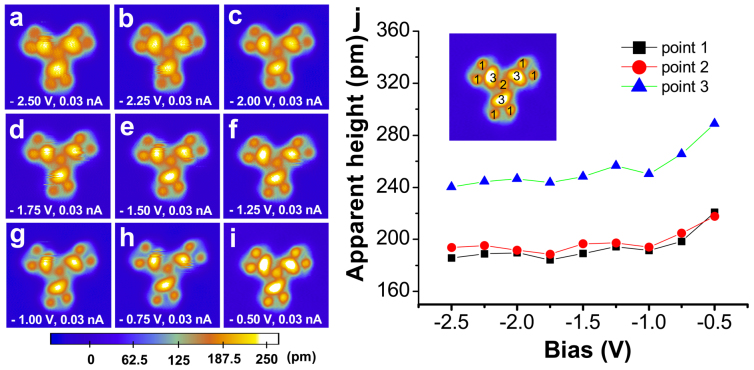
As previously discussed, the binding of water molecules accounts for the differences between the STM patterns of valinomycin and its hydrates. However, we should exclude the possibility that the changes in the STM patterns attribute to the diversity of molecular orbital contributions at the various imaging biases31. Therefore, the influence of the imaging conditions on the STM patterns was also evaluated. Valinomycin tetramer, which has a distinct, well-defined three-fold symmetrical adsorption configuration, adsorbed onto the Cu(111) surface and was easily observed and confirmed in the experiment. Therefore, a series of image conditions were applied to image an individual valinomycin tetramer in the control experiments. Figures 5a to 5i display a series of STM images for the same valinomycin tetramer collected at different imaging biases. The STM patterns of the valinomycin tetramer under the various biases are essentially the same even though the brightness and size of the protrusions are slightly different. All of the protrusions in the STM images were classified into three categories of points, as shown in Fig. S4 in the supplementary information. The observed height–bias curves (Figure 5j) illustrate that the contributions from different molecular orbitals cause the brightness diversification of the protrusions in the STM images. However, the new additional protrusions that appear in Figures 2b, 3b and 4b were not observed in these control experiments. This result suggests that the potential changes in the molecular orbital contributions under various imaging conditions do not lead to the observed change in the STM patterns in our studies.
Figure 5. Bias dependence of the apparent height of the valinomycin tetramer.
(a–i) A series of STM images (8 nm × 8 nm) of an individual valinomycin tetramer collected at the various biases. The STM imaging conditions are illustrated in the corresponding STM images. (j) Apparent height–bias curve of the valinomycin tetramer. Every apparent height value is the average value for all of the same types of protrusions on the same valinomycin tetramer.
Discussion
In our study, two types of sample preparation methods were employed to investigate the hydration of valinomycin. The “one-step dosing” method allows the water and valinomycin molecules to fully mix and interact. The intervention of water completely alters the adsorbed structure of valinomycin. The valinomycin–water interactions affect the adsorption conformation of valinomycin. However, the “two-step dosing” method provides a better platform for investigating the valinomycin–water interactions at the single-molecule level, which provides more precise information regarding the number of binding water molecules, binding sites and interaction energies. In the “two-step dosing” experiments, both the valinomycin molecules and their hydrates exhibited well-defined, nearly three-fold symmetrical STM patterns when they adsorbed onto the perfect Cu(111) terraces, as shown in Figures 2–4. These symmetric monomers and tetramers are the primary adsorbates, which were observed in our experiments. However, surface defects or impurities will inevitably capture some adsorbates because they are favorable anchoring sites32. Although these cases seldom occur, we recorded them in Fig. S5 in the supplementary information. However, only those adsorbates anchored on the perfect Cu(111) terrace would be a valuable platform for studying the binding of water molecules because their well-defined structure allows for analysis and comparison without the influence of other factors (such as surface defects). Therefore, we focused our single-molecule-level investigation of the hydration of valinomycin on the nearly three-symmetrical adsorbates.
