Abstract
The intron 22 inversion found in up to 50% of severe hemophilia A patients results from a recombination between three intron 22 homologous copies (int22h). This study evaluated the implication of these copies in the formation of extended duplications comprising exons 1–22 of the factor 8 (F8) gene and their association with hemophilia and mental retardation. Two hemophilic patients with moderate and severe phenotypes and a third nonhemophilic patient with developmental delay were studied. All exhibited a duplication of F8 gene exons 1–22 identified by multiplex ligation-dependent probe amplification along with abnormal patterns on Southern blotting and unexpected long-range PCR amplification. Breakpoint analysis using array comparative genomic hybridization was performed to delimit the extent of these rearrangements. These duplications were bounded on one side by the F8 intragenic int22h-1 repeat and on the other side by extragenic int22h-2 or int22h-3 copies. However, the simultaneous identification of a second duplication containing F8 gene exons 2–14 for the moderate patient and the classical intron 22 inversion for the severe patient are considered in this study as the genetic causal defects of hemophilia. This study shows that the well-known int22h copies are involved in extended duplications comprising F8 gene exons 1–22. These specific duplications are probably not responsible for hemophilia and intellectual disability, but should be carefully considered in genetic counseling, while continuing to investigate the causal mutation of hemophilia.
Keywords: hemophilia, F8 gene, genomic duplication, CGH array, int22h repeats
Introduction
The X-linked coagulation disorder hemophilia A (HA) is caused by mutations in the factor 8 (F8) gene, which encodes coagulation factor VIII (FVIII) and is located at Xq28, the telomeric end of the long arm of the X chromosome. The most recurrent mutations associated with severe HA are intron 1 and 22 inversions in the F8 gene. During male gametogenesis, both inversions are mediated by intrachromosomal homologous recombination between the highly homologous copies located in intron 1 or 22 and other extragenic copies positioned more telomerically outside the gene.1, 2, 3, 4
Point mutations, deletions, and insertions were detected in all 26 exons of the F8 gene in HA patients, causing phenotypes of variable severity characterized by complete or partial deficiency of circulating FVIII. Large deletions in the F8 gene involving one or more exons account for about 5% of all severe HA cases,5, 6 (HADB (aka HAMSTeRS) the Hemophilia A Database, http://hadb.org.uk/). The identification of a large duplication comprising one or more F8 gene exons was recently made possible following the introduction of multiplex ligation-dependent probe amplification (MLPA) technology, with an estimated occurrence of 1%.7, 8 Large duplications are associated with different severity of HA depending on the localization, length of exons involved, and on whether the duplications generate an in-frame or out-frame protein.9, 10
In a large HA cohort from Belgium undergoing diagnostic analysis,11 two male patients with moderate and severe phenotypes were found with an abnormal band pattern of intron 22 inversion on Southern blotting (SB) and large duplications involving F8 gene exons 1–22 using MLPA. These cases were further analyzed using high-resolution custom array comparative genomic hybridization (CGH). As supported by a third nonhemophilic patient with intellectual disability but the same duplication of F8 gene exons 1–22 by MLPA, a comprehensive mechanistic model was proposed to explain these complex rearrangements.
Materials and methods
Case histories
Case 1
This 25-year-old male was diagnosed with HA at 4 months of age. FVIII concentrations measured by one-stage and chromogenic assays were between 2 and 4%, and thus considered as moderate HA. The patient had a history of recurrent knee, elbow, and ankle hemarthroses since infancy, without inhibitor development during replacement therapy. HA was observed in at least three generations. DNA samples from his obligate carrier mother and cousins of his maternal obligate carrier were tested.
Case 2
This 18-year-old patient was the only son of nonconsanguineous parents without a family history of coagulation disorders. Severe HA was diagnosed at the age of 1 year on account of large hematomas. The patient had a history of recurrent knee, elbow, and ankle hemarthroses since infancy. No inhibitor development occurred during replacement therapy. His mother was diagnosed as a carrier female.
Case 3
This 21/2-year-old patient was prematurely born at 342/7 weeks of gestation. At birth, Pierre Robin sequence with micrognathia, cleft palate, glossoptosis, and upper airway obstruction was diagnosed. The patient was developmentally delayed: at 11 months he sat upright and at 21/2 years started walking, saying just a few words. Facial features comprised a high forehead, long face, upper eyelid fullness, and convergent strabismus. The nasal bridge was broad, and anteversion of the nostrils was observed together with a long philtrum, open mouth, thin upper lip, and microrethrognatia. Large ears, fetal fingertip pads, rocker bottom feet, and pes valgus were noted. Array CGH revealed a large duplication at Xq28 located in the intron 22 homologous region (int22h) between int22h-1 and int22h-2 repeats. The patient had no coagulation disorder, with normal FVIII activity.
FVIII activity and FVIII inhibitor screening
FVIII activity levels (FVIII:C) were determined by standard one-stage coagulation and chromogenic assays. FVIII inhibitor screening and titration were performed according to Bethesda/Nijmegen modification assays.
DNA collection
A blood sample (5–10 ml) was collected in an EDTA tube, and genomic DNA purification from peripheral leukocytes was performed using salting out procedures.12
Detection of intron 1 and 22 inversions
The intron 22 inversion was detected using both SB11 and a modified long-range PCR (LR-PCR) adapted from Bagnall et al.13 Details of this method are available upon request. To interpret the abnormal band patterns obtained using SB, a combination of primers was analyzed in separate reactions: H1-F and H1-R for detecting int22h-1; H2-F and H2-3 R for int22h-2; H3-F and H2-3 R for the int22h-3 repeat; H1-F and H2-3 R in addition to H3-F and H1-R for detecting rearrangements between int22h copies.
The method for the detection of intron 1 inversion was performed as previously described.4
Multiplex ligation-dependent probe amplification
MLPA was performed as previously described.11
Array CGH, hybridization procedures, and analysis
High-resolution custom array CGH (Agilent 8 × 15 K, Santa Clara, CA, USA) targeting duplications was designed using e-arrays (Agilent). A backbone set of 2247 probes was selected on chromosome X, whereas 10 384 probes were chosen in a targeted region between position 153 346 981 Mb and the q-telomeric end of chromosome X (hg19 (GRCh37) assembly). Array CGH was performed according to the manufacturer's instructions using 350 ng of genomic DNA in a random priming labeling reaction (Bioprime Total Genomic Labeling Module, Invitrogen, Paisley, UK). Expected log ratio values for duplication in males and females were 1 and 0.58, respectively. Aberration calls were performed using ADM2, with a threshold of 6.0 in Genomic Work Bench. Incorporation of nonunique probes in the analysis may slightly modify the aberration breakpoints. These alternative breakpoints are represented as light gray boxes in Figure 2.
Quantitative PCR to validate duplication breakpoints
Amplifications distal to repeat int22h-3 were not easily assessed using microarray analysis given the highly repetitive nature of the genomic interval between int22h-3 and the TMLHE gene. To confirm the array results, a quantitative PCR (qPCR) system was developed. It was impossible to design a single-site amplification site. The selected primers (forward: 5′-TGTGCCAAAGTCCAGAAATAGT-3′ reverse: 5′-CCAACAGAGAAAGTAGCAGGAA-3′) amplified two loci, namely chrX: 154 575 182–154 575 278 (proximal to repeat a2) and chrX: 154 724 935–154 725 031 (distal to repeat a3). Consequently, in a male, if the duplication encompassed a single site, a genome equivalent to 0.75 was expected (three copies vs four in normal females); if both sites were duplicated, a genome equivalent to 1 was expected. In a female carrier with duplication at one or two sites, a genome equivalent to1.25 or 1.5, respectively, was expected (vs normal female DNA). Normalization and qPCR data analysis were conducted according to Hoebeeck et al.14 using two reference genes. An additional gene on chromosome X (MECP2) was used as negative control.
Multiplex PCR assays
Multiplex amplicon quantification (MAQ) is a robust, rapid, and easy-to-perform method for analyzing specific copy-number variations. The method, performed according to the manufacturer's protocol, consists of fluorescently labeled multiplex PCR with amplicons in the copy-number variations (target amplicons) and amplicons with a stable copy number. Two series of five primer pairs extending from position 154 147 798 to 154 344 960 on chromosome X for the first series, and from 154 441 804 to 154 664 293 on chromosome X for the second were designed to target F8 locus (Table 1). The first amplicon was located in F8 gene intron 14, and the last between int22h-2 and int22h-3 repeats.
Table 1. Localization, primer sequences, and sizes of PCR products for studying copy number changes at the F8 locus.
| Amplicon | Size (bp)a | Startb | Stopb | Primer forward (5′-3′)c | Primer reverse (5′-3′)d |
|---|---|---|---|---|---|
| MAQ01 assay | |||||
| amp1_1 | 91+30 | 154 147 798 | 154 147 888 | AGGAAACTTGGGCAGCAGA | AACAATGCCACCAAGCAGA |
| amp1_2 | 180+30 | 154 213 190 | 154 213 369 | CAGCAAAGTAGGCACACAGC | CCTGATGCCTCAAGCTAACC |
| amp1_3 | 143+30 | 154 282 491 | 15 428 2633 | GAAACAGAATTGCCTCTGTAACC | CCTGATGGGAAGGAGGATG |
| amp1_4 | 70+30 | 154 318 777 | 154 318 846 | AATAAGTTTGTCAGCTTTCAGGATG | CAAACACAAGAACCAGGAACC |
| amp1_5 | 295+30 | 154 344 666 | 154 344 960 | ATCTTTCAAGGGCAAGGATG | GTGGCAAAGCAGCACTGA |
|
Amplicon |
Size (pb)a |
Startb |
Stopb |
Primer forward (5′-3′) |
Primer reverse (5′-3′) |
| MAQ02 assay | |||||
| amp2_1 | 136+30 | 154 441 669 | 154 441 804 | GTGGAGATTGGGACAGAACC | GATATCCTTAGCAACTTCCAGAACC |
| amp2_2 | 188+30 | 154 470 742 | 154 470 929 | GATTTGAAACACCTCACTCATAACC | TCACCTAACCCAGACAGAAATG |
| amp2_3 | 86+30 | 154 563 105 | 154 563 190 | CTCCTCACATAACAAGCACACAG | CTTCTACAATGGACAAACATCACAG |
| amp2_4 | 165+30 | 154 630 587 | 154 630 751 | GAGTGCTTGAGGAAAGAGGCTA | TGGCAAACACATCCATGC |
| amp2_5 | 107+30 | 154 664 187 | 154 664 293 | TTTAAGCTAGAGCAGCCGTGT | AGGGAAGAGGAGAGGCAGA |
F8, factor 8; MAQ, multiplex amplicon quantification.
The size of amplicons corresponds to the sum of the distance between primer pairs and the length of the tail added to the locus-specific primer pairs for PCR amplification (30 bp in total).
