Abstract
Brugada syndrome is an inherited arrhythmogenic disorder leading to sudden death predominantly in the 3–4 decade. To date the only reliable treatment is the implantation of a cardioverter defibrillator; however, better criteria for risk stratification are needed, especially for asymptomatic subjects. Brugada syndrome genetic bases have been only partially understood, accounting for <30% of patients, and have been poorly correlated with prognosis, preventing inclusion of genetic data in current guidelines. We designed an observational study to identify genetic markers for risk stratification of Brugada patients by exploratory statistical analysis. The presence of genetic variants, identified by SCN5A gene analysis and genotyping of 73 candidate polymorphisms, was correlated with the occurrence of major arrhythmic events in a cohort of 92 Brugada patients by allelic association and survival analysis. In all, 18 mutations were identified in the SCN5A gene, including 5 novel, and statistical analysis indicated that mutation carriers had a significantly increased risk of major arrhythmic events (P=0.024). In addition, we established association of five polymorphisms with major arrhythmic events occurrence and consequently elaborated a pilot risk stratification algorithm by calculating a weighted genetic risk score, including the associated polymorphisms and the presence of SCN5A mutation as function of their odds ratio. This study correlates for the first time the presence of genetic variants with increased arrhythmic risk in Brugada patients, representing a first step towards the design of a new risk stratification model.
Keywords: arrhythmia, Brugada syndrome, sudden death, genetic variation, risk stratification
Introduction
Brugada syndrome (BrS) is a cardiac disorder with estimated prevalence of 1:5000 in western countries, characterized by electrical ventricular instability leading to sudden cardiac death (SCD) predominantly in the 3–4 decade, although cases of infant death have also been reported.1 Diagnosis is based on the presence of type I ECG, characterized by right bundle branch block and ST elevation in right precordial leads.2, 3 To date the only reliable treatment is defibrillator (ICD) implantation.4 Current guidelines select higher risk patients based mainly on the presence of syncope and spontaneous ECG.5 However, predisposition of BrS patients to develop ventricular arrhythmias still cannot be easily predicted and new criteria for prognostic risk stratification are needed, especially for asymptomatic subjects.
BrS is genetically heterogeneous, SCN5A gene mutations accounting for <25%.6 Accordingly, the emerging concept of arrhythmia genomics supports the idea of a complex disorder, where the co-segregation of different mutations and common genetic variants can contribute to the clinical phenotype.7
The aim of our study was to evaluate the contribution of candidate genetic variants to the predisposition to malignant arrhythmias in a cohort of BrS patients. We identified disease-modifying variants associated with arrhythmic phenotype and elaborated an innovative model of risk stratification based on these genetic markers.
Materials and methods
Patients
The investigation conformed to principles outlined in the Declaration of Helsinki. Written informed consent for genetic analysis was obtained. In all, 92 consecutive Italian BrS patients were diagnosed based on the presence of type I ECG, either spontaneous (n=43) or induced by flecainide infusion (n=49).8 Patients were subjected to personal and family history acquisition, evaluation of blood electrolytes, 24 h Holter-ECG, echocardiography and, whenever possible, cardiac magnetic resonance imaging to exclude the presence of other conditions associated with ST elevation. Electrophysiological study was performed when possible, including measurement of conduction intervals and programmed ventricular stimulation from the right ventricular apex with a maximum of three ventricular premature beats with three basic pacing cycles (600–500 and 400 ms); the shortest coupling interval of the premature beats was limited to 200 ms. A patient was considered inducible when a sustained ventricular arrhythmia (defined as one lasting >30 s or requiring intervention to terminate) was induced. Therapeutic options and ICD implantation were evaluated according to current guidelines5 and clinical judgment. Patients were followed at least yearly by clinical assessment, ECG, echocardiography and ICD check. Documented ventricular tachycardia/fibrillation, SCD or appropriate ICD shock were defined as major arrhythmic events (MAE). All relevant clinical and genetic data were collected in an electronic database.
Genetic analysis
Mutation detection
Genomic DNA was extracted from peripheral blood with Maxwell16 (Promega, Milano, Italy) and SCN5A gene-coding exons, including adjacent intronic regions, were screened by PCR, DHPLC (Transgenomic, Glasgow, UK) and automated DNA sequencing (ABI 3730, Applied Biosystems, Monza, Italy). For primer sequences, see Supplementary Table S1. Variants were described using the Human Genome Variation Society guidelines for nomenclature.9 SCN5A reference sequence was NCBI NM_000335. Mutations placement was assigned using a combination of Swissprot (http://ca.expasy.org/uniprot/) and studies of linear topologies for sodium channel pore-forming alpha subunit.10
SNPs selection and analysis
Candidate single nucleotide polymorphisms (SNPs) were selected among those previously described as modulating ECG intervals11, 12, 13, 14, 15, 16, 17 or predisposition to arrhythmic events.18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 In particular, most variants were located in genes encoding cardiac voltage-dependent channels (KCNQ1, KCNH2, SCN5A, KCNJ11, KCNE1, KCNE2, KCNJ2), structural proteins (ANK2, GJA5, PALLD), sympathetic system modulators (ADRB1, ADRB2, ADRB3), blood pressure regulators (NOS1AP, PAI-1), modulators of ionic flows (PLN, CERKL, SLCO3A1), transcription factors (BRUNOL4, LITAF), regulators of cell morphology (NRG3) and other genes known to be associated to ECG intervals modulation (MDR1, CASQ2, NDRG4, NUBPL, RNF207; Supplementary Table S2).
