Abstract
Extensive programmed rearrangement of DNA, including DNA elimination, chromosome fragmentation, and DNA descrambling, takes place in the newly developed macronucleus during the sexual reproduction of ciliated protozoa. Recent studies have revealed that two distant classes of ciliates use distinct types of non-coding RNAs to regulate such DNA rearrangement events. DNA elimination in Tetrahymena is regulated by small non-coding RNAs that are produced and utilized in an RNAi-related process. It has been proposed that the small RNAs produced from the micronuclear genome are used to identify eliminated DNA sequences by whole-genome comparison between the parental macronucleus and the micronucleus. In contrast, DNA descrambling in Oxytricha is guided by long non-coding RNAs that are produced from the parental macronuclear genome. These long RNAs are proposed to act as templates for the direct descrambling events that occur in the developing macronucleus. Both cases provide useful examples to study epigenetic chromatin regulation by non-coding RNAs.
Introduction: Big discoveries from a peculiar organism
The Nobel prize-winning discoveries of ribozymes (Chemistry, 1989) and telomeres (Physiology or Medicine, 2009) illustrate the utility of Tetrahymena thermophila as a eukaryotic model for the study of non-coding RNAs. But why was Tetrahymena used for these studies? It turns out that these major achievements relied on a peculiarity of ciliate chromosomes.
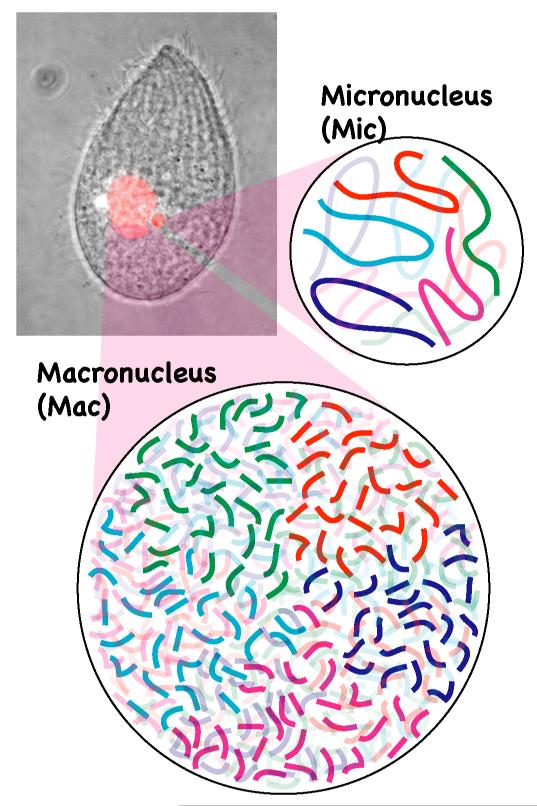
Like most other ciliates, Tetrahymena has two different nuclei, a micronucleus and a macronucleus, within a single cell (Fig. 1). The micronucleus is diploid and contains 10 chromosomes (five per haploid genome). However, the macronucleus has over 20,000 chromosomes in Tetrahymena. There are several reasons for this extremely high number of chromosomes in the macronucleus: 1) there are approximately 250 different chromosomes per haploid genome, 2) most chromosomes attain ploidy levels of around 50, and 3) the rDNA “minichromosome” is amplified to 10,000 copies [1]. Therefore, a single Tetrahymena cell has over 400 times more chromosome ends (i.e., telomeres) than a human cell. This difference was one of the key reasons why telomere structure was first determined in Tetrahymena [2]. Telomerase activity, which elongates telomere repeats, was also first identified in Tetrahymena [3]. Later studies have revealed that the telomerase complex consists of telomerase RNA, a non-coding RNA that acts as a template for the elongation of telomeres, and proteins including telomerase reverse transcriptase [4]. The unique minichromosome structure of the rDNA and its extensive amplification also allowed researchers to determine the primary structure and a detailed transcriptional map of rDNA by the late 1970s [5]. This led to the unexpected discovery of a self-splicing intron in the Tetrahymena rDNA [6].
Figure 1. Nuclear dimorphism in Tetrahymena.