The positions of primers on chromosome X are based on the hg19 assembly.
A tail used for PCR amplification was added at the 5′ end of these locus-specific primers (5′-AGCGGATAACAATTTCACACAGG-3′).
A tail used for PCR amplification was added at the 5′ end of these locus-specific primers (5′-GTTTCTT-3′).
MAQ analysis was performed according to the manufacturer's protocol. Details of this method are available upon request. Raw data generated by fragment analysis on a capillary sequencer were analyzed using MAQ-S (Multiplicom (http://www.multiplicom.com)), a specially designed software program, to calculate and visualize the normalized peak area or dosage quotient, which reflected the copy number of each target amplicon.
Results
For the three patients with chromosomal rearrangements affecting gene F8, data including gene dosage analysis obtained by SB and LR-PCR (Figure 1), MLPA (data not shown), targeted array CGH, and MAQ (Figure 2) are described below case-by-case. The extent of the duplication was confirmed using qPCR. Owing to the highly repetitive genomic sequence around the int22h-2 and int22h-3 repeats, alternative quantification methods were required to validate the extent of the rearranged regions that were not clearly defined using array CGH (Figure 2).
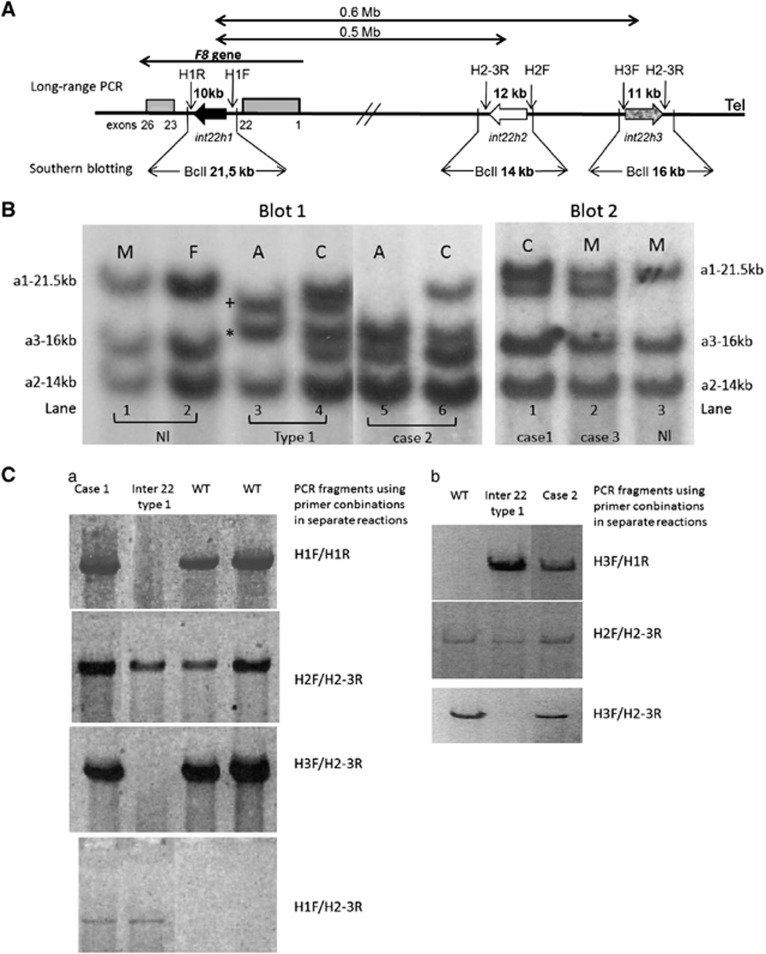
Figure 1.
Results of SB and LR-PCR. (A) Schematic representation of int22h copies at the Xq28 locus according to Bagnall et al.13 (figure not to scale). The large arrows indicate the position of the int22h copies. The arrowhead represents the orientation of the DNA sequence. Gray boxes correspond to exons 1–22 and 23–26 of the F8 gene. The location of primer-binding sites and expected LR-PCR product size are drawn above, while the expected BclI restriction fragments sizes obtained by SB are indicated below. (B) Results of SB. Blot 1 lanes 1 and 2 correspond to a normal pattern. The probe hybridized with three fragments. The largest 21.5-kb BclI fragment contains 9.5 kb segment of intron 22 (int22h-1 or a1). The int22h-3 repeat or a3, considered the most telomeric, is located in the 16-kb BclI fragment. The Int22h-2 repeat or a2 is positioned in the 14-kb BclI fragment. Lanes 3 and 4: classical inversion of intron 22 type 1 in a male (A) and in a female carrier (C). +Indicates the rearranged 20 kb int22h-1/3; *is associated with the rearranged 17.5-kb int22-3/1 centromeric repeat. For further details, see Figure 3A. Lanes 5 and 6: case 2 patient with his carrier mother. Fragments corresponding to int22h-2 and int22h-3 repeats have normal size. However, the band associated with int22h-2 copies or a2 appears duplicated. The fragment corresponding to 17.5 kb (also duplicated) indicates the presence of the classical inversion of intron 22 type 1. However, the absence of the 20-kb fragment suggests a more complex rearrangement. Blot 2 lane 1: carrier mother of case 1 patient. Normal pattern with an additional 20-kb fragment. Lane 2: case 3 without the HA phenotype. Case 1 patient presented the same pattern as case 3 (data not shown). Lane 3 corresponds to a normal pattern. A, HA affected patient; C: carrier HA female; F, female; M, male. (C) Results of LR-PCR. As shown in a wild-type patient (WT), PCR amplification was only obtained using combinations of primers: H1-F/H1-R for detecting int22h-1; H2-F/H2-3R for int22h-2; and H3-F/H2-3R for int22h-3 repeat. For a patient harboring the classical inversion of intron 22 type 1, PCR fragments could be only obtained using primers H3-F/H1-R and H1-F/H2-3R, indicating the recombination between the intragenic int22h-1 repeat with its extragenic int22h-3 homologs. As shown in Figure 3A, amplification of the int22h-2 copy with primers H2-F/H2-3R is conserved. For case 1 (a), PCR amplification of the three int22 copies was obtained as the WT patient. However, PCR obtained with H1-F/H2-3R indicates a rearrangement between int22h copies. For case 2 (b), amplification with primers H3-F/H1-R and H1-F/H2-3R indicates the inversion of intron 22 type 1. However, PCR obtained with primers H3-F/H2-3R shows a more complex recombination.
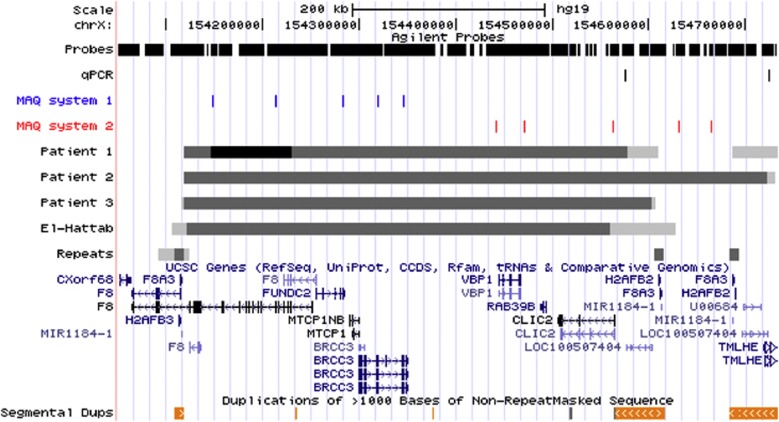
Figure 2.
Fine-mapping and breakpoint analyses using array CGH or multiplex PCR assays. Illustration of the rearrangements of the Xq28 region using custom tracks in the UCSC browser. The scale is provided above. The section ‘probes' shows the distribution of probes on the targeted custom array in the selected region (6526 probes in the selected window). Each vertical line corresponds to a probe. Sections ‘qPCR' and ‘MAQ systems 1 and 2' illustrate the localization of the qPCR and PCR systems used to confirm copy number changes (see article and Table 1 for details). Thereafter follow the rearrangement intervals as determined from the array CGH results for patients 1, 2, and 3 and from the 2011 publication by El-Hattab et al. (patients with duplication or deletion in fetus and carrier females). Clearly duplicated (or deleted) intervals are depicted in dark gray, whereas regions not clearly rearranged are shown in light gray. The region with three copies (‘triplication') in female patient 1 is shown in black. The deletion identified by Pegoraro et al. is not provided as no fine-mapping information is available. The section ‘repeats' show the int22h-1, int22h-2, and int22h-3 repeats in dark gray as well as intron 22 of F8 (gray rectangle). Genes and segmental duplications (>99% similarity) are depicted using information provided by the University of California, Santa Crus (University of California, Santa Crus genome browser. http://genome.ucsc.edu).
Case 1
A normal profile with an additional 20-kb fragment was observed using SB. The normal amplification of int22h copies was obtained in LR-PCR using repeat-specific primers pairs. However, amplification was also obtained with the combination of H1-F and H2-3R primers. A large duplication of F8 gene exons 1–22 was identified using MLPA assay, along with a second duplication of exons 2–14. Owing to insufficient patient DNA, DNA samples from his obligate carrier mother, presenting the same SB and MLPA patterns, were used for array CGH analysis. A 0.5-Mb duplication (log ratio=0.520) extending from intron 22 of factor F8 (position 154 117 967 bp on genome build hg19) to repeat sequences int22h-2 (position 154 609 974 bp) was revealed in the vicinity of the H2AFB2 and F8A2 genes, which was confirmed by MAQ assay. Higher log ratios between positions 154 146 529 and 154 229 174 bp (0.870) suggested that this region within the duplication was present in three copies (i.e., ‘second duplication'). The breakpoints of this second 82-kb duplication fell within intron 14 and intron 1 of F8 gene, respectively.
Case 2
An abnormal pattern was observed using SB, with normal fragments containing int22h-2 and int22h-3 repeats as well as a 17.5-kb band (probably duplicated), which was associated with one of the two rearranged fragments in an intron 22 type 1 inversion. This classic inversion was confirmed by the int22h-1 rearranged 11.5-kb fragment via LR-PCR. A large duplication of F8 gene exons 1–22 was identified using MLPA. Array CGH analysis revealed a large duplication of 0.6 MB extending from the int22h-1 to int22h-3 repeats, or even further distally and so nearer the TMLHE gene. The region between the int22h-2 and int22h-3 repeats was clearly duplicated, but the highly repetitive nature of the region distal to h3 did not clarify whether this duplication extended more distally, given the lack of a unique probe in this interval. Thus, the duplication ranged from 154 118 907 to 154 722 370 or 154 731 487 bp on chromosome X. qPCR using this male patient's DNA with primers hybridizing to both sides of int22h-2 and int22h-3 repeats provided a genome equivalent of 0.83 vs normal female DNA. This suggested that the region close to the int22h-2 repeat was duplicated, whereas the duplication did probably not extend beyond the int22h-3 repeat in the TMLHE gene region (two copies close to int22h-2+1 copy close to int22h-3 vs four copies in the reference female).
The patient's mother had the same MLPA and SB profiles.
Case 3
The patient's SB results were the same as case 1. A large duplication of F8 gene exons 1–22 was identified using MLPA. His mother presented the same SB and MLPA patterns. Array CGH for both mother and son showed a large duplication between positions 154 116 088 and 154 606 719, which was located between int22h-1 and int22h-2 repeats (confirmed by MAQ analysis), whereas the interval between int22h-2 and int22h-3 repeats was not duplicated. qPCR performed using patient DNA provided a genome equivalent of 0.79, in line with a duplication in the proximal region of int22h-2 repeat, but not distal to int22h-3.
MAQ analysis on normal DNA controls
Multiplex PCR assays were developed to detect the same 0.5- or 0.6-Mb duplication extending from int22h-1 to int22h-2 or -3 repeats on 420 X chromosomes (84 females and 252 males). No duplication between int22h-1 and extragenic int22h sequences was found. However, one 0.5-Mb heterozygous deletion between int22h-1 and int22h-2 repeats was identified in a normal female. This deletion including F8 gene exon 1–22 was confirmed via MLPA.
Discussion
In a large cohort of 148 Belgian HA families analyzed for F8 gene mutations, we identified a duplication of F8 gene exons 1–22 in two male patients with moderate and severe HA.11 A third duplication of F8 gene exons 1–22 in a nonhemophilic patient suffering from intellectual disability and Pierre Robin syndrome (case 3) was added to this study. Case 1 patient with moderate HA presented a second duplication of F8 gene exons 2–14, whereas case 2 with severe HA harbored the classic inversion of intron 22 type 1. Breakpoint array CGH analyses revealed that rearrangements were delimited by the intragenic int22h-1 repeat on one side and on the other side, by int22h-2 for cases 1 and 3, and int22h-3 copies for case 2. These results demonstrate that partial F8 gene segments may be duplicated by mechanisms involving the same DNA repeats as those involved in the classical intron 22 inversion. Based on these results, we propose a comprehensive modeling of the mechanisms involved.
Xq28 locus is considered a region at high risk of genomic instability, with micro-rearrangements (deletion or duplication) described, for example, in adrenoleukodystrophy,15 X-linked mental retardation syndrome (MECP2),16 and HA.1, 2 Indeed, the F8 gene intron 22 contains a 9.5-kb region known as copy int22h-1, which is outside the gene and situated ∼0.5 and 0.6 Mb more telomerically to the two other highly homologous copies, namely int22h-2 and int22h-3.1, 2 The int22h-1 and int22h-3 copies lying in opposite directions are responsible for recurrent intrachromosomal rearrangements involving the F8 ‘distal' inversion or inversion type 1. Recently, int22h-1 and int22h-2 sequences were reported to display the same orientation,17 which may cause, theoretically, highly deleterious 0.5 Mb deletions or duplications in the case of recombination between misaligned sister chromatids or homologous chromosomes. It has been suggested that they are either extremely rare or nonexistent13, 18, 19 and could cause problems in mutation testing in HA.20 However, a very recent publication21 reported novel 0.5 Mb duplications between int22h-1 and int22h-2 copies in three unrelated males, presenting cognitive impairment, behavioral abnormalities, recurrent infections, and characteristic facial features.
Our report describes the same rearrangement in two patients (cases 1 and 3), both presenting a large duplication of F8 gene exons 1–22 extending from int22h-1 to int22h-2 repeats. The presence of an extra 20-kb BclI band detected using SB is likely to be the consequence of genomic rearrangement, which produced the altered int22h restriction fragment as confirmed by LR-PCR (Figure 3). Additionally, we describe a more complex not previously described rearrangement in a severe HA patient (case 2) presenting a large duplication estimated at 0.6 Mb extending from int22h-1 to int22h-3 repeats. If the large duplication in the F8 locus in cases 1 and 3 could be the consequence of int22h-1 misaligning with a similarly oriented and more telomeric int22h repeat from the sister chromatid via a nonallelic homologous recombination mechanism, we presume that numerous different duplications are possible depending on the initial alignments. In case 2, our results suggest a nonallelic recombination in the female carrier between a normal X chromosome and intron 22 inversion of the sister X chromosome, resulting in a large duplication between int22h-1 and int22h-3 copies as well as fragments compatible with an intron 22 type 1 (Figure 3). This explains why this duplication extended between repeats int22h-1 and int22h-3.
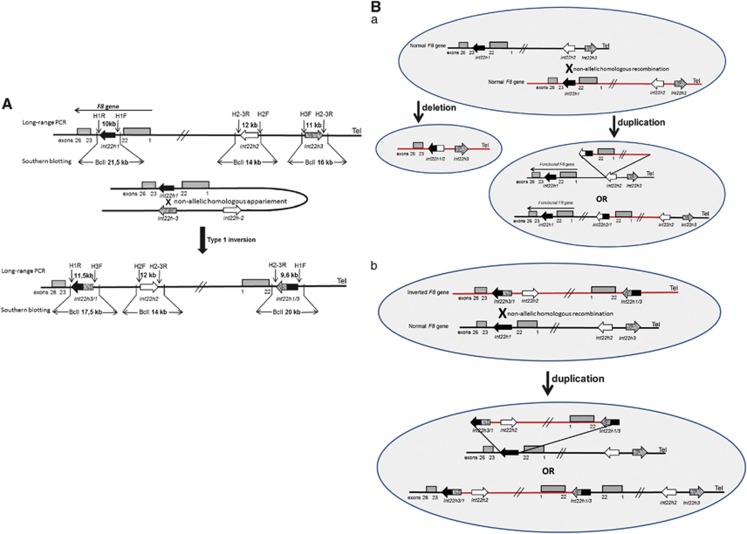
Figure 3.
Nonallelic homologous recombination (NAHR) events between int22h copies (figures not to scale). (A) Schematic mechanism of the recurrent int22h inversion type 1 in severe HA. This mode of rearrangement is facilitated in male meiosis, as this mechanism involves flipping the end of the X chromosome, which is inhibited in female germ cells by X chromosome pairing. (B) Proposed mechanisms for the formation of complex duplication/deletion in cases 1 and 3 (a) and in case 2 (b). (a) Misalignment of two normal maternal X chromatids during meiosis, which may result in a NAHR mechanism in an unequal crossing over that provides copies with the duplication of 0.5 Mb between the int22h-1 and int22h-2 copies or the deletion of 0.5 Mb. The duplication maintains a normal and functional copy of the F8 gene. Normal LR-PCR products and BclI restriction fragment sizes using SB are obtained for the three int22h copies. However, the duplication generates an additional fragment of 20 kb observed on SB as well as amplification with the combination of primers, H1-F and H2-3R. (b) Misalignment between one normal chromatid and another chromatid with an inversion of intron 22 in a carrier female during meiosis. Unequal crossing over by a NAHR mechanism produces duplication between int22h-1 and int22h-3 copies instead of duplication between int22h-1 and int22h-2 in the mechanism proposed in B (a) of this figure. This rearrangement involves the presence of two incomplete sequences of exons 1–22 of the F8 gene, two copies of int22h-2, one copy of the int22h-3 repeats, one copy associated with the rearranged fragment of 17.5 kb in the int22-3/1 repeat, and one copy associated with another 17.5 kb rearranged fragment corresponding to the newly produced int22-1/3 repeat.
Whether these 0.5/0.6 Mb duplications mediated by int22h-1/int22h-2 or -3 rearrangement are associated with HA is uncertain. After excluding other causal mutations in F8 gene exons via bidirectional sequencing, we assume that the HA phenotype of case 1 was associated with a second duplication of F8 gene exons 2–14. Numerous publications have reported the causality of large intragenic duplications, especially in HA patients with different phenotypes.7, 22, 23 Despite using different primer combinations, it was impossible to characterize the duplication breakpoints for this patient and his relative. In case 2, the recombination between the normal X chromosome and intron 22 inversion of the sister X chromosome produced a more complex rearrangement without maintaining a complete copy of the F8 gene (Figure 3). Again, after complete gene sequencing we concluded that the severe HA phenotype of case 2 was caused by classical intron 22 inversion. In the study by El-Hattab et al.21, the index case of family 3 presented, along with his intellectual disability, a minor HA type with FVIII levels estimated at 24%. The complete sequencing of the F8 gene revealed hemizygosity for the point mutation c.6089G>A (p.Ser2030Asn), which was associated with mild HA (HADB (aka HAMSTeRS) http://hadb.org.uk/). According to Figure 3Ba and as suggested by Bagnall et al.13, a complete copy of the F8 gene is preserved after the formation of the 0.5-Mb duplication mediated by int22h-1/int22h-2 rearrangement, thus producing normal FVIII levels. We propose that males or females with such an extending 0.5-Mb duplicated rearrangement would not be at risk of HA, as they maintain a normal copy of the F8 gene unless if a causal mutation, such as the large duplication of exons 2–14 in case 1 or point mutation in family 3 of El-Hattab et al.'s study, occurs in the normal F8 copy.
El-Hattab et al. suggested that the 0.5-Mb duplication mediated by int22h-1/int22h-2 rearrangement was associated with a novel X-linked intellectual disability. However, several arguments are in contradiction with this conclusion. Indeed, no intellectual disability was observed in our hemophilic cases nor was there any developmental delay noted in the mothers and their relatives (obligate carrier grandmother, affected uncle, and his two obligate carrier cousins for the first case; carrier grandmother for the third case), whereas in El-Hattab et al.'s study, carrier mothers exhibited learning difficulties despite the abnormal X chromosome being preferentially inactivated. In our study, chromosome X-inactivation analysis at the androgen receptor24, 25 showed skewed chromosome X inactivation for the case 1 mother (ratio 87:13) but no skewed chromosome X inactivation for the case 3 mother (ratio 80:20). Case 3 displayed dysmorphic features identical to those described in the three El-Hattab families, with several dissimilar features. Additionally, none of the six known genes localized in the 0.5/0.6-Mb duplicated intervals (Figure 2) were shown to be associated with X-linked intellectual disability. These genes exhibit low-expression levels in the brain with the exception of the RAB39B gene for which two different hemizygous mutations have recently been identified in male patients presenting autism, spectrum disorder, epileptic seizures, and macrocephaly.26
These 0.5/0.6 Mb duplications mediated by int22h-1/int22h-2 or -3 rearrangement must be very rare, as they are not reported in the Copy-Number Variation Database (database of genomic variants http://projects.tcag.ca/variation/). No new duplication was found in a screen of 420 X chromosomes via specific multiplex PCR. However, a new 0.5-Mb reciprocal deletion between int22h-1 and int22h-2 copies was reported in a normal female. As suggested by Pegoraro et al.27 and El-Hattab et al.21, these deletions may be lethal for males in utero but have no phenotypic consequences in females, as they result in completely skewed chromosome X inactivation. As CGH is an increasingly used screening method in prenatal diagnosis and patients with cognitive and behavioral abnormalities, this 0.5 Mb deletion or the corresponding duplication is likely to be increasingly found in the future. Clinical interpretation of large rare rearrangements identified by arrays is an important problem in current genetic testing and require guidelines to assess their clinical relevance.28
In the literature, we found three studies29, 30, 31 describing patients with severe HA phenotypes associated with the same SB combination of fragments of 21.5, 20, 16, and 14 kb as found in our cases 1 and 3. The study of Zimmermann et al.32 identified an aberrant 20 kb band associated with a duplication of exons 7–22 in a patient with severe HA resulting from rearrangement with int22h repeats. Furthermore, two other studies33, 34 reported the same abnormal SB profiles associated with severe HA (17.5, 16, and 14 kb) as in case 2. Only further analyses (sequencing and MLPA) are likely to reveal whether these abnormal SB patterns are the real cause of HA, thus improving genetic counseling.
The inversion of intron 22 occurs almost exclusively in male germ cells,35 as the mechanism for intron 22 involves flipping the end of the X chromosome, which is facilitated in male meiosis but inhibited in female germ cells by X-chromosome pairing. Thus, the 0.5/0.6-Mb deletion or duplication mediated by int22h rearrangement could occur in maternal meiosis, as these rearrangements require the pairing of two homologous chromatids (Figure 3).
All duplications described in this article were maternally inherited. However, the parental origin of the arrangement could not be determined, as DNA from the ascending generation was not available. For case 1, it is currently impossible to determine which duplication (0.5 MB or F8 exons 2–14) occurred first or to identify the parental origin.
Conclusion
Our data provide additional proof for the genomic instability at Xq28, showing that the int22h copies responsible for the well-known intr22 inversion are implicated in extended duplications (and deletion) involving F8 gene exons 1–22. These duplications are probably not associated with HA, as they coexisted with other molecular defects clearly responsible for HA. Contrary to a previous report and based on our study results,21 these duplications do not appear to be associated with intellectual disability.
Acknowledgments
We are indebted to the subjects participating in this study and their families. We thank Mrs I Abinet, Mr A Bosmans, and Mr V Kayndaszyk for their expert technical assistance, and Mrs Pascale Hilbert for chromosome-X-inactivation analysis. These studies were partially supported by a grant from Bayer Schering Pharma.
The authors declare no conflict of interest.
References
- Lakich D, Kazazian HH, Antonarakis DE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of hemophilia A. Nat Genet. 1993;5:236–241. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- Naylor JA, Brinke A, Hassock S, Green PM, Giannelli F. Characteristic mRNA abnormality found in half the patients with severe hemophilia A is due to large DNA inversion. Hum Mol Genet. 1993;2:1773–1778. doi: 10.1093/hmg/2.11.1773. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, The international consortium study Factor VIII gene inversions in severe hemophilia A: results of an international consortium study. Blood. 1995;86:2206–2212. [PubMed] [Google Scholar]
- Bagnall RD, Waseem N, Green PM, Giannelli F. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe haemophilia A. Blood. 2002;99:168–174. doi: 10.1182/blood.v99.1.168. [DOI] [PubMed] [Google Scholar]
- Oldenburg J, Ananyeva NM, Saenko EL. Molecular basis of haemophilia A. Haemophilia. 2004;10 (Suppl 4:133–139. doi: 10.1111/j.1365-2516.2004.01005.x. [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Markoff A, Pollmann H, et al. Spectrum of molecular defects and mutation detection rate in patients with severe hemophilia. Hum Mutat. 2005;26:249–254. doi: 10.1002/humu.20208. [DOI] [PubMed] [Google Scholar]
- Rost S, Löffler S, Pavlova A, Müller CR, Oldenburg J. Detection of large duplications within the factor VIII gene by MLPA. J Thromb Haemost. 2008;6:1996–1999. doi: 10.1111/j.1538-7836.2008.03125.x. [DOI] [PubMed] [Google Scholar]
- Lannoy N, Abinet I, Dahan K, Hermans C. Identification of de novo deletion in the factor VIII gene by MLPA technique in two girls with isolated factor VIII deficiency. Haemophilia. 2009;15:797–801. doi: 10.1111/j.1365-2516.2008.01974.x. [DOI] [PubMed] [Google Scholar]
- Acquila M, Pasino M, Lanza T, Bottini F, Molinari AC, Bicocchi MP. Duplication of exon 13 causes one third of the cases of mild hemophilia A in northern Italy. Haematologica. 2004;89:758–759. [PubMed] [Google Scholar]
- Zimmermann MA, Oldenburg J, Müller CR, Rost S. Characterization of duplication breakpoints in the factor VIII gene. J Thromb Haemost. 2010;8:2696–2704. doi: 10.1111/j.1538-7836.2010.04040.x. [DOI] [PubMed] [Google Scholar]
- Lannoy N, Abinet I, Bosmans A, Lambert C, Vermylen C, Hermans C. Computational and molecular approaches for predicting unreported causal missence mutations in Belgian patients with haemophilia A. Haemophilia. 2012;18:e331–e339. doi: 10.1111/j.1365-2516.2011.02640.x. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall RD, Giannelli F, Green PM. Int22h-related inversions causing hemophilia A: a novel insight into their origin and a new more discriminant PCR test for their detection. J Thromb Haemost. 2006;4:591–598. doi: 10.1111/j.1538-7836.2006.01840.x. [DOI] [PubMed] [Google Scholar]
- Van der Luijt R, Poppe B, De Smet E, et al. Rapid detection of VHL exon deletions using real-time quantitative PCR. J Lab Invest. 2005;85:24–33. doi: 10.1038/labinvest.3700209. [DOI] [PubMed] [Google Scholar]
- Kutsche K, Ressler B, Katzera HG, et al. Characterization of breakpoint sequences of five rearrangements in L1CAM and ABCD1 (ALD) genes. Hum Mutat. 2002;19:526–535. doi: 10.1002/humu.10072. [DOI] [PubMed] [Google Scholar]
- Bauters M, Van Esch H, Friez MJ, et al. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 2008;18:847–858. doi: 10.1101/gr.075903.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, et al. The DNA sequence of the human X chromosome. Nature. 2005;5:236–241. [Google Scholar]
- De Brasi CD, Bowen DJ. Molecular characteristics of the intron 22 homologs of the coagulation factor VIII gene: an update. J Thromb Haemost. 2008;6:1822–1824. doi: 10.1111/j.1538-7836.2008.03094.x. [DOI] [PubMed] [Google Scholar]
- Bagnall RD, Giannelli F, Green PM. Polymorphism and hemophilia A causing inversions in distal Xq28: a complex picture. J Thromb Haemost. 2005;3:2598–2599. doi: 10.1111/j.1538-7836.2005.01566.x. [DOI] [PubMed] [Google Scholar]
- Green P, Hill M, Bowen DJ, UK Haemophilia Centre Doctors' Organisation Haemophilia Genetics Network Duplications involving int22h-1 of the factor VIII gene: a cause for concern in genetic testing for hemophilia A. Thromb Haemost. 2007;5:2155–2156. doi: 10.1111/j.1538-7836.2007.02697.x. [DOI] [PubMed] [Google Scholar]
- El-Hattab AW, Fang P, Jin W, et al. Int22h-1/int22h-2-mediated Xq28 rearrangements: intellectual disability associated with duplications and in utero male lethality with deletions. J Med Genet. 2011;48:840–850. doi: 10.1136/jmedgenet-2011-100125. [DOI] [PubMed] [Google Scholar]
- Hu XY, Burghes AH, Ray PN, Thompson MW, Murphy EG, Worton RG. Partial gene duplication in Duchenne and Becker muscular dystrophies. J Med Genet. 1988;25:369–376. doi: 10.1136/jmg.25.6.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murru S, Casula L, Pecorara M, Mori P, Cao A, Pirastu M. Illegitimate recombination produced a duplication within the FVIII gene in a patient with mild hemophilia A. Genomics. 1990;7:115–1158. doi: 10.1016/0888-7543(90)90526-z. [DOI] [PubMed] [Google Scholar]
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human Androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1999;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Sharp A, Robinson D, Jacobs P. Age-and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet. 2000;107:343–349. doi: 10.1007/s004390000382. [DOI] [PubMed] [Google Scholar]
- Giannandrea M, Bianchi V, Mignogna ML, et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet. 2010;86:185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro E, Whitaker J, Mowery-Rushton P, Surti U, Lanasa M, Hoffman EP. Familial skewed X inactivation: a molecular trait associated with high spontaneous abortion rate maps to Xq28. Am J Hum Genet. 1997;61:160–170. doi: 10.1086/513901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Pfundt R, de Leeuw N, et al. Genomic microarrays in mental retardation: a practical workflow for diagnostic applications. Hum Mutat. 2009;30:283–292. doi: 10.1002/humu.20883. [DOI] [PubMed] [Google Scholar]
- Andrikovics H, Klein I, Bors A, et al. Analysis of large structural changes of the factor VIII gene, involving intron 1 and 22, in severe hemophilia A. Haematologica. 2003;88:778–784. [PubMed] [Google Scholar]
- Arruda VR, Pieneman WC, Reitsma PH, et al. Eleven novel mutations in the factor VIII gene from Brazilian hemophilia A patients. Blood. 1995;86:3015–3020. [PubMed] [Google Scholar]
- Windsor S, Taylor SA, Lillicrap D. Direct detection of a common inversion mutation in the genetic diagnosis of severe hemophilia A. Blood. 1994;84:2202–2205. [PubMed] [Google Scholar]
- Zimmermann MA, Oldenburg J, Müller CR, Rost S. Unusual genomic rearrangements in introns 1 and 22 of the F8 gene. Hamostaseologie. 2011;31 (Suppl 1:S69–S73. [PubMed] [Google Scholar]
- Yamazaki E, Mohri H, Inaba H, Harano H, Kanamori H, Okubo T. Variant of intron 22 inversions in the factor VIII gene in severe hemophilia A. Blood Coagul Fibrinolysis. 1997;8:445–449. doi: 10.1097/00001721-199710000-00011. [DOI] [PubMed] [Google Scholar]
- Enayat MS, Theophilus BD, Williams MD, Hill FG. Another unique variant pattern of intron 22 rearrangement in factor Vlll gene seen in a hemophilia A family. Blood. 1995;85:2639. [PubMed] [Google Scholar]
- Rosslier JP, Young M, Kimberland ML, et al. Factor VIII gene inversions causing severe hemophilia A originate almost exclusively in male germ cell. Hum Mol Genet. 1994;3:1035–1039. doi: 10.1093/hmg/3.7.1035. [DOI] [PubMed] [Google Scholar]