Our studies were focused on valinomycin–water interactions. However, the presence of water on the Cu(111) surface is necessary to be discussed. The physical images of water on the various metal surfaces have been investigated because of the importance of water–metal interfaces33,34,35,36,37,38,39. Morgenstern et al. have demonstrated that the deposition of water (D2O) onto Cu(111) surfaces at cryogenic temperatures (<85 K) leads to three-dimensional amorphous clusters33,40,41,42,43,44. In our study, amorphous water clusters coexisted with valinomycin on the 78 K Cu(111) surface without annealing, as shown in Fig. S6 (see the supplementary information). By comparing the three continuously collected images, we found that the water clusters diffused across the 78 K Cu(111) surface. Fig. S6 also reveals that the three-fold symmetrical patterns became disordered, and the image became fuzzy after the sample was dosed with water molecules. This result suggests that the water molecules bind to the adsorbed valinomycin molecules and result in the formation of valinomycin–water complexes. After the sample was annealed at the RT carousel for 2 min, the water clusters disappeared from the Cu(111) surface, and the STM patterns of the valinomycin hydrate were clearly observed (Fig. S5 in the supplementary information).
Our DFT calculations indicate that the main contribution to the valinomycin–water interaction energy arises from the hydrogen bonds between the carbonyl groups of valinomycin and water rather than from van der Waals interactions, electrostatic energy and torsion energy. By analyzing the chemical structure (Figure 1a), we determined that valinomycin contains two types of carbonyls: ester carbonyls and amide carbonyls. As shown in Tables 1 and 2, the carbonyl oxygen is the energetically favorable binding site and has the most potential to participate in the formation of hydrogen bonds with water molecules15,16,18. In addition, water molecules have the ability to bind with ester oxygens and amide nitrogens. The results of theoretical calculations indicate that ML and MR contain 15 and 18 possible binding sites for water, respectively. For the formation of hydrates, water molecules occupy specific sites, which are determined by the surfactant nature of the adsorbed valinomycin molecules and the steric factor of valinomycin's exposed face15.
In summary, valinomycin can be hydrated via both the “one-step dosing” and “two-step dosing” experiments. The results of the “one-step dosing” experiments show that the valinomycin–water interactions strongly affect the conformation of valinomycin molecules, which is critical for the transport of K+ ions at water/membrane interfaces. The results from DFT calculations reveal that the formation of valinomycin hydrates result from hydrogen-bond interactions between valinomycin and water. The binding of water molecules might occur at carbonyl oxygen, ester oxygen and amide nitrogen sites in the valinomycin molecule. The detailed information regarding the valinomycin–water interactions obtained at the single-molecule level in the “two-step dosing” experiments is valuable for future studies of the role of water in the conformational transition of valinomycin under physiological conditions.
Our work provides an effective solution based on UHV-LT-STM to investigate the interaction between water and valinomycin, which exhibits low water solubility. Our work also demonstrates the application of STM for probing the high-resolution structures and properties of biological macromolecules at the single-molecule level. UHV conditions provide an ideal in vitro environment for investigating pure valinomycin–water interactions by avoiding the influence of other complex physiological factors, such as K+ ions and the cell environment. Our study reveals the potential for valinomycin hydration and a single-molecule perspective of the valinomycin–water interactions. These results provide fresh molecular insight for understanding the mechanism of K+ transport with the participation of water under physiological conditions.
Methods
Experimental Details
All of the experiments were performed in an Omicron scanning tunneling microscope ultra-high vacuum chamber with a base pressure of less than 2 × 10−10 mbar. A Cu(111) single crystal was cleaned in a preparation chamber by repeated cycles of neon-ion bombardment at room temperature and annealing at approximately 850 K. Valinomycin molecules (Aldrich, 90%) were vapor-deposited onto the Cu(111) surface at 78 K from a heated crucible and were subsequently flash-annealed at the RT carousel for 2 min to obtain their stable adsorption configurations. Then, the sample was placed back on the STM sample stage and cooled to 78 K for STM imaging. The valinomycin hydrates were prepared using two different methods, including the “one-step dosing” and “two-step dosing” methods. Ultra-pure water (H2O) was further treated under vacuum with freeze–pump–thaw cycles and was dosed onto the 78 K Cu(111) surface via a high-precision leak valve. In the “one-step dosing” experiments, valinomycin hydrates (valinomycin–water complexes) were prepared by the simultaneous co-evaporation of valinomycin and water onto the 78 K Cu(111) surface. Then, the sample was flash-annealed at the RT carousel for 2 min and cooled to 78 K on the STM sample stage overnight prior to the STM experiments being performed. In the “two-step dosing” experiments, uncomplexed valinomycin adsorbates (including monomers and tetramers) were first prepared as previously described. In the second step, gaseous water was dosed onto the 78 K Cu(111) surface, where valinomycin molecules had been previously anchored. The sample was flash-annealed at the RT carousel for 2 min and was subsequently placed on the 78 K STM sample stage and cooled overnight prior to the STM experiments being performed on the hydrates. All of the STM experiments were performed at 78 K, and all of the imaging conditions are reported in the figure captions. All of the STM images were collected and processed using the SPIP software.
Calculation details
The theoretical calculations were performed using density functional theory (DFT) as implemented in the DMol3 code45. In DMol3, the electronic wave function was expanded in a localized atom-centered basis set with each basis function defined numerically on a dense radial grid. We employed the double-numeric polarized (DND) basis sets46,47. The Perdew and Wang parameterization of the local exchange-correlation energy was applied in the local spin density approximation (LSDA) to describe the exchange and correlation48. The inner core electrons for Cu are represented by the DFT semilocal pseudopotential (DSPP), which was specifically developed for the DMol3 calculations, and the 19 electrons in Cu (i.e., those corresponding to the atomic levels 3s, 3p, 3d and 4s) were explicitly treated. Each basis function was restricted to within a cutoff radius of Rcut = 5.5 Å. In addition, spin-restricted wave functions were employed. A self-consistent field procedure was performed with a convergence criterion of 10−5 a.u. for the energy and electron density.
To evaluate the interaction energy between valinomycin and water on the Cu (111) surface, we adopted a three-layer Cu slab model as a simplified model. An optimized Cu lattice parameter (i.e., 3.6145 Å) was used in the calculation to reduce the effect of stress. Our calculated value is nearly equal to the experimental value of 3.6149 Å, which indicates that our DFT methods are suitable for this system. First, we optimized the system consisting of valinomycin adsorbed onto the Cu (111) surface. We then investigated the interactions between valinomycin and the water molecules. During the structural optimizations, we only allowed the valinomycin and water molecules to move until all of the forces decreased to within 1.0 × 10−4 hartrees per Bohr, and the Cu atoms in the slab were fixed in their bulk configurations. The interaction energy is defined as Ead = ECu–valinomycin-H2O − (ECu–valinomycin + EH2O).
Author Contributions
Y.C. and C.W. designed the project; Y.C. performed the experiments; K.D. performed the DFT calculations. Y.C., K.D., X.Q. and C.W. discussed the results and wrote the manuscript.
Supplementary Material
supplementary information
Acknowledgments
The authors thank Prof. Yanlian Yang, Dr. Rong Yang, Mr. Yuqun Xu and Dr. Xiaobo Mao (NCNST) for their discussion. This work was supported by the National Basic Research Program of China (2012CB933001, 2009CB930100) and National Natural Science Foundation of China (20933008, 91127043, 21261130090). Financial support from the National Basic Research Program of China (2011CB932800) and the Natural Science Foundation of Fujian Province, China (2012J05032) is also gratefully acknowledged. CAS Key Laboratory for Biological Effects of Nanomaterials and Nanosafety is also gratefully acknowledged.
References
- Gouaux E. & MacKinnon R. Principles of selective ion transport in channels and pumps. Science Signalling 310, 1461–1465 (2005). [DOI] [PubMed] [Google Scholar]
- Payandeh J., El-Din T. M. G., Scheuer T., Zheng N. & Catterall W. A. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486, 135–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil F. I., Leonetti M., Muir T. W. & MacKinnon R. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science Signalling 314, 1004–1007 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosteson D. C., Cook P., Andreoli T. & Tieffenberg M. The effect of valinomycin on potassium and sodium permeability of HK and LK sheep red cells. The Journal of general physiology 50, 2513–2525 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan S., Nair C., Easwaran K. & Vijayan M. A novel conformation of valinomycin in its barium complex. Nature 286, 640–641 (1980). [DOI] [PubMed] [Google Scholar]
- Karle I. L. & Flippen-Anderson J. L. New conformation exhibiting near-threefold symmetry for uncomplexes valinomycin in crystals from dimethyl sulfoxide. Journal of the American Chemical Society 110, 3253–3257 (1988). [Google Scholar]
- Karle I. L. Conformation of valinomycin in a triclinic crystal form. Journal of the American Chemical Society 97, 4379–4386 (1975). [DOI] [PubMed] [Google Scholar]
- Duax W., Hauptman H., Weeks C. & Norton D. Valinomycin crystal structure determination by direct methods. Science 176, 911–914 (1972). [DOI] [PubMed] [Google Scholar]
- Wang F., Zhao C. & Polavarapu P. L. A study of the conformations of valinomycin in solution phase. Biopolymers 75, 85–93 (2004). [DOI] [PubMed] [Google Scholar]
- Feigenson G. & Meers P. 1H NMR study of valinomycin conformation in a phospholipid bilayer. Nature 283, 313–314 (1980). [DOI] [PubMed] [Google Scholar]
- Grossman M. et al. Correlated structural kinetics and retarded solvent dynamics at the metalloprotease active site. Nature Structural & Molecular Biology 18, 1102–1108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin M. Do we underestimate the importance of water in cell biology? Nature Reviews Molecular Cell Biology 7, 861–866 (2006). [DOI] [PubMed] [Google Scholar]
- Angel T. E., Chance M. R. & Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proceedings of the National Academy of Sciences 106, 8555–8560 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester T. R., Smith W. & Clarke J. H. R. Antibiotic activity of valinomycin Molecular dynamicssimulations involving the water/membrane interface. J. Chem. Soc., Faraday Trans. 93, 613–619 (1997). [Google Scholar]
- Khutorskii V., Kamenchuk A. & Shcheckhin I. Theoretical investigation of the stereochemical structure of valinomycin and its hydration. Theoretical and Experimental Chemistry 23, 334–339 (1987). [Google Scholar]
- Gupta S., D'Mello R. & Chance M. R. Structure and dynamics of protein waters revealed by radiolysis and mass spectrometry. Proceedings of the National Academy of Sciences 109, 14882–14887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester T., Smith W. & Clarke J. Molecular dynamics simulations of valinomycin and its potassium complex in homogeneous solvents. Biophysical journal 71, 544–553 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell E., Funck T. & Eggers F. Structure and dynamic properties of ion-specific antibiotics. Membranes 3, 1–126 (1975). [PubMed] [Google Scholar]
- Mao X. et al. Sequence effects on peptide assembly characteristics observed by using scanning tunneling microscopy. Journal of the American Chemical Society 135, 2181 (2013). [DOI] [PubMed] [Google Scholar]
- Uchihashi T., Iino R., Ando T. & Noji H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–758 (2011). [DOI] [PubMed] [Google Scholar]
- Kodera N., Yamamoto D., Ishikawa R. & Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 468, 72–76 (2010). [DOI] [PubMed] [Google Scholar]
- Mao X. B. et al. Beta structure motifs of islet amyloid polypeptides identified through surface-mediated assemblies. Proceedings of the National Academy of Sciences 108, 19605–19610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. et al. Chaperon-mediated single molecular approach toward modulating Aβ peptide aggregation. Nano letters 9, 4066–4072 (2009). [DOI] [PubMed] [Google Scholar]
- Deng Z. et al. A Close Look at Proteins: Submolecular Resolution of Two-and Three-Dimensionally Folded Cytochrome c at Surfaces. Nano letters 12, 2452–2458 (2012). [DOI] [PubMed] [Google Scholar]
- Otero R. et al. Elementary structural motifs in a random network of cytosine adsorbed on a gold (111) surface. Science 319, 312–315 (2008). [DOI] [PubMed] [Google Scholar]
- Tanaka H. & Kawai T. Partial sequencing of a single DNA molecule with a scanning tunnelling microscope. Nature nanotechnology 4, 518–522 (2009). [DOI] [PubMed] [Google Scholar]
- Uchihashi T., Kodera N. & Ando T. Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nature Protocols 7, 1193–1206 (2012). [DOI] [PubMed] [Google Scholar]
- Hahn J. R. & Ho W. Single Molecule Imaging and Vibrational Spectroscopy with a Chemically Modified Tip of a Scanning Tunneling Microscope. Physical Review Letters 87, 196102 (2001). [DOI] [PubMed] [Google Scholar]
- Gross L., Mohn F., Moll N., Liljeroth P. & Meyer G. The chemical structure of a molecule resolved by atomic force microscopy. Science 325, 1110–1114 (2009). [DOI] [PubMed] [Google Scholar]
- Sautet P. & Bocquet M.-L. Shape of molecular adsorbates in STM images: A theoretical study of benzene on Pt (111). Physical Review B 53, 4910 (1996). [DOI] [PubMed] [Google Scholar]
- Chen Y. et al. Defect-Induced tert-Butylamine Molecular Chains on Cu (111) Surface Observed with Scanning Tunneling Microscopy. The Journal of Physical Chemistry C 113, 6725–6729 (2009). [Google Scholar]
- Hinch B. & Dubois L. Stable and metastable phases of water adsorbed on Cu (111). The Journal of chemical physics 96, 3262 (1992). [Google Scholar]
- Mitsui T., Rose M., Fomin E., Ogletree D. F. & Salmeron M. Water diffusion and clustering on Pd (111). Science 297, 1850–1852 (2002). [DOI] [PubMed] [Google Scholar]
- Salmeron M., Verdaguer A., Bluhm H. & Sacha G. Molecular structure of water at interfaces: Wetting at the nanometer scale. Chemical reviews 106, 1478–1510 (2006). [DOI] [PubMed] [Google Scholar]
- Carrasco J., Hodgson A. & Michaelides A. A molecular perspective of water at metal interfaces. Nature Materials 11, 667–674 (2012). [DOI] [PubMed] [Google Scholar]
- Carrasco J. et al. A one-dimensional ice structure built from pentagons. Nature Materials 8, 427–431 (2009). [DOI] [PubMed] [Google Scholar]
- Yamada T., Tamamori S., Okuyama H. & Aruga T. Anisotropic water chain growth on Cu (110) observed with scanning tunneling microscopy. Physical Review Letters 96, 036105 (2006). [DOI] [PubMed] [Google Scholar]
- Morgenstern K. & Rieder K.-H. Dissociation of water molecules with the scanning tunnelling microscope. Chemical physics letters 358, 250–256 (2002). [Google Scholar]
- Yamamoto S. et al. Hydroxyl-induced wetting of metals by water at near-ambient conditions. The Journal of Physical Chemistry C 111, 7848–7850 (2007). [Google Scholar]
- Michaelides A. & Morgenstern K. Ice nanoclusters at hydrophobic metal surfaces. Nature Materials 6, 597–601 (2007). [DOI] [PubMed] [Google Scholar]
- Mehlhorn M., Carrasco J., Michaelides A. & Morgenstern K. Local investigation of femtosecond laser induced dynamics of water nanoclusters on Cu (111). Physical Review Letters 103, 026101 (2009). [DOI] [PubMed] [Google Scholar]
- Stähler J. et al. Impact of Ice Structure on Ultrafast Electron Dynamics in D2O Clusters on Cu (111). Physical Review Letters 98, 206105 (2007). [DOI] [PubMed] [Google Scholar]
- Morgenstern K. & Rieder K.-H. Formation of the cyclic ice hexamer via excitation of vibrational molecular modes by the scanning tunneling microscope. The Journal of chemical physics 116, 5746 (2002). [Google Scholar]
- Becke a. A. A multicenter numerical integration scheme for polyatomic molecules. The Journal of chemical physics 88, 2547 (1988). [Google Scholar]
- Delley B. From molecules to solids with the DMol approach. The Journal of chemical physics 113, 7756 (2000). [Google Scholar]
- Delley B. An all-electron numerical method for solving the local density functional for polyatomic molecules. The Journal of chemical physics 92, 508 (1990). [Google Scholar]
- Perdew J. P. & Wang Y. Accurate and simple analytic representation of the electron-gas correlation energy. Physical Review B 45, 13244 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information