Like most ciliates, Tetrahymena thermophila (picture) has two different types of nuclei (highlighted in red) in a single cell: a macronucleus (Mac) and a micronucleus (Mic). The micronucleus has five chromosomes per haploid genome whereas the macronucleus has over 20,000 chromosomes.
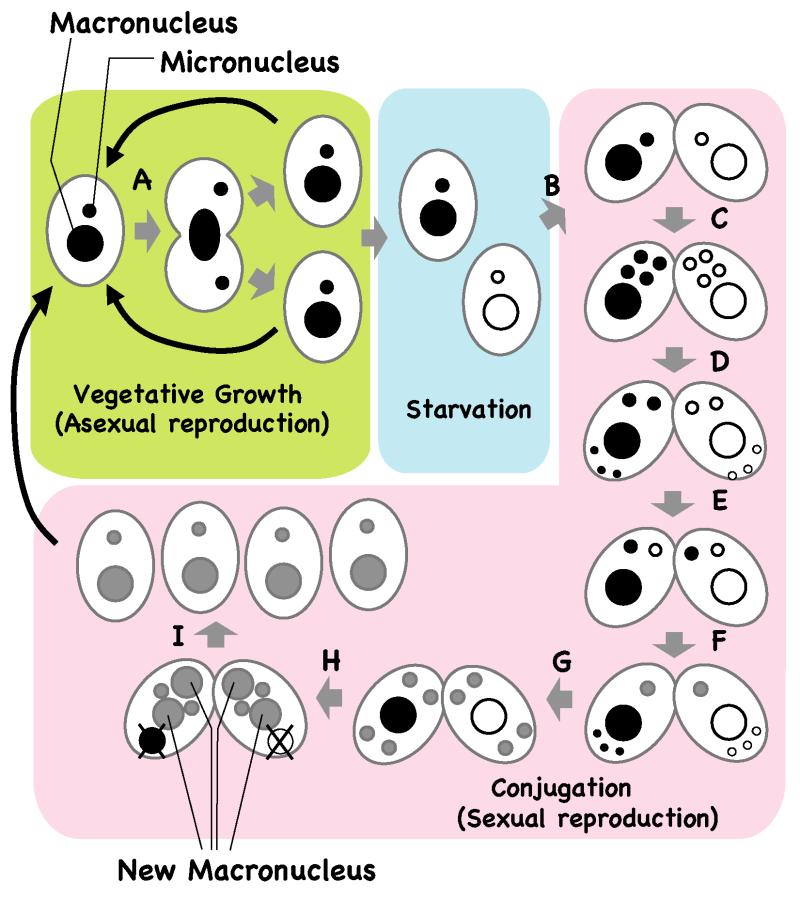
Despite the drastic differences in their chromosomal content, the micronucleus and macronucleus are produced from a common zygotic nucleus during sexual reproduction. The life cycle of Tetrahymena is presented in Figure 2. When enough nutrients are available, Tetrahymena proliferates asexually by binary fission. The micronucleus divides mitotically whereas the macronucleus divides by amitosis (Fig. 2A). In the absence of sufficient nutrients, Tetrahymena undergoes the sexually reproductive process of conjugation (Fig. 2B-F). In this process, two cells of complementary mating types partially fuse to form a pair (Fig. 2B) and their micronuclei undergo meiosis (Fig. 2C). One of the four meiotic products in each cell is selected to undergo one mitotic division and yield two haploid pronuclei; the other three meiotic products are degraded (Fig. 2D). The paired cells exchange one of their pronuclei (Fig. 2E), and the stationary and migratory pronuclei fuse into a zygotic nucleus (Fig. 2F). The zygotic nucleus undergoes two mitotic divisions (Fig. 2G). The two anteriorly positioned daughter nuclei differentiate into macronuclei whereas the two posterior nuclei become micronuclei, and the parental macronucleus is eliminated by an apoptosis-like process (Fig. 2H). The pairing then dissolves and the progeny resume vegetative growth when nutrients are available (Fig. 2I).
Figure 2. Life cycle of Tetrahymena.
When enough nutrients are available, vegetative Tetrahymena cells multiply by binary fission and the macronucleus and micronucleus divide independently (A). After prolonged starvation, two cells of complementary mating types fuse to start the sexual reproduction process of conjugation (B). Their micronuclei undergo meiosis (C) and one of the meiotic products survives and divides mitotically, giving rise to two gametic nuclei, one of which is stationary and the other migratory (D). The migratory gametic nucleus crosses the conjugation bridge (E) and the gametic nuclei fuse to produce a diploid zygotic nucleus (F). The zygotic nucleus undergoes two mitotic divisions (G), and two of the products differentiate as macronuclei while the other two differentiate as micronuclei (H). The parental macronucleus becomes pyknotic and is resorbed. Finally, the pairing dissolves and the progeny resume vegetative growth when nutrients are available again (F).
During the differentiation of the new macronucleus in Tetrahymena, micronuclear chromosomes are fragmented by chromosome breakage, which is followed by the formation of new telomeres (Fig. 3A). In addition, approximately 6,000 internal DNA segments are eliminated and the macronuclear chromosomes are endoreplicated to around 50 copies (Fig. 3A). In parallel, the single micronuclear rDNA locus is excised, rearranged into an inverted repeat, telomeres are formed at both ends, and endoreplicated to upward of 10,000 copies (Fig. 3B). Although the patterns of rDNA rearrangement vary between different ciliate species, chromosome fragmentation and DNA elimination are common traits in most ciliates [7] [8]. Moreover, DNA descrambling (detailed below) occurs in some spirotrich ciliates such as Oxytricha and Stylonychia (Fig. 3C).
Figure 3. DNA rearrangement and endoreplication in ciliates.
(A) DNA elimination and chromosome fragmentation in Tetrahymena. Numerous internal eliminated sequences (IESs, green lines marked with the letter “i”) are removed and two flanking macronuclear-destined sequences are re-ligated. In parallel, chromosome breakage at the Cbs (chromosome breakage sequence, marked with an arrow and the letter “c”) occurs and new telomeres (red triangles) are formed. The macronuclear chromosomes are eventually endoreplicated to around 50 copies.
(B) rDNA rearrangement in Tetrahymena. The single micronuclear rDNA locus (blue arrow) is excised and rearranged into an inverted repeat. Telomeres are formed at both ends de novo and endoreplicated to approximately 10,000 copies.
(C) DNA descrambling in spirotrich ciliates (e. g., Oxytricha). Many genes are fragmented, “scrambled” (that is, not 1-2-3-4-5-6 but 1-3-2-4-5-6, in this example) and some segments are inverted (segment 5 in this example) and separated by IESs (green lines marked with “i”) in the micronucleus (Mic). They are joined and assembled (descrambled) into the proper order and direction in the macronucleus (Mac).
In the past decade, we have come to realize that DNA elimination and DNA descrambling are regulated by non-coding RNAs. In this review, I will first overview how short non-coding RNAs regulate programmed DNA elimination in Tetrahymena. Then, I will describe long non-coding RNA-mediated DNA descrambling in Oxytricha.
DNA elimination in Tetrahymena is epigenetically regulated by short RNAs
During macronuclear development in Tetrahymena, approximately 6,000 different internal DNA segments are removed and the flanking sequences are religated (Fig. 3A). A stretch of DNA destined to be eliminated is called an internal eliminated sequence (IES). IESs are 0.5-20 kb in size and a total of around 20 Mbp (~15%) of DNA in the micronuclear genome is removed from the newly formed macronuclear chromosomes. IESs have not been found in gene-coding sequences in Tetrahymena, although some are located in introns [9]. Many of the known IESs are transposon-like repeats or other repeated sequences (Fig. 4) [10] [11] [12], although some are single-copy sequences [13].
Figure 4. Elimination of transposon repeats from the macronucleus.
Tetrahymena cells in a vegetative state were used to detect two distinct transposon-related sequences, Tlr1 (top) and REP (bottom), by fluorescent in situ hybridization (FISH, green, left). DNA was stained with DAPI (magenta, middle). The micronucleus (i) and the macronucleus (a) are marked.
IES elimination in Tetrahymena can occur reproducibly at a specific site or at a limited number of alternative sites. The boundaries of IESs are relatively precise (within a few nucleotides) when compared between different progeny [14]. However, no consensus sequence motif has been identified in or around IESs. The only common elements analyzed to date that are associated with IESs are short (1-8 bp) direct repeats of varied sequences at the ends of the IESs; these sequences have no known function in IES removal [7,8]. In addition, the flanking regions of some IESs have cis-acting sequences required for IES removal but no sequence homology has been observed across different elements [7,8]. Therefore, we can identify IESs only by directly comparing micronuclear and macronuclear DNA sequences. It has long been known that IES elimination is regulated by an epigenetic mechanism, such that the new macronucleus copies the sequence pattern of the parental macronucleus [7,8] [15,16]. To achieve such epigenetic regulation, some sequence-specific information must be transferred from the parental macronucleus to the new macronucleus. Several lines of evidence indicate that this trans-nuclear information is carried by 28- to 29-nucleotide small RNAs that are made by an RNAi-related pathway.
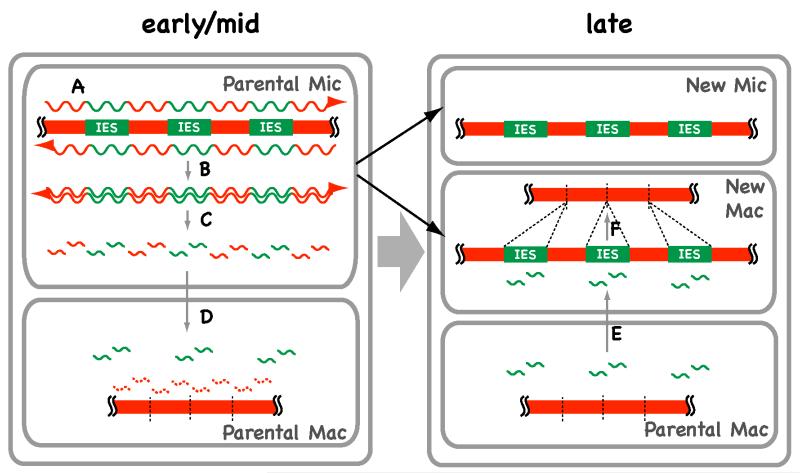
To provide a general idea of how these small RNAs epigenetically support the precise elimination of IESs without any common DNA sequence, I first describe here a model termed the scan RNA model (Fig. 5), which we have proposed previously [17,18]. The pathway starts by bi-directional transcription of the entire micronuclear genome during early conjugation (Fig. 5A). The transcripts form double-stranded RNAs (dsRNAs) (Fig. 5B) that are processed to short RNAs by RNAi-related machinery (Fig. 5C). We call these small RNAs scan (scn) RNAs because they scan macronuclear and micronuclear DNA to identify IESs. The scnRNAs then localize to the (parental) macronucleus, where those that have homologous macronuclear DNA sequences are degraded (Fig. 5D). As a result, only scnRNAs homologous to micronucleus-specific sequences (i.e., IESs) remain in the parental macronucleus. Finally, these IES-specific scnRNAs move to the developing macronucleus in later stages (Fig. 5E). There, sequences homologous to the scnRNAs are identified and targeted for elimination (Fig. 5F). In this way, IESs can be identified purely by the scnRNA-mediated comparison of micronuclear and macronuclear sequences, just as we do in silico.
Figure 5. A model for small RNA-directed DNA rearrangement in Tetrahymena.
In the early conjugation stages, the genome of the micronucleus (Mic), including the IESs, is transcribed bi-directionally (A) and the resulting transcripts form dsRNA molecules (B). The dsRNAs are processed into small RNAs (scnRNAs) (C). The scnRNAs are transferred to the parental macronucleus (Mac) and any scnRNAs homologous to DNA sequences in the parental Mac are degraded in the mid-conjugation stages (D). In late conjugation stages, the scnRNAs that were not degraded in the parental Mac (those homologous to IESs) are transferred to the developing new Mac (E), where they target IESs to be eliminated by base pairing (F).
Small RNA-directed DNA elimination in Tetrahymena
RNAi-related pathways are unified by their dependence on base-pairing interaction s between small (~20-30 nt) RNAs and target sequences and their common use of Argonaute family proteins [19]. Small RNAs can be produced by Dicer family proteins from various dsRNAs or by a Dicer-independent mechanism from single-stranded RNAs [20]. Some classes of small RNAs in mammals and plants are 2′-O-methylated at their 3′ termini by conserved homologues of the RNA methyltransferase Hen1 [21] [22]. The Tetrahymena genome encodes 12 Argonaute proteins that interact with different classes of small RNAs [23] [24], three Dicer proteins that have distinct biological roles [25] [23], and a single Hen1 homologue [26].
The 28- to 29-nucleotide small RNAs known as scnRNAs are expressed exclusively during sexual reproduction [17]. They interact with the Argonaute protein Twi1p [27], are produced by the Dicer protein Dcl1p [28] [25], and are stabilized by 2′-O-methylation at their 3′-termini by the RNA methyltransferase Hen1p [26]. Twi1p and Dcl1p are indispensable for DNA elimination [17] [28] [25] and Hen1p is required for efficient DNA elimination [26], indicating that scnRNAs play a pivotal role in DNA elimination. Consistent with this idea, injection of dsRNAs designed to be homologous to a macronuclear-destined sequence (MDS) into conjugating cells induced the elimination of the target MDS sequence from newly formed macronuclei in the same manner as an IES [29]. Thus, dsRNAs, and probably the small RNAs processed from them, are the primary signal for DNA elimination. Dicer-dependent accumulation of scnRNAs and the requirement for this protein in DNA elimination have also been reported in Paramecium [30]. Therefore, this scnRNA-directed DNA elimination mechanism is probably conserved among oligohymenophorean ciliates.
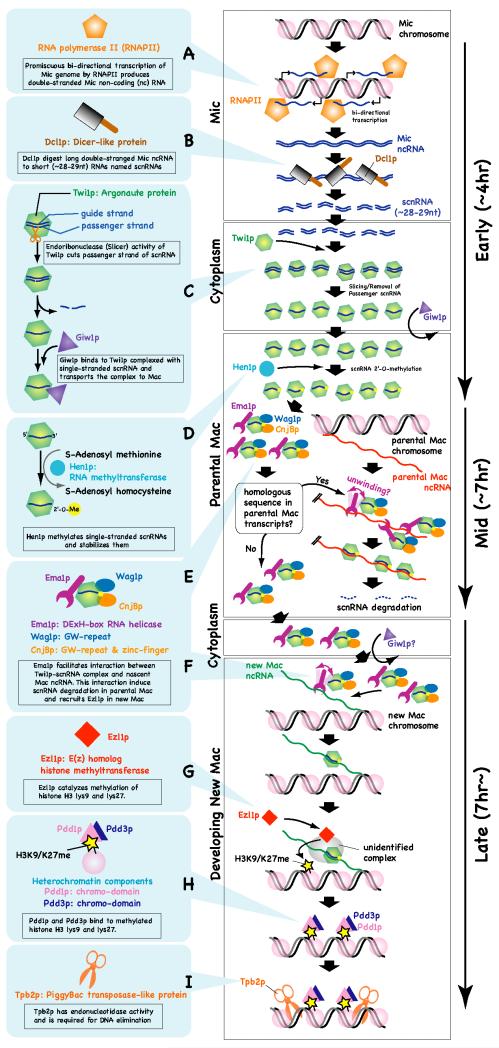
Because scnRNAs can hybridize to IES sequences in vitro, it was expected that they were derived from the micronucleus [17] [27]. Although the micronucleus is transcriptionally inert in most of the life stages of Tetrahymena (and of other ciliates as well), it becomes transcriptionally active during prophase of meiosis [31] [32]. In this stage, at least some reagions of the micronuclear genome are transcribed bidirectionally [33]. Although the exact mechanism that produces these micronuclear transcripts is not clear, temporal micronuclear localization of TATA-binding protein [34] and RNA polymerase II (RNAPII) [35] during prophase of meiosis indicates that they are transcribed by RNAPII (Fig. 6A). The Dicer protein Dcl1p also localizes to the micronucleus during meiosis and disruption of the DCL1 gene results in loss of scnRNA production and over-accumulation of micronuclear RNA [25]. These results indicate that Dcl1p processes micronuclear transcripts into scnRNAs in the micronucleus during meiosis (Fig. 6B).
Figure 6. Small RNA-directed DNA rearrangement in Tetrahymena.
Events occurring sequentially are shown from top to bottom. The approximate stages at which the events occur are indicated on the right by arrows. See text for details. Mic: micronucleus, Mac: Macronucleus.
On the other hand, the Argonaute protein Twi1p localizes to the cytoplasm in the meiotic stages of Tetrahymena development [17] (Mochizuki et al. 2002). Thus, scnRNAs produced in the micronucleus are probably transported to the cytoplasm where they complex with Twi1p (Fig. 6C). The molecular mechanism involved in the micronuclear export of scnRNAs has not yet been elucidated. In the mid-conjugation stages, Twi1p-scnRNA complexes localize to the parental macronucleus [17]. Because Dcl1p is required for the macronuclear localization of Twi1p [36], Twi1p must be complexed with an scnRNA to be imported into the macronucleus. In addition, the endoribonuclease activity of Twi1p, which is vital for removing one of the two strands of an scnRNA from the Twi1p-scnRNA complex, and the Twi1p-associated protein Giw1p are essential for the macronuclear localization of Twi1p [36]. Because the endoribonuclease activity of Twi1p is required for its association with Giw1p, Giw1p probably senses the state of Twi1p-associated scnRNAs and selectively transports mature Twi1p-siRNA complexes into the nucleus (Fig. 6C) [36].
Tetrahymena Hen1p only methylates single-stranded RNAs [26]. Therefore, scnRNAs can be 2′-O-methylated by Hen1p only after the Twi1p-scnRNA complex releases one of the two scnRNA strands. Because Hen1p is localized to the parental macronucleus [26], 2′-O-methylation of scnRNA probably occurs after the mature Twi1p-siRNA complex is imported into the parental macronucleus (Fig. 6D). Loss of Hen1p causes complete abolishment of scnRNA methylation, a gradual reduction in the accumulation and the length of scnRNAs, and a defect in DNA elimination [26]. Therefore, Hen1p-mediated 2′-O-methylation stabilizes scnRNAs and ensures DNA elimination in Tetrahymena.
Although scnRNAs complementary to both macronuclear-destined sequences and micronuclear-specific sequences (mostly IESs in Tetrahymena) are produced in the early conjugation stage, only the second class of scnRNAs becomes gradually enriched during the mid-conjugation stages [27] [37]. As described above, Twi1p-scnRNA complexes are localized to the parental macronucleus in the mid-conjugation stages. Therefore, it is reasonable to expect that some mechanism specifically degrades scnRNAs complementary to the genomic DNA in the parental macronucleus.
The three Twi1p-binding proteins Ema1p (RNA helicase), CnjBp (GW repeat and Zinc-finger protein), and Wag1p (GW repeat protein) play important roles in the selective degradation of scnRNAs [37] [38]. Ema1p mediates the interaction between nascent non-coding transcripts and scnRNA-Twi1p complexes in the parental macronucleus [37]. Therefore, it has been proposed that scnRNA degradation is induced by a base-pairing interaction between scnRNAs and nascent non-coding transcripts from the parental macronucleus (Fig. 6E). The importance of parental macronuclear transcripts for proper DNA elimination has also been demonstrated in studies of another ciliate, Paramecium tetraurelia, in which down-regulation of the macronuclear non-coding transcripts by RNAi blocked the scanning process in targeted regions and induced ectopic DNA elimination [39]. CnjBp and Wag1p have redundant roles in the selective degradation of scnRNAs [38], although the molecular mechanisms underlying their involvement remain unclear.
When the new macronucleus is developed, the localization of Twi1p changes from the parental macronucleus to the new macronucleus (Fig. 6F) [17]. In the newly formed macronucleus, Twi1p is required for the methylation of histone H3 at lysines 9 (H3K9me) and 27 (H3K27me), reactions catalyzed by the enhancer-of-zeste homologue Ezl1p (Fig. 6G) [40] [41]. These histone modifications are bound by the chromodomain proteins Pdd1p and Pdd3p (Fig. 6H) [42] [41]. During the process of DNA elimination, IESs are found in dense heterochromatic regions at the nuclear periphery [43]. Twi1p, Ezl1p, and Pdd1p are required for the formation of this heterochromatic structure and for DNA elimination [44] [40,41]. Therefore, IESs are wrapped in heterochromatