Abstract
Spinal muscular atrophy (SMA) is a monogenic disorder that is subdivided into four different types and caused by survival motor neuron gene 1 (SMN1) deletion. Discordant cases of SMA suggest that there exist additional severity modifying factors, apart from the SMN2 gene copy number. Here we performed the first genome-wide methylation profiling of SMA patients and healthy individuals to study the association of DNA methylation status with the severity of the SMA phenotype. We identified strong significant differences in methylation level between SMA patients and healthy controls in CpG sites close to the genes CHML, ARHGAP22, CYTSB, CDK2AP1 and SLC23A2. Interestingly, the CHML and ARHGAP22 genes are associated with the activity of Rab and Rho GTPases, which are important regulators of vesicle formation, actin dynamics, axonogenesis, processes that could be critical for SMA development. We suggest that epigenetic modifications may influence the severity of SMA and that these novel genetic positions could prove to be valuable biomarkers for the understanding of SMA pathogenesis.
Keywords: SMA, DNA methylation profile, CpG sites
Introduction
Proximal spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disorder caused by degeneration of motor neurons in the anterior horns of the spinal cord. SMA is subdivided into four types depending on the age of onset and the severity of symptoms.1, 2 Mutations in the survival motor neuron gene 1 (SMN1) are responsible for all forms of SMA. The SMN1 gene encodes for the ubiquitously expressed SMN protein, which is involved in snRNP biogenesis. SMN appears to fulfill special functions in motor neurons or participates in mechanisms that are critical for their survival.3, 4 Particularly, SMN has been identified in β-actin mRNA transport granules and it might be involved in actin cytoskeleton dynamics via Rho GTPases–ROCK pathway.5, 6 But the precise role of SMN in the degradation of motor neurons is still unknown. The SMN2 gene is an identical centromeric copy of SMN1 and is suggested to be the main modifier of disease manifestation. A strong correlation between the SMN2 gene copy number and the clinical severity of SMA has been confirmed in different populations.7, 8 However, rare cases where siblings with identical SMN1 mutations, identical SMN2 copy numbers show marked clinical discrepancies were reported. As well as observed differences in SMN2 gene copy number between asymptomatic individuals, suggests the existence of additional genetic or epigenetic factors modifying the phenotype.9, 10 Investigation of these modifiers is important for understanding the details of the molecular mechanisms involved in SMA pathogenesis, the development of efficient therapies and identification of new molecular biomarkers. In previous studies, the influence of plastin 3 and profilin IIa proteins level, as well as c.859G>C substitution in the SMN2 gene, on the severity of SMA has been revealed.11, 12, 13 However, these findings can only be applied to a limited number of SMA cases.
DNA methylation was shown to be an important epigenetic modification altering gene expression pattern. Changes of this methylation pattern are associated with various disease processes.14 Several studies have reported differences in DNA methylation level in leukocytes between cancer patients and healthy individuals, supporting the possibility for using this characteristic as a disease marker.15, 16 A differential methylation level of CpG sites near the transcriptional start site of SMN2 was demonstrated between SMA patients of type I and III in DNA samples isolated from leukocyte and fibroblast cell lines.17 However, to date no previous studies have assessed the whole-genome methylation status in SMA patients compared with healthy individuals. Meanwhile, such analysis could be relevant not only to identify new possible SMA modifying factors but also to predict the changes in genomic DNA methylation induced by potential SMA therapy agents, as most of them possess DNA-demethylase activity.17, 18
In this study, we have carried out whole-genome methylation pattern analysis in DNA samples from peripheral whole blood leukocytes from 12 male SMA patients with severe and mild forms of the disease and 11 male healthy individuals of same age.
Materials and methods
Ethics statement
All healthy individuals, adult patients and parents of all children gave written informed consent to the diagnostic procedures. The analysis was approved by the ethics committee at Ott's Institute of Obstetrics and Gynecology RAMS.
Subjects
In all, 12 patients with SMA and 11 healthy individuals from the North-Western region of Russia were selected for this study (Table 1). Only male subjects were included in this study in order to eliminate gender effects. All SMA patients were tested for the deletion in the SMN1 gene and SMN2 gene copy number as described previously.10
Table 1. Information about SMA patients and control individuals.
|
Subjects | ||||||
|---|---|---|---|---|---|---|
| Patients | Type of disease | Age | Age of onset | SMN2 copy numbers | Controls | Age |
| 1 | I | 5 Months | 2 Months | 2 | 1 | 1 Year |
| 2 | I | 3 Months | 3 Months | 2 | 2 | 2 Years |
| 3 | I | 6 Months | Before 6 months | 2 | 3 | 2.5 Years |
| 4 | II | 10 Months | 2.5 Months | 3 | 4 | 2.5 Years |
| 5 | II | 2 Years | Before 1 year | 3 | 5 | 3–4 Years |
| 6 | II | 1.5 Year | Before 1 year | 2 | 6 | 19 Years |
| 7 | IIIb | 18 Years | 15 Years | 4 | 7 | 11 Years |
| 8 | IIIb | 12 Years | 10 Years | 4 | 8 | 19 Years |
| 9 | IIIb | 29 Years | 19 Years | 3 | 9 | 20 Years |
| 10 | IIIb | 19 Years | 5 Years | 4 | 10 | 19 Years |
| 11 | IIIb | 19 Years | 5 Years | 4 | 11 | 20 Years |
| 12 | IV | 21 Years | 21 Years | 4 | ||
DNA methylation profiling
The Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA, USA), which allow assessment of the methylation status of 485 577 cytosines distributed over the whole genome,19 was used to determine the methylation profile of genomic DNA. All genomic DNA was isolated from peripheral blood leukocytes by phenol–chloroform extraction.20 Bisulfite conversion of genomic DNA and sample preparation for the platform analysis was performed as described earlier.21 The BeadChip was read on an Illumina iScan scanner and processed using the GenomeStudio 2009.2 software (Illumina).
Data processing and statistical analysis
All downstream data processing and statistical analyses were performed with the statistical software R (www.r-project.org) together with the lumi, limma and IMA packages of the Bioconductor project.22
Two measurements, β-value and M-value, have been proposed to measure the methylation level.19 The β-value indicates the percentage of methylated alleles of a site and is a biologically more intuitive measurement than the M-value, which is a logarithmic ratio between unmethylated and methylated probes.23 However, the M-value has better statistical properties and was used for conducting the differential methylation analysis, although results were reported using β-values.
Data preprocessing
Quality control
The data were imported and submitted to quality control using the IMA.methy450PP function of the IMA package, resulting in the removal of CpG sites with missing β-values, detection P-value <0.05 and sites where <75% of the samples had a detection P-value <10−5. In addition, samples with missing β-values, with average detection P-values >10−05 and samples having <75% of sites with detection P-value <10−5 were also removed at the sample level. Overall 469 977 CpG sites and all samples passed the quality control successfully.
All downstream analyses were performed separately for two groups, first group including the six SMA patients with type I and II of the disease and the five corresponding control individuals while second group contained the six SMA patients with III and IV type of disease and six the corresponding control individuals (Table 1).
Normalization
Quantile normalization was performed on the M-values of all the 469 977 CpG sites using the lumiMethyN function of the lumi package.
Data processing
Modelization of the methylation level of each CpG site
The relationships between methylation level and variables of interest were determined using limma's robust regression method (lmFit command with setting method=‘robust', maxit=10 000) to fit the following linear model (1) for each CpG site k:
where Mk is the log2 transformed methylation level of CpG site k, H is the dichotomized health state (control=0 and patient=1), A is the age and ɛk is the unexplained variability. The coefficients bkx summarizes the correlation between the methylation level and the variables of interest. Moderated t-statistics for each variable and CpG site were created using an empirical Bayes model as implemented in limma (eBayes command), in order to efficiently correct batch effects.24 To control for false positives, P-values were adjusted for multiple comparisons as proposed by Benjamini and Hochberg (BH).25 Adjusted P-values >0.05 were considered nonsignificant.
Modelization of the methylation level of each region
The similar linear model (2) was implemented for region-based differential methylation analysis for each of the 11 regions included in the 450k BeadChip annotation:
where Mk is the average log2 transformed methylation level of gene/CpG island k. More detailed description can be found in Supplementary 1.
Gene ontology (GO) enrichment analysis
In each group, the gene or genes associated with each gene-based site was identified. The web-based DAVID functional annotation tool (www.david.abcc.ncifcrf.gov) was used to determine the functional groups to which the genes belonged to, and to extract the functional groups most represented in each of the gene lists. All genes from the 450k BeadChip annotation file were used as the background list. The classification stringency was set as ‘medium' (default).
Results
Which genes are differentially methylated between SMA patients and controls?
We aimed to identify genes with distinct methylation patterns between SMA patients and healthy individuals. To this end, the link between the methylation profile and SMA was first identified by fitting a linear model that explains the methylation level for each CpG site and for each of the 11 annotated regions. For each of the linear models, we conducted two separate differential methylation analyses: one in first analyzed group, and another in second analyzed group. Then, we tallied both analyses with each other and looked for similarities between them.
Analysis at the CpG sites level
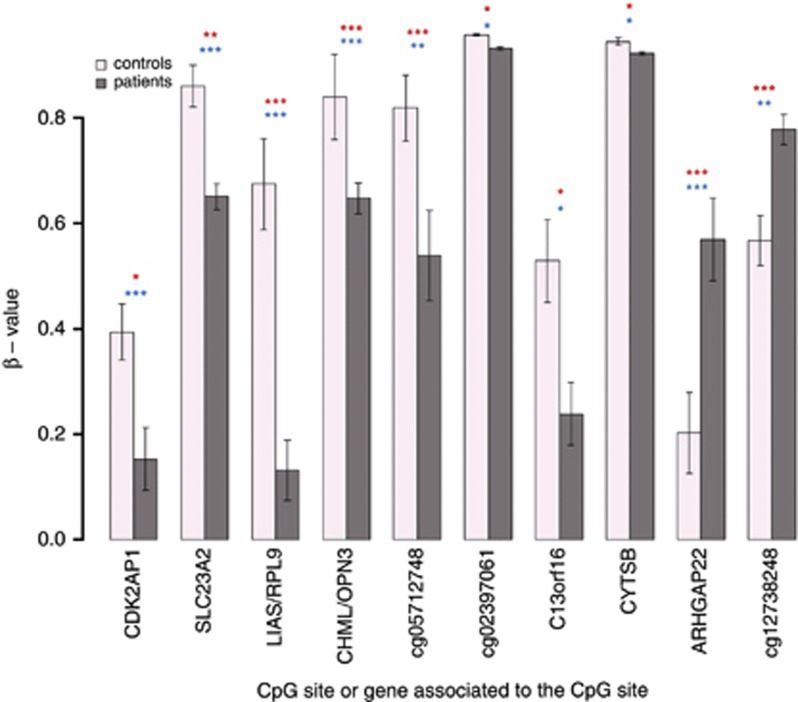
Overall, 1512 and 873 CpG sites were found to be significantly differentially methylated between patients and controls, in first and second groups, respectively. The significant CpG sites were separated into two groups: ones considered significantly more methylated in patients than controls (positive fold change), and ones significantly less methylated in patients than in controls (negative fold change; Figure 1). Only genes associated with sites displaying the same fold change direction in both groups were considered for further analysis. In total, 10 common CpG sites were found to be significantly differentially methylated between patients and controls in both first and second analyzed groups two with a positive fold change, and the remaining 8 having a negative fold change (Figure 2). Seven of these sites were associated with nearby genes (Table 2). In addition, two sites located in the gene body of ARHGAP22 were found to have a significant negative fold change exclusively in second analyzed group.
Figure 1.
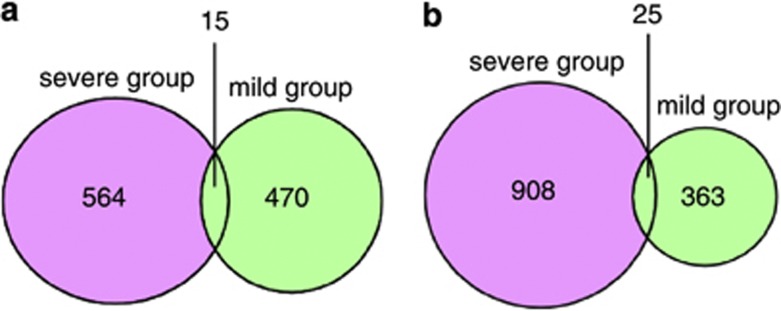
Venn diagrams of the significant CpG sites/genes, which are more methylated in patients than in controls (a) and less methylated in patients than in controls (b). (a) Among 15 CpG sites there were 2 common significant CpG sites for two analyzed groups and 13 common genes with at least one significant CpG sites but not the same for 2 analyzed groups. (b) Among 25 CpG sites there were 8 common significant CpG sites for 2 analyzed groups and 17 common genes with at least one significant CpG sites but not the same for 2 analyzed groups.
Figure 2.
Bar plot of the β-values of 10 CpG sites significantly different methylated in patients than in controls for both analyzed group. First and second analyzed groups are pooled. The vertical black lines are the standard errors of the means. The significance level is presented as * if 0.05>P-value>0.01, ** if 0.01>P-value>0.001 and *** if P-value<0.001 in blue for first analyzed group and in red for second analyzed group.
Table 2. Information about 10 common significantly different methylated CpG sites or genes associated with these CpG sites.
| Gene/CpG site | Annotated region | Genomic location | Average β-value (I group controls) | Average β-value (I group patients) | Adjusted P-value (I group) | Average β-value (II group controls) | Average β-value (II group patients) | Adjusted P-value (II group) |
|---|---|---|---|---|---|---|---|---|
| CDK2AP1 | TSS1500 | chr12:123757860 | 0.39 | 0.193 | 1.24E–02 | 0.396 | 0.113 | 1.80E–08 |
| SLC23A2 | 5′UTR | chr20:4983091 | 0.922 | 0.661 | 2.39E–03 | 0.809 | 0.642 | 3.66E–04 |
| RPL9/LIAS | TSS1500 | chr4:39460490 | 0.816 | 0.0485 | 5.80E–07 | 0.557 | 0.215 | 2.53E–06 |
| CHML/OPN3 | TSS1500 | chr1:241800323 | 0.946 | 0.615 | 4.09E–05 | 0.752 | 0.682 | 5.82E–04 |
| cg05712748 | Genomic | chr1:43472312 | 0.942 | 0.572 | 2.01E–07 | 0.716 | 0.505 | 3.79E–03 |
| cg02397061 | Genomic | chr6:42528725 | 0.957 | 0.932 | 3.11E–02 | 0.957 | 0.931 | 2.21E–02 |
| C13orf16 | Body | chr13:111980537 | 0.541 | 0.364 | 1.72E–02 | 0.517 | 0.114 | 1.51E–02 |
| CYTSB | Body | chr17:20006054 | 0.958 | 0.932 | 3.59E–02 | 0.933 | 0.912 | 3.98E–02 |
| ARHGAP22 | 3′UTR | chr10:49654342 | 0.277 | 0.521 | 3.01E–08 | 0.141 | 0.619 | 8.75E–05 |
| cg12738248 | Genomic | chr7:5156928 | 0.55 | 0.754 | 6.63E–06 | 0.582 | 0.801 | 3.14E–03 |
Annotated region: region where the site is located.
TSS1500=1500-bp upstream of transcription start site (TSS). I group includes SMA patients with severe form of disease and their corresponding controls. II group includes SMA patients with mild form of disease and their corresponding controls.
Apart from these 10 sites, we identified 30 other genes for which there was at least one significant CpG site with the same fold change in both groups. However, the concerned sites were not the same in the two groups, and often in different regions (Supplementary Table). Description of methylation level for some of these genes can be found in Supplementary 1.
The P-values calculated for the 10 common sites belonged to the lowest P-values of all the significant gene/CpG sites found in common between both groups (Supplementary Figure 1), thus supporting the significance of these sites. In addition, when applying a more stringent P-value adjustment (Bonferroni correction instead of BH correction), only the CpG sites associated with LIAS/RPL9 and ARHGAP22 remained significant in both groups.
Analysis at the region level
Description of results of the analysis at the region level can be found in Supplementary 1.
To what GO terms do the genes belong?
Gene enrichment analysis revealed eight GO clusters in first analyzed group that displayed an enrichment score >5, including three clusters involved in the regulation of transcription (enrichment scores 10.3, 7.7 and 6.6). In second analyzed group, only two clusters displayed an enrichment score >5, including one cluster involved in cancer pathways (enrichment score 6.5).
Discussion
In this study, we performed the first genome-wide methylation analysis between patients with severe and mild forms of SMA and healthy individuals of the same age, in order to identify SMA modifying methylation changes. We found 10 CpG sites (Table 2) having significantly different methylation levels both between severe form SMA patients and the corresponding controls, and SMA patients with the mild form and the corresponding controls (Table 1).
It is clear that axon and synaptic pathology has an important role in the development of SMA as a significant reduction in number together with morphological and functional alterations of neuromuscular junctions (NMJs) and neurofilament aggregation have been found in severe SMA mice.26, 27 Moreover, defects in motor neuron axon outgrowth and path finding as well as decrease in the growth cone area, have been observed in different SMA animal models and cell cultures.28, 29, 30 All these processes are connected with disturbances in the regulation of actin filament polymerization and disassembly performed by actin-binding proteins.31 The decreased methylation level at one CpG site in the CHML gene and increased methylation level at another CpG site in the ARHGAP22 gene, between patients and controls in both first and second analyzed groups, is of particular interest in this context. CHML encodes an Rab Escort Protein 2 (REP2) that assists in the geranylgeranylation of most Rab GTPases, whereas the ARHGAP22 gene encodes a Rho GTPase-activating protein (RhoGAP). It is known that Rab and Rho GTPases are important regulators of actin dynamics.32 Using the String 9.0 database,33 we found that the product of the CHML gene, REP2 interacts with Rab5A, Rab3A, Rab1A, Rab6A GTPases and ARHGAP22 protein interacts with RhoB, RhoD, RhoF, RhoG, RhoH, RhoU, RhoQ GTPases (string-db.org, medium confidence score 0.400).
The isoprenol modification of the Rab C-terminus, catalyzed by the REP2 protein, is necessary for Rab GTPases to interact with the membrane and to regulate vesicular trafficking.34, 35 Rab proteins are involved in processes of membrane trafficking, including vesicle formation, vesicle movement along actin and tubulin networks, and membrane fusion.32, 36 The Rab3A GTPase is involved in targeting synaptic vesicles to the active zones and in neurotransmitter release in nerve terminals.37 The number of Rab3A-depleted NMJs was about 25% higher in P0-1 SMA mice than in healthy control mice. A marked decline in Rab3A expression was also observed at P5-6 SMA mice.38 Several Rabs appear to be linked to microtubule- or actin-based motor proteins.36 Indeed, Rab6A interacts with myosin II, dynaein–dynactin complex in microtubule-dependent transport pathways from and to Golgi complex.39, 40 According to the FunCoup database, Rab6A interacts with profilin II (PFN2) pfc-value of 0.552 (Fun-Coup database). Profilin II, which has been confirmed to interact directly with SMN, is a regulator of actin dynamics and is required for actin polymerization in the synapse.6, 41
The product of the ARHGAP22 gene, RhoGAP, converts Rho GTPases to an inactive GDP-bound state. Rho GTPases are known as important regulators of initiating, growth, guidance and branching of axons.32, 42 Cooperation of Rho GTPase effectors, ROCK and Formin family of proteins, is supposed to form actin filaments and actomyosin bundles.43 On one hand inactivation of the Rho/ROCK pathway increases the amount of dephospohorylated profilin IIa, leading to F-actin destabilization.32 On other hand, Rho/ROCK action may negatively regulate the early axon outgrowth development in cultured neurons.32 It is important to note that increased activation of RhoA GTPase was identified in the spinal cord of intermediate SMA mice and the inhibition of the main RhoA target ROCK contributes to their lifespan extension.44 We found increased methylation levels at one CpG site in the ARHGAP22 gene in both severe and mild SMA patients compared with the corresponding controls that may explain the decreased expression level of this gene and consequently the lower level of Rho GTPase inhibition in the patients.
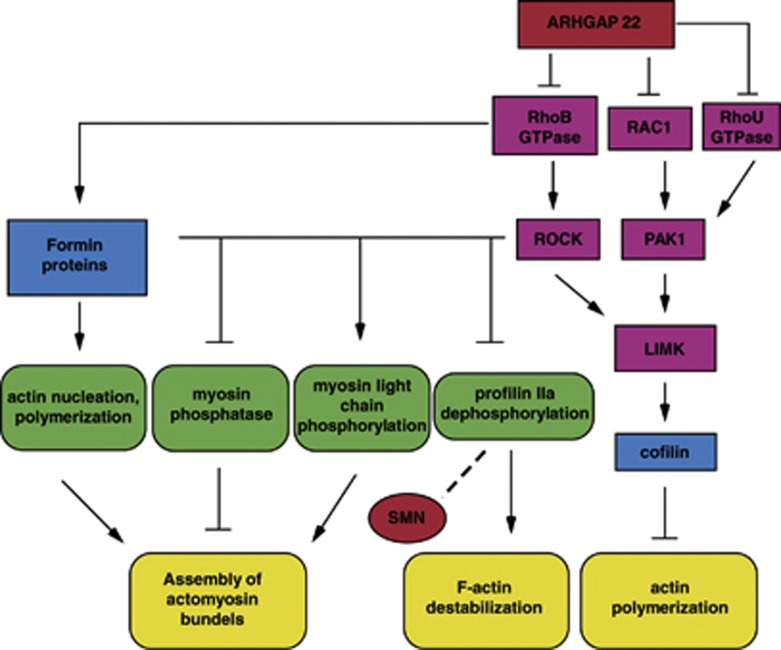
This allows us to hypothesize that proteins that influence Rab and Rho GTPase activity could be SMA severity modifying factors. We propose an hypothetical simplified scheme showing the possible involvement of the ARHGAP22 gene in actin dynamics regulation and the potential connection of this process to SMN (Figure 3).
Figure 3.
Simplified scheme reflecting involvement of ARHGAP22 gene in actin dynamics and cytoskeleton regulation and possible connection of SMN with this process. ARHGAP22 may influence on the activity of RhoB, RhoU, RAC1 GTPases. RhoB GTPase affects ROCK and Formin family of proteins. ROCK is known to phosphorylate myosin light chain, myosin phosphatase and LIM kinase (LIMK). LIMK in its turn phosphorylate and inactivates actin-depolymerizing and severing factor, cofilin. Formin proteins catalyze actin nucleation and polymerization. These processes lead to actin filaments and actomyosin bundles assembly. RAC1 and RhoU GTPase affect actin polymerization through LIMK/cofilin pathway. But the activation of Rho/ROCK pathway may contribute to profilin IIa dephosphorylation and accordingly F-actin destabilization. SMN interacting with profilin IIa may influence on its association with ROCK.
Another differently methylated CpG site is linked to the CDK2AP1 gene. This gene encodes cyclin-dependent kinase 2 associated protein 1, a negative regulator of CDK2. Through downregulation of CDK2 kinase CDK2AP1 protein inhibits G1/S transition of cell cycle.45 Moreover, CDK2AP1 is implicated in apoptosis stimulation and cell proliferation reduction.46 In the context of SMA pathogenesis, CDK2AP1 could be involved in apoptosis initiation of motor neurons. Apoptosis has been demonstrated to have a crucial role in SMA motor neuron degeneration.47, 48
A difference in methylation level was found for a CpG site related to the CYTSB gene (sperm antigen with calponin homology and coiled-coil domains 1). NSP5a3b and NSP5b3b protein isoforms encoded by CYTSB are predicted to have a CH (Calponin-like) domain, which is similar to the CH domain of actin-binding proteins.49 The presence of this domain suggests that these proteins might interact with actin.49 NSP5a3a isoform is possibly involved in RNA metabolism and RNA processing as it interacts with nuclear phosphoprotein B23 and the heterogeneous ribonucleoprotein hnRNP-L. Also, the NSP5a3a isoform could be involved in the induction of apoptosis through an unknown p73-dependent process.50 We therefore speculate that the CYTSB gene may have an effect on SMA severity through its interaction with actin or participation in apoptosis processes.
One differentially methylated CpG site is associated with the SLC23A2 gene, which encodes sodium/ascorbate cotransporter.51 This transporter provides high ascorbate concentration in most tissues. In the CNS, ascorbate has several functions including antioxidant protection, peptide amidation, myelin formation, synaptic potentiation and protection against glutamate toxicity.52 Ascorbate has neuroprotective properties against the oxidative damage associated with different neurodegenerative diseases such as Alzheimer's, Parkinson's and Huntington's disease.52 Thus, the decreased methylation level of CpG site in the SLC23A2 gene in both groups of SMA patients, assuming there is increased expression level of this gene, might denote the SLC23A2 gene could fulfill a certain compensatory function for motor neurons and aid in their survival in SMA patients.
Differences between SMA patients and healthy individuals in the methylation level of a CpG site in the RPL9 gene are not clearly associated to SMA. The RPL9 gene encodes a protein belonging to the L6P family of ribosomal proteins. Putative SMN function in local translational regulation has been hypothesized previously.53 SMN was also found in stress granules of neurons where the translation of some proteins can be blocked during stress reaction.5 It was also demonstrated that SMN is associated with polyribosomes in dendrites.54 The decreased methylation level that we identified in one CpG site in the RPL9 gene in both severe and mild SMA patients might suggest the increased level of the RPL9 gene expression and accordingly the increased amount of RPL9 protein. It is of interest to note that more than a twofold upregulation of RPL32 (ribosomal protein L32), RPL18 (ribosomal protein L8) and more than a twofold downregulation of Rps7 (ribosomal protein S7) has been found in the rostral LAL (levator auris longus) muscle from P1 SMA mice, compared with littermate controls.55
For the remaining four CpG sites strong association with any gene was not demonstrated (Table 2). Additional description and discussion about these sites can be found in Supplementary 1.
The additionally 30 genes (Supplementary Table 1), which contain two or more CpG sites differently methylated between patients and the corresponding controls, but not the same for two analyzed groups, could also be considered as possible candidates as severity modulators of SMA. The group of genes, which are involved in transcription and gene regulation processes: KCNQ1, ATF7IP, WWTR1, NCOR2, PPP1R13L, DIP2C, CD3EAP, WHSC1 (string-db.org, GO enrichment analysis, medium confidence score 0.400) should be noted here. Also among the differentially methylated CpG sites in first analyzed group, gene enrichment analysis revealed three GO clusters of genes that are also involved in transcription regulation. The putative role of SMN in transcription regulation has been previously discussed.56, 57 Discussion for the genes not brought up here can be found in Supplementary 1.
To conclude, we found several differentially methylated candidate loci containing genes that could be possible modifiers of the SMA severity. Several of these identified genes appear to be associated with the cytoskeleton system, processes of neuronal development and maintenance, apoptosis and transcriptional regulation. Further studies are needed to confirm if the peculiarities in the methylation pattern of these genes in blood cells is a distinctive feature, as well as for motor neurons, as the methylation profile of somatic cells may be different.58 The association between the methylation profile and gene expression levels should also be explored as genes may display a relatively poor correlation between CpG methylation status and transcriptional activity and even hypomethylation in TSS may anticorrelate with gene expression.14, 58 Thus, further research will help to make clear the contribution of these candidate genes in the molecular basis of SMA pathology.
Acknowledgments
The methylation array was performed at the Genotyping Technology Platform, Uppsala, Sweden (http://www.genotyping.se) with support from Uppsala University and the Knut and Alice Wallenberg foundation at the Uppsala Genome Center. The study was supported by the Swedish Research Council, Brain Research Foundation, Novo Nordisk, Tore Nilsons foundation, Åhlens foundation, the scholarship within the Swedish Institute Baltic Sea Region Exchange Program, Russian President Scholarship for study abroad and Federal program ‘Scientific and pedagogical manpower of innovative Russia' (2009–2013). We are thankful to Dr Josefin A Jacobsson and Dr Tatyana V Nikitina for helpful advices. Also, we are grateful to Dr Michael Williams for English correction of the article.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Munsat TL, Davies KE. International SMA consortium meeting. Neuromuscul Disord. 1992;2:423–428. doi: 10.1016/s0960-8966(06)80015-5. [DOI] [PubMed] [Google Scholar]
- Zerres K, Rudnik-Schöneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol. 1995;52:518–523. doi: 10.1001/archneur.1995.00540290108025. [DOI] [PubMed] [Google Scholar]
- Briese M, Esmaeili B, Sattelle DB. Is spinal muscular atrophy the result of defects in motor neuron processes. Bioessays. 2005;27:946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- Kong L, Wang X, Choe DW, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572:69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Nölle A, Zeug A, van Bergeijk J, et al. The spinal muscular atrophy disease protein SMN is linked to the Rho-kinase pathway via profilin. Hum Mol Genet. 2011;20:4865–4878. doi: 10.1093/hmg/ddr425. [DOI] [PubMed] [Google Scholar]
- Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuscó I, Barceló MJ, Rojas-García R, et al. SMN2 copy number predicts acute or chronic spinal muscular atrophy but does not account for intrafamilial variability in siblings. J Neurol. 2006;253:21–25. doi: 10.1007/s00415-005-0912-y. [DOI] [PubMed] [Google Scholar]
- Jedrzejowska M, Borkowska J, Zimowski J, et al. Unaffected patients with a homozygous absence of the SMN1 gene. Eur J Hum Genet. 2008;16:930–934. doi: 10.1038/ejhg.2008.41. [DOI] [PubMed] [Google Scholar]
- Zheleznyakova GY, Kiselev AV, Vakharlovsky VG, et al. Genetic and expression studies of SMN2 gene in Russian patients with spinal muscular atrophy type II and III. BMC Med Genet. 2011;12:96. doi: 10.1186/1471-2350-12-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea GE, Kröber S, McWhorter ML, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman M, Anderson CL, Beauvais A, Boyl PP, Witke W, Kothary R. SMN, profilin IIa and plastin 3: a link between the deregulation of actin dynamics and SMA pathogenesis. Mol Cell Neurosci. 2009;42:66–74. doi: 10.1016/j.mcn.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Prior TW, Krainer AR, Hua Y, et al. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet. 2009;85:408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- Rouprêt M, Hupertan V, Catto JWF, et al. Promoter hypermethylation in circulating blood cells identifies prostate cancer progression. Int J Cancer. 2008;122:952–956. doi: 10.1002/ijc.23196. [DOI] [PubMed] [Google Scholar]
- Hauke J, Riessland M, Lunke S, et al. Survival motor neuron gene 2 silencing by DNA methylation correlates with spinal muscular atrophy disease severity and can be bypassed by histone deacetylase inhibition. Hum Mol Genet. 2009;18:304–317. doi: 10.1093/hmg/ddn357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Pr. 1989;3:e3–e4. [Google Scholar]
- Almén MS, Jacobsson JA, Moschonis G, et al. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–137. doi: 10.1016/j.ygeno.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang C-C, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Chai H, Wu Y, et al. Batch effect correction for genome-wide methylation data with Illumina Infinium platform. BMC Med Genomics. 2011;4:84. doi: 10.1186/1755-8794-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- Bowerman M, Murray LM, Beauvais A, Pinheiro B, Kothary R. A critical smn threshold in mice dictates onset of an intermediate spinal muscular atrophy phenotype associated with a distinct neuromuscular junction pathology. Neuromuscul Disord. 2012;22:263–276. doi: 10.1016/j.nmd.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Kariya S, Mauricio R, Dai Y, Monani UR. The neuroprotective factor Wld(s) fails to mitigate distal axonal and neuromuscular junction (NMJ) defects in mouse models of spinal muscular atrophy. Neurosci Lett. 2009;449:246–251. doi: 10.1016/j.neulet.2008.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadi M, Sleigh JN, Walker A, et al. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6:e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AHM, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel TL, McWhorter ML, Workman E, et al. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau PC. Actin in axons: stable scaffolds and dynamic filaments. Results Probl Cell Differ. 2009;48:65–90. doi: 10.1007/400_2009_15. [DOI] [PubMed] [Google Scholar]
- Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Strom M, Godfrey RF, Seabra MC. Structural determinants of Rab and Rab Escort Protein interaction: Rab family motifs define a conserved binding surface. Biochem Biophys Res Commun. 2003;301:92–97. doi: 10.1016/s0006-291x(02)02963-7. [DOI] [PubMed] [Google Scholar]
- Larijani B, Hume AN, Tarafder AK, Seabra MC. Multiple factors contribute to inefficient prenylation of Rab27a in Rab prenylation diseases. J Biol Chem. 2003;278:46798–46804. doi: 10.1074/jbc.M307799200. [DOI] [PubMed] [Google Scholar]
- Hammer JA, Wu XS. Rabs grab motors: defining the connections between Rab GTPases and motor proteins. Curr Opin Cell Biol. 2002;14:69–75. doi: 10.1016/s0955-0674(01)00296-4. [DOI] [PubMed] [Google Scholar]
- Tuvim MJ, Adachi R, Hoffenberg S, Dickey BF. Traffic control: Rab GTPases and the regulation of interorganellar transport. News Physiol Sci. 2001;16:56–61. doi: 10.1152/physiologyonline.2001.16.2.56. [DOI] [PubMed] [Google Scholar]
- Dachs E, Hereu M, Piedrafita L, Casanovas A, Calderó J, Esquerda JE. Defective neuromuscular junction organization and postnatal myogenesis in mice with severe spinal muscular atrophy. J Neuropathol Exp Neurol. 2011;70:444–461. doi: 10.1097/NEN.0b013e31821cbd8b. [DOI] [PubMed] [Google Scholar]
- Short B, Preisinger C, Schaletzky J, Kopajtich R, Barr FA. The Rab6 GTPase regulates recruitment of the dynactin complex to golgi membranes. Curr Biol. 2002;12:1792–1795. doi: 10.1016/s0960-9822(02)01221-6. [DOI] [PubMed] [Google Scholar]
- Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- Pilo Boyl P, Di Nardo A, Mulle C, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine-Bertrand J, Villemure J-F, Lamarche-Vane N. Implication of rho GTPases in neurodegenerative diseases. Curr Drug Targets. 2011;12:1202–1215. doi: 10.2174/138945011795906543. [DOI] [PubMed] [Google Scholar]
- Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr. 2011;5:170–180. doi: 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman M, Beauvais A, Anderson CL, Kothary R. Rho-kinase inactivation prolongs survival of an intermediate SMA mouse model. Hum Mol Genet. 2010;19:1468–1478. doi: 10.1093/hmg/ddq021. [DOI] [PubMed] [Google Scholar]
- Wong DTW, Kim JJ, Khalid O, Sun HH, Kim Y. Double edge: CDK2AP1 in cell-cycle regulation and epigenetic regulation. J Dent Res. 2012;91:235–241. doi: 10.1177/0022034511420723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno Y, Patel V, Kim Y, et al. Apoptosis, proliferation and p12doc-1 profiles in normal, dysplastic and malignant squamous epithelium of the Syrian hamster cheek pouch model. Oral Oncol. 2002;38:274–280. doi: 10.1016/s1368-8375(01)00055-0. [DOI] [PubMed] [Google Scholar]
- Tsai MS, Chiu YT, Wang SH, Hsieh-Li HM, Lian WC, Li H. Abolishing Bax-dependent apoptosis shows beneficial effects on spinal muscular atrophy model mice. Mol Ther. 2006;13:1149–1155. doi: 10.1016/j.ymthe.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Okouchi M, Ekshyyan O, Maracine M, Aw TY. Neuronal apoptosis in neurodegeneration. Antioxid Redox Signal. 2007;9:1059–1096. doi: 10.1089/ars.2007.1511. [DOI] [PubMed] [Google Scholar]
- D'Agostino L, Giordano A. Possible functional role of NSPs in cancer. Cell Cycle. 2008;7:1810–1827. doi: 10.4161/cc.7.12.6023. [DOI] [PubMed] [Google Scholar]
- D'agostino L, Giordano A. NSP 5a3a: a potential novel cancer target in head and neck carcinoma. Oncotarget. 2010;1:423–435. doi: 10.18632/oncotarget.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WJ, Johnson D, Jarvis SM. Vitamin C transport systems of mammalian cells. Mol Membr Biol. 2001;18:87–95. doi: 10.1080/09687680110033774. [DOI] [PubMed] [Google Scholar]
- May JM. Vitamin C transport and its role in the central nervous system. Subcell Biochem. 2012;56:85–103. doi: 10.1007/978-94-007-2199-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazzon N, Rage F, Schlotter F, Moine H, Branlant C, Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J Biol Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- Béchade C, Rostaing P, Cisterni C, et al. Subcellular distribution of survival motor neuron (SMN) protein: possible involvement in nucleocytoplasmic and dendritic transport. Eur J Neurosci. 1999;11:293–304. doi: 10.1046/j.1460-9568.1999.00428.x. [DOI] [PubMed] [Google Scholar]
- Mutsaers CA, Wishart TM, Lamont DJ, et al. Reversible molecular pathology of skeletal muscle in spinal muscular atrophy. Hum Mol Genet. 2011;20:4334–4344. doi: 10.1093/hmg/ddr360. [DOI] [PubMed] [Google Scholar]
- Strasswimmer J, Lorson CL, Breiding DE, et al. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum Mol Genet. 1999;8:1219–1226. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- Campbell L, Hunter KMD, Mohaghegh P, Tinsley JM, Brasch MA, Davies KE. Direct interaction of Smn with Dp103, a putative RNA helicase: a role for Smn in transcription regulation. Hum Mol Genet. 2000;9:1093–1100. doi: 10.1093/hmg/9.7.1093. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands—‘a rough guide'. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.