SNP genotyping was performed with GenomeLab SNPstream platform (Beckman Coulter, Brea, CA, USA), based on 48-plex PCR reaction followed by single base extension and tag-driven hybridization. Two-color fluorescence was detected by SNPstream and intensity signals converted into genotypes. Primers for multiplex PCR were designed by Autoprimer software (www.autoprimer.com; Supplementary Table S3). Manufacturer protocols were followed for reaction conditions and data analysis.
Statistical analysis
Association analyses were performed using PLINK-v1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/),32 StataSE 11 (StataCorp LP, College Station, TX, USA) and SPSS version 13.0 software (SPSS, Bologna, Italy). Quality control criteria were applied for SNP exclusion (genotype call rate <0.9; minor allele frequency <0.01 and Hardy–Weinberg equilibrium P-value ≤0.001 in controls represented by non-MAE patients). Each SNP was tested for allelic association using χ2-test and odds ratio and relative 95% confidence intervals were measured. Genotypic association analysis was performed testing several inheritance models (dominant, additive and recessive).
Kaplan–Meier curves were exploited for survival analysis, defining survival time as the interval between birth and occurrence of MAE or last follow-up information. Log-rank test was used to compare survival distribution across groups according to the presence/absence of SCN5A mutation and SNPs genotypes. An uncorrected P-value of 0.05 was considered as statistically significant as these analyses were considered as exploratory.
A weighted genetic risk score algorithm was calculated based on the presence of SCN5A mutations and selected SNPs by multiplying the number of risk alleles found to be significantly associated at either association or survival analysis by the weight of that SNP, representing the natural log of the OR of each allele, and then taking the sum across the included SNPs.33 Mann–Whitney test was used for weighted genetic risk score comparisons. The contribution of weighted genetic risk score to MAE prediction was evaluated generating a receiver operating characteristic curve by plotting the sensitivity of continuous weighted genetic risk score against 1−specificity and calculating the area under the curve as measure of performance.
Results
Patients clinical profile
Ninety-two consecutive Italian BrS patients (age 47.5±13.7, 85.8% males) were considered (Table 1). In all, 40 were symptomatic for cardiac arrest, ventricular tachycardia/fibrillation or syncope, while in 52 asymptomatic patients diagnosis of BrS was suspected during routine cardiac ECG or family screening. Twenty-nine patients showed family history for SCD and/or BrS ECG and 13 first-degree relatives suffered from SCD. Twenty-seven patients had supraventricular arrhythmias and 17 documented nonsustained ventricular tachycardia. Also, 46/82 tested patients resulted positive to electrophysiological study and 62 received ICD (Table 1). Mean follow-up was 26±16.7 months; 12 patients experienced lifetime MAE, with mean age at the event 42±12.4 years (Table 2).
Table 1. Clinical features of Brugada patients.
| Description | N (%) |
|---|---|
| Number of patients | 92 |
| Follow-up (months, mean and SD) | 26±16.7 |
| Age (years, mean and SD) | 47.5±13.7 |
| Males | 79 (85.8) |
| Family history for SCD and/or BrS | 29 (31.5) |
| Asymptomatic | 52 (56.5) |
| Spontaneous type I ECG | 43 (46.7) |
| Syncope | 34 (36.9) |
| EPS performed | 82 (89.1) |
| Inducible VT/VF at EPS | 46/82 (56.1) |
| ICD implanted | 62 (67.4) |
| Supraventricular arrhythmias | 27 (29) |
| Documented nonsustained ventricular tachycardia | 17 (18.5) |
| MAE | 12 (13) |
| Age at MAE (years, mean and SD) | 42±12.4 |
| SCN5A mutation carriers | 17 (18.5) |
Abbreviations: EPS, electrophysiologic study; ICD, implantable cardioverter defibrillator; MAE, major arrhythmic event; SCD, sudden cardiac death; VF, ventricular fibrillation; VT, ventricular tachycardia.
Table 2. Clinical features of 12 BrS patients experiencing lifetime MAE.
| ID | Year of birth | Age at MAE | Sex | Family history for SCD | s-Type I ECG | Result EPS | Syncope | SVA | MAE | SCN5A mutation |
|---|---|---|---|---|---|---|---|---|---|---|
| 071603P | 1957 | 50 | M | − | + | nd | + | − | CA | − |
| 09193P | 1974 | 35 | M | − | + | + | − | − | ICD shock | − |
| 062293P | 1976 | 30 | M | − | − | + | − | + | VT/VF | − |
| 0711P | 1954 | 54 | M | + | + | + | + | − | ICD shock | − |
| 061352P | 1981 | 25 | M | − | − | − | + | − | VFL/VF | p.E1225K |
| 05256P | 1958 | 47 | M | − | − | − | − | + | CAa | − |
| 081712P | 1953 | 49 | M | − | − | − | + | + | VT/VF, CA | p.R104Q |
| 063003P | 1980 | 27 | M | − | + | nd | − | − | VF | p.A1428S |
| 092941P | 1969 | 40 | M | − | − | − | + | − | VF | − |
| 073671P | 1974 | 31 | M | − | + | + | + | + | VT/VF | p.L276Q |
| 062107P | 1945 | 61 | M | + | − | + | + | − | VT/VF | − |
| 053609P | 1949 | 56 | M | − | − | + | + | + | VF | − |
Abbreviations: CA, cardiac arrest; EPS, electrophysiological study; M, male; MAE, major arrhythmic event; nd, not done; s-Type I, spontaneous type I ECG, SVA, supraventricular arrhythmias; VF, ventricular fibrillation; VFL, ventricular flutter; VT/VF, polymorphic ventricular tachycardia degenerating in fibrillation.
After propafenone administration.
SCN5A mutation analysis
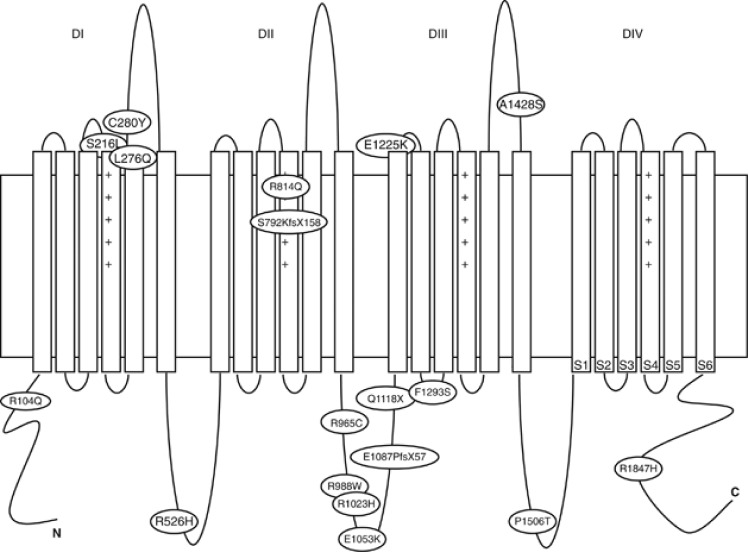
18 SCN5A mutations were identified in 17 BrS patients, as one carried two different variants (p.E1087PfsX57 and p.F1293S). All the mutations were private and included 15 missense substitutions, 1 nonsense mutation, 1 frameshift deletion and 1 insertion. Five variants had not been previously described (Figure 1, Table 3). Pathogenicity of these unreported variants was assessed by evaluating their absence in at least 200 control Italian chromosomes, in 1000 genomes (http://browser.1000genomes.org) and Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) databases and analyzing intra-familiar segregation. In addition, all the new variants were classified as potentially damaging protein function by in silico prediction softwares Polyphen 2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT (http://sift.jcvi.org/).
Figure 1.
SCN5A mutations identified in BrS patients. Domains of the Nav1.5 protein are depicted.
Table 3. SCN5A gene mutations identified in BrS patients.
| cDNA variation | Protein effect | Mutation type | Location (exon, domain, segment) |
|---|---|---|---|
| c.311 G>A | p.R104Q | Missense | Exon 3, N-term |
| c.647 C>T | p.S216L | Missense | Exon 6, IS3-S4 |
| c.827 T>A | p.L276Q | Missense | Exon 7, IS5 |
| c.839 G>A | p.C280Y | Missense | Exon 7, IS5-S6 |
| c.1577 G>A | p.R526H | Missense | Exon 12, IS6-IIS1 |
| c.2374_2375insA | p.S792KfsX158 | Frameshift | Exon 15, IIS3 |
| c.2441 G>A | p.R814Q | Missense | Exon 16, IIS4 |
| c.2893C>T | p.R965C | Missense | Exon 17, IIS6-IIIS1 |
| c.2962C>T | p.R988W | Missense | Exon 17, IIS6-IIIS1 |
| c.3068 G>A | p.R1023H | Missense | Exon 17, IIS6-IIIS1 |
| c.3157 G>A | p.E1053K | Missense | Exon 17, IIS6-IIIS1 |
| c.3258-3261del4 | p.E1087PfsX57 | Frameshift | Exon 18, IIS6-IIIS1 |
| c.3352C>T | p.Q1118X | Nonsense | Exon 18, IIS6-IIIS1 |
| c.3673 G>A | p.E1225K | Missense | Exon 21, III S1-S2 |
| c.3878 T>C | p.F1293S | Missense | Exon 22, IIIS2-S3 |
| c.4282 G>T | p.A1428S | Missense | Exon 24, IIIS5-S6 |
| c.4516C>A | p.P1506T | Missense | Exon 26, IIIS6-IVS1 |
| c.5540 G>A | p.R1847H | Missense | Exon 28, C-term |
Bold indicates previously unreported variants.
SCN5A mutations were distributed along the entire predicted topology of the Nav1.5 protein: one in N-terminus and one in C-terminus, three in transmembrane spanning segments, nine in intracellular and four in extracellular loops (Figure 1). The functional effect of three of these mutations (p.S216L, p.E1087PfsX57, p.F1293S) on biophysics properties of the sodium channel Nav 1.5 has been previously evaluated and reported by our group.34, 35
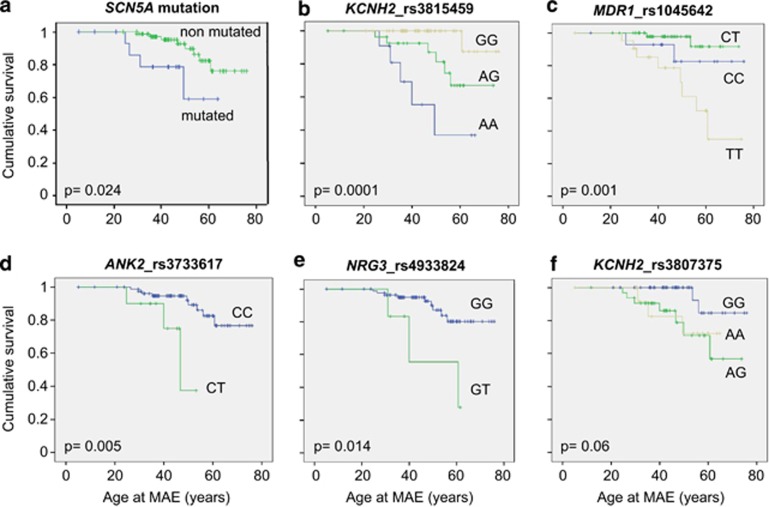
Kaplan–Meier curves were drawn considering lifetime occurrence of MAE as endpoint, demonstrating that SCN5A mutation carriers had an increased risk of MAE compared with non-carriers (P=0.024; Figure 2a).
Figure 2.
Kaplan–Meier survival curves. Lifetime risk of MAE according to the presence of the following genetic variants: (a) SCN5A mutations; (b) SNP KCNH2_ rs3815459; (c) SNP MDR1_rs1045642; (d) SNP ANK2_rs3733617; (e) SNP NRG3_rs4933824; (f) SNP KCNH2_rs3807375. Censored values at different time-points are indicated by vertical lines.
SNPs analysis and risk stratification
In order to explain the variability of clinical phenotype, we subsequently studied the association of genetic variants with MAE manifestation by comparing patients experiencing lifetime MAE vs non-MAE patients. We focused on 73 candidate SNPs previously associated to changes in ECG intervals or modulation of arrhythmic risk (Supplementary Table S2). Nineteen SNPs were not polymorphic (minor allele frequency <0.01), 18 were filtered out on the basis of genotype call rates (<0.9) and one for Hardy–Weinberg disequilibrium (P=1.2 × 10−6). After frequency and genotyping pruning, 35 SNPs were included in allelic association analysis by χ2-test, showing four SNPs significantly associated with MAE occurrence: KCNH2_rs3815459A (P=4.9 × 10−6), NRG3_rs4933824T (P=0.006), MDR1_rs1045642T (P=0.025) and KCNH2_rs3807375A (P=0.027) (Table 4).
Table 4. SNPs composing the weighted genetic risk score and weight assigned to each marker.
| SNP | Ch | Bp | RA | RA-MAE | RA-non-MAE | P-value (χ2-test) | Allelic OR | 95% CI | Ln OR Weight |
|---|---|---|---|---|---|---|---|---|---|
| NRG3 rs4933824 | 10 | 83819125 | T | 3/24 (0.125) | 3/160 (0.02) | 0.006 | 7.48 | 1.416–39.47 | 2.01 |
| KCNH2 rs3815459 | 7 | 150644394 | A | 16/24 (0.67) | 35/160 (0.22) | 4.9 × 10−6 | 7.14 | 2.825–18.06 | 1.97 |
| ANK2 rs3733617 | 4 | 114278277 | T | 3/24 (0.125) | 7/160 (0.04) | 0.102 | 3.12 | 0.749–13.01 | 1.14 |
| MDR1 rs1045642 | 7 | 8713865 | T | 18/24 (0.75) | 81/160 (0.51) | 0.025 | 2.93 | 1.104–7.75 | 1.07 |
| KCNH2 rs3807375 | 7 | 150667210 | A | 13/24 (0.54) | 49/160 (0.31) | 0.027 | 2.60 | 1.09–6.204 | 0.95 |
| SCN5A mutation | 3 | NA | y | 4/12 (0.33) | 13/80 (0.16) | 0.166 | 2.58 | 0.675–9.833 | 0.95 |
Abbreviations: Ch, chromosome; Bp, SNP location; RAMAE, risk allele counts and (frequency) in MAE patients; RA-non-MAE: risk allele counts and (frequency) in patients not experiencing MAE; OR, odds ratio; CI, confidence interval; NA, not applicable; y, presence.
All ORs used in the algorithm are referenced to the risk allele of each SNP, which can be the minor or major allele.
Kaplan–Meier curves were subsequently drawn considering lifetime MAE and plotted for each of the 35 SNPs, showing an association with the time to MAE in four SNPs by Log-rank test (KCNH2_rs3815459A, P=0.0001; MDR1_rs1045642T, P=0.001; ANK2_rs3733617T, P=0.005; NRG3_rs4933824T, P=0.014) and a trend for KCNH2_rs3807375A (P=0.06). These risk alleles were associated with an increased susceptibility to develop life-threatening arrhythmic events (Figure 2b–f).
In addition, the trend of Kaplan–Meier curves allowed us to infer on possible SNPs inheritance traits, suggesting a dominant mechanism for KCNH2_rs3807375 (the presence of a single copy of the risk allele A was sufficient to give the risk phenotype) and co-dominance for KCNH2_rs3815459 (the heterozygous genotype had an intermediate risk between the two homozygous). MDR1_rs1045642 was compatible with recessive inheritance as only the presence of two T risk alleles was associated with arrhythmic phenotype. No patient was homozygous for the risk allele in NRG3_rs4933824 and ANK2_rs3733617. There was no evidence of epistasis among these risk loci and between SNPs and the presence of mutations (not shown).
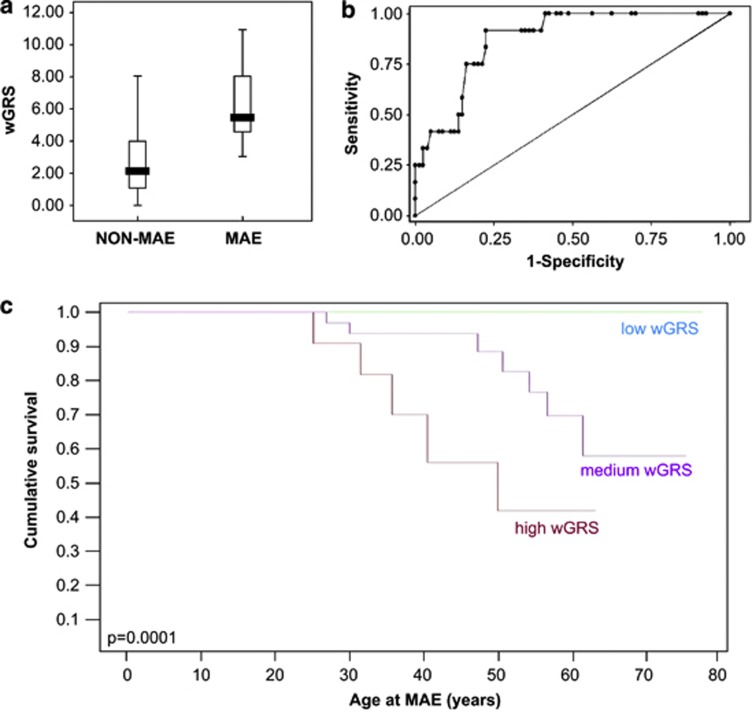
We finally calculated a weighted genetic risk score to evaluate the potential of a pilot risk stratification model. The weight of each risk allele and the presence of SCN5A mutation was calculated as function of its odds ratio obtained with χ2-test33 (Table 4). Using this model, patients experiencing MAE had mean genetic risk score=6.36±2.5, while event-free patients=2.87±1.99 (Mann–Whitney P=1.5 × 10−5; Figure 3a). To further examine weighted genetic risk score discriminatory power, we performed receiver operating characteristic curve analysis showing an area under the curve=0.873 (CI 95% 0.791–0.956, P=3.24 × 10−5; Figure 3b). A weighted genetic risk score cutoff value ≥4.06 was considered to classify MAE BrS patients (sensitivity: 91,67% specificity: 77,50% accuracy: 0,84585). Risk stratification according to weighted genetic risk score value was also represented by Kaplan–Meier curves (P=0.0001, Figure 3c).
Figure 3.
Weighted genetic risk score (wGRS). (a) Value of the wGRS in MAE vs non-MAE patients. Median values are indicated in boxplot. (b) ROC curve analysis of the discriminatory power of wGRS in predicting MAE occurrence. The graph is generated by plotting the sensitivity of continuous wGRS against 1−specificity. The area under ROC curve is 0.873 (CI 95% 0.791–0.956, P=3.24 × 10−5). (c) Kaplan–Meier survival curves representing MAE lifetime risk by stratifying patients according to wGRS value: high (>6), medium (3–6), and low (<3).
Correlation of weighted genetic risk score with MAE occurrence was validated on 37 independent BrS patients, consecutively enrolled after the 92 index patients (age 40.4±13.9, 86.5% males, 35% SCN5A mutation carriers) and never experiencing MAE. Their weighted genetic risk score was 3.23±1.58, significantly different from that of MAE patients of the original cohort (Mann–Whitney P<0.0001) and not from non-MAE patients (Mann–Whitney P=0.172).
Discussion
We propose for the first time a role of genetics in BrS risk stratification, correlating the presence of SCN5A gene mutations and common SNPs with increased arrhythmic risk. Indeed, in our cohort, SCN5A mutation carriers experienced significantly increased risk of lifetime MAE compared with non-carriers (Figure 2a). Although still controversial, this observation agrees with recent studies suggesting association of SCN5A mutations with arrhythmic phenotypes. Probst et al36 described conduction defects in SCN5A mutation carriers. Other studies observed association between severe phenotypes and inactivating SCN5A mutations compared with milder variants.37, 38 Recent studies suggested that the presence of SCN5A mutation was a significant predictor of cardiac events39 and could be associated with recurrence of ventricular fibrillation.40
Genetic and clinical heterogeneity is relevant in BrS. Indeed, the classical concept of BrS as a mendelian disease has been recently modified according to the idea of arrhythmia genomics, suggesting that different variants may contribute to the phenotype and sodium channel defects may be one of the contributory causes and not necessarily the main.7, 41 Inter-individual genetic differences may modulate excitation–contraction physiology, influencing arrhythmia susceptibility and may even explain differences in clinical phenotype presentation among distinct ethnicities.42, 43 Ion channels carry out their function as part of multiproteic complexes, interacting with accessory subunits and regulatory proteins. Variants impairing function or expression of one of these proteins or influencing indirectly cardiac contraction, such as vagal tone or vascular pressure, may thus create clinically detectable phenotypical differences.44 Hence, the identification of genetic variants modulating the phenotype in BrS patients may be useful for risk stratification.
In this pilot study, we evaluated whether we could establish an association between MAE occurrence and the presence of selected genetic variants. Candidate SNPs were chosen based on the idea that a change in proteins involved in the modulation of cardiac excitability and ECG phenotype may influence vulnerability to arrhythmias. This study considered all BrS probands in our cohort independently from the presence of SCN5A mutations, according to the concept that patients should have common pathogenetic mechanisms underlying ST segment elevation.45 As we did not include relatives, our cohort reflects the heterogeneity of a population referring to an arrhythmology department for various reasons (cardiac arrest, syncope, asymptomatic ECG alterations, family history for SCD), and it is thus not enriched for asymptomatic individuals identified only by intra-familiar genetic screening.
Allelic association and Kaplan–Meier survival analyses identified five variants significantly associated with arrhythmic phenotype (Figure 2b–f). These SNPs were located on different linkage disequilibrium blocks, as results from HapMap analysis, and correlated with arrhythmic risk in an independent way, even if at present we cannot predict whether the modulator effect is linked to the SNP itself or to other variants in the same linkage disequilibrium block. HERG potassium channel seems to be important for arrhythmia susceptibility in our cohort. Indeed, we identified two SNPs in KCNH2 gene associated with MAE occurrence: KCNH2_rs3815459 and KCNH2_rs3807375, previously involved in QT interval modulation.13 The association of these SNPs with arrhythmic events therefore underlines the importance of the depolarization/repolarization balance in BrS arrhythmogenesis. ANK2_rs3733617 is located in the gene encoding the structural protein ankyrin B, linking integral membrane proteins to the underlying spectrin–actin cytoskeleton and required for targeting and stability of Na/Ca exchanger, Na/K ATPase and IP3 receptor in cardiomyocytes. Mutations in this gene cause defects in sodium and calcium regulation and have been associated with sinus node dysfunction, atrial fibrillation and long QT syndrome type 4.46 NRG3_rs4933824 localizes in the gene encoding the cardiac growth factor neuregulin 3, important during cardiac development and morphogenesis and previously associated to a prolonged QT.12, 28 The reason why we did not observe any homozygous individual for the risk allele in NRG3_rs4933824 and ANK2_rs3733617 may be ascribed to their low minor allele frequency (<5%). MDR1_rs1045642 lays in the MDR1-ABCB1 gene, belonging to the superfamily of ATP-binding cassette transporters and recently described as a marker of resident cardiac stem/progenitor cell subpopulations, thus governing cardiac remodeling and regeneration.47
In order to elaborate a pilot model for risk stratification based on genetic data, we calculated a weighted genetic risk score for each patient, considering genetic variants associated with MAE (presence of SCN5A mutation and the five significant SNPs). This score, calculated by weighting each genetic variant based on the association effect size (OR) with MAE (Table 4), was significantly higher in MAE vs event-free patients (Figure 3). These results support the emerging ‘Multi-Hit Hypothesis', suggesting that particular combinations of common genetic variations in biologically relevant genes can affect electrogenesis and propagation and therefore modulate arrhythmogenic risk.48, 49 This concept has also been proposed in a recent work studying genetic complexity in sporadic idiopathic epilepsy, suggesting that oligogenic electrical relationship may likely have a role even in ‘monogenic' channelopathies, as carriers of such mutations often show a spectrum of clinical phenotypes, including the absence of disease, even within the same pedigree.50
Indeed, in our cohort 5 out of 7 individuals carrying ≥3 risk genotypes had experienced MAE, while only 1 out of 29 patients not carrying any risk genotype suffered life-threatening arrhythmias. This observation corroborates the association between at-risk genotypes and severe phenotype and underline the possibility that the contemporary presence of these alleles may have a cumulative effect on the phenotype.
Study limitations and conclusions
As the present study is based on data from a population of 92 patients, with 17 SCN5A mutation carriers and 12 MAE, statistical analysis may suffer from reduced power. Although discriminatory power of weighted genetic risk score was validated in 37 independent BrS patients, this model will need confirmation in larger cohorts. Moreover, other genetic variants may be involved in predisposition to MAE, prompting future genome-wide association genotyping or deep sequencing studies for their identification. Nevertheless, this study, for the first time, was able to correlate the presence of genetic variants with increased arrhythmic risk in BrS patients, representing a proof of principle that genetics can indeed have a role. Our data may thus constitute a first step towards the design of a new risk stratification model, which in the future might also be integrated with other variables associated with increased arrhythmic risk in BrS patients.
Acknowledgments
We are grateful to all the patients and their families. We acknowledge all personnel of the Arrhythmology Unit of San Raffaele Hospital, in particular, Francesco Sacco and Francesca Zuffada for collaboration in BrS patients' characterization and Vincenzo Santinelli for critically revising the manuscript. We also thank Dr Amarild Cuko, Dr Ciro Indolfi, Dr Massimo Zoni Berisso, Dr Stefano Favale and Dr Luca Rocchetti for collaboration in identifying patients. We thank Alessandra Foglio and Chiara Redaelli for help in SCN5A genetic analysis and setup of SNP genotyping. This study was supported by grants from Medtronic Italy (to MF) and Italian Istituto Superiore di Sanità (ISS-526D/55 to CP).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Brugada R. Sudden death: managing the family, the role of genetics. Heart. 2011;97:676–681. doi: 10.1136/hrt.2009.185884. [DOI] [PubMed] [Google Scholar]
- Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- Wilde AA, Antzelevitch C, Borggrefe M, et al. Proposed diagnostic criteria for the Brugada syndrome: consensus report. Circulation. 2002;106:2514–2519. doi: 10.1161/01.cir.0000034169.45752.4a. [DOI] [PubMed] [Google Scholar]
- Antzelevitch C, Brugada P, Borggrefe M, et al. Brugada syndrome: report of the second consensus conference. Heart Rhythm. 2005;2:429–440. doi: 10.1016/j.hrthm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Epstein AE, DiMarco JP, Ellenbogen KA, et al. ACC/AHA/HRS 2008 Guidelines for Device-Based Therapy of Cardiac Rhythm Abnormalities: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Revise the ACC/AHA/NASPE 2002 Guideline Update for Implantation of Cardiac Pacemakers and Antiarrhythmia Devices): developed in collaboration with the American Association for Thoracic Surgery and Society of Thoracic Surgeons. Circulation. 2008;117:e350–e408. doi: 10.1161/CIRCUALTIONAHA.108.189742. [DOI] [PubMed] [Google Scholar]
- Cummings S, Priori S. Genetics of cardiac arrhythmias. Minerva Med. 2011;102:209–222. [PubMed] [Google Scholar]
- Roden DM. Human genomics and its impact on arrhythmias. Trends Cardiovasc Med. 2004;14:112–116. doi: 10.1016/j.tcm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Brugada R, Brugada J, Antzelevitch C, et al. Sodium channel blockers identify risk for sudden death in patients with ST-segment elevation and right bundle branch block but structurally normal hearts. Circulation. 2000;101:510–515. doi: 10.1161/01.cir.101.5.510. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li Z, Shen J, Keating MT. Genomic organization of the human SCN5A gene encoding the cardiac sodium channel. Genomics. 1996;34:9–16. doi: 10.1006/geno.1996.0236. [DOI] [PubMed] [Google Scholar]
- Pfeufer A, Sanna S, Arking DE, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi S, Heaton C, Mack K, et al. Whole genome association study identifies polymorphisms associated with QT prolongation during iloperidone treatment of schizophrenia. Mol Psychiatry. 2009;14:1024–1031. doi: 10.1038/mp.2008.52. [DOI] [PubMed] [Google Scholar]
- Gouas L, Nicaud V, Chaouch S, et al. Confirmation of associations between ion channel gene SNPs and QTc interval duration in healthy subjects. Eur J Hum Genet. 2007;15:974–979. doi: 10.1038/sj.ejhg.5201866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarnoudse AJ, Newton-Cheh C, de Bakker PI, et al. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- Newton-Cheh C, Eijgelsheim M, Rice KM, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Salisbury BA, Reed CR, Ackerman MJ. Pharmacogenetic issues in thorough QT trials. Mol Diagn Ther. 2006;10:153–162. doi: 10.1007/BF03256454. [DOI] [PubMed] [Google Scholar]
- Roden DM, Viswanathan PC. Genetics of acquired long QT syndrome. J Clin Invest. 2005;115:2025–2032. doi: 10.1172/JCI25539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman MJ, Splawski I, Makielski JC, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1:600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Kaab S, Schulze-Bahr E. Susceptibility genes and modifiers for cardiac arrhythmias. Cardiovasc Res. 2005;67:397–413. doi: 10.1016/j.cardiores.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Schulze-Bahr E. Arrhythmisa Predisposition. Between rare disease paradigms and common ion channel gene variants. J Am Coll Cardiol. 2006;48:67–78. [Google Scholar]
- Iwasa H, Itoh T, Nagai R, Nakamura Y, Tanaka T. Twenty single nucleotide polymorphisms (SNPs) and their allelic frequencies in four genes that are responsible for familial long QT syndrome in the Japanese population. J Hum Genet. 2000;45:182–183. doi: 10.1007/s100380050207. [DOI] [PubMed] [Google Scholar]
- Niu DM, Hwang B, Hwang HW, et al. A common SCN5A polymorphism attenuates a severe cardiac phenotype caused by a nonsense SCN5A mutation in a Chinese family with an inherited cardiac conduction defect. J Med Genet. 2006;43:817–821. doi: 10.1136/jmg.2006.042192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelzing S, Forleo C, Samodell M, et al. SCN5A polymorphism restores trafficking of a Brugada syndrome mutation on a separate gene. Circulation. 2006;114:368–376. doi: 10.1161/CIRCULATIONAHA.105.601294. [DOI] [PubMed] [Google Scholar]
- Groenewegen WA, Firouzi M, Bezzina CR, et al. A cardiac sodium channel mutation cosegregates with a rare connexin40 genotype in familial atrial standstill. Circ Res. 2003;92:14–22. doi: 10.1161/01.res.0000050585.07097.d7. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xia M, Jin Q, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzina CR, Shimizu W, Yang P, et al. Common sodium channel promoter haplotype in asian subjects underlies variability in cardiac conduction. Circulation. 2006;113:338–344. doi: 10.1161/CIRCULATIONAHA.105.580811. [DOI] [PubMed] [Google Scholar]
- Mango R, Vecchione L, Raso B, et al. Pharmacogenomics in cardiovascular disease: the role of single nucleotide polymorphisms in improving drug therapy. Expert Opin Pharmacother. 2005;6:2565–2576. doi: 10.1517/14656566.6.15.2565. [DOI] [PubMed] [Google Scholar]
- Weiss J, Ten Hoevel MM, Burhenne J, et al. CYP2C19 genotype is a major factor contributing to the highly variable pharmacokinetics of voriconazole. J Clin Pharmacol. 2009;49:196–204. doi: 10.1177/0091270008327537. [DOI] [PubMed] [Google Scholar]
- Mank-Seymour AR, Richmond JL, Wood LS, et al. Association of torsades de pointes with novel and known single nucleotide polymorphisms in long QT syndrome genes. Am Heart J. 2006;152:1116–1122. doi: 10.1016/j.ahj.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Makita N, Mochizuki N, Tsutsui H. Absence of a trafficking defect in R1232W/T1620M, a double SCN5A mutant responsible for Brugada syndrome. Circ J. 2008;72:1018–1019. doi: 10.1253/circj.72.1018. [DOI] [PubMed] [Google Scholar]
- Anson BD, Ackerman MJ, Tester DJ, et al. Molecular and functional characterization of common polymorphisms in HERG (KCNH2) potassium channels. Am J Physiol Heart Circ Physiol. 2004;286:H2434–H2441. doi: 10.1152/ajpheart.00891.2003. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Chibnik LB, Cui J, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni S, Di Resta C, Rocchetti M, et al. A Brugada syndrome mutation (p.S216L) and its modulation by p.H558R polymorphism: standard and dynamic characterization. Cardiovasc Res. 2011;91:606–616. doi: 10.1093/cvr/cvr142. [DOI] [PubMed] [Google Scholar]
- Sommariva E, Vatta M, Xi Y, et al. Compound heterozygous SCN5A gene mutations in asymptomatic Brugada syndrome child. Cardiogenetics. 2012;2:53–58. [Google Scholar]
- Probst V, Allouis M, Sacher F, et al. Progressive cardiac conduction defect is the prevailing phenotype in carriers of a Brugada syndrome SCN5A mutation. J Cardiovasc Electrophysiol. 2006;17:270–275. doi: 10.1111/j.1540-8167.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- Meregalli PG, Tan HL, Probst V, et al. Type of SCN5A mutation determines clinical severity and degree of conduction slowing in loss-of-function sodium channelopathies. Heart Rhythm. 2009;6:341–348. doi: 10.1016/j.hrthm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Benito B, Brugada J, Brugada R, Brugada P. Brugada syndrome. Rev Esp Cardiol. 2009;62:1297–1315. doi: 10.1016/s1885-5857(09)73357-2. [DOI] [PubMed] [Google Scholar]
- Makimoto H, Nakagawa E, Takaki H, et al. Augmented ST-segment elevation during recovery from exercise predicts cardiac events in patients with Brugada syndrome. J Am Coll Cardiol. 2010;56:1576–1584. doi: 10.1016/j.jacc.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Nishii N, Ogawa M, Morita H, et al. SCN5A mutation is associated with early and frequent recurrence of ventricular fibrillation in patients with Brugada syndrome. Circ J. 2010;74:2572–2578. doi: 10.1253/circj.cj-10-0445. [DOI] [PubMed] [Google Scholar]
- Probst V, Wilde AA, Barc J, et al. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circ Cardiovasc Genet. 2009;2:552–557. doi: 10.1161/CIRCGENETICS.109.853374. [DOI] [PubMed] [Google Scholar]
- Suh Y, Vijg J. SNP discovery in associating genetic variation with human disease phenotypes. Mutat Res. 2005;573:41–53. doi: 10.1016/j.mrfmmm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Tateyama M, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297:1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- Meadows LS, Isom LL. Sodium channels as macromolecular complexes: implications for inherited arrhythmia syndromes. Cardiovasc Res. 2005;67:448–458. doi: 10.1016/j.cardiores.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Wilde AA, Postema PG, Di Diego JM, et al. The pathophysiological mechanism underlying Brugada syndrome: depolarization versus repolarization. J Mol Cell Cardiol. 2010;49:543–553. doi: 10.1016/j.yjmcc.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman MJ, Mohler PJ. Defining a new paradigm for human arrhythmia syndromes: phenotypic manifestations of gene mutations in ion channel- and transporter-associated proteins. Circ Res. 2010;107:457–465. doi: 10.1161/CIRCRESAHA.110.224592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister O, Oikonomopoulos A, Sereti KI, et al. Role of the ATP-binding cassette transporter Abcg2 in the phenotype and function of cardiac side population cells. Circ Res. 2008;103:825–835. doi: 10.1161/CIRCRESAHA.108.174615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner PM. Sudden cardiac death: The larger problem... The larger genome. J CardiovascElectrophysiol. 2009;20:585–596. doi: 10.1111/j.1540-8167.2008.01419.x. [DOI] [PubMed] [Google Scholar]
- Prutkin JM, Sotoodehnia N. Genetics of sudden cardiac arrest. Prog Cardiovasc Dis. 2008;50:390–403. doi: 10.1016/j.pcad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Klassen T, Davis C, Goldman A, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145:1036–1048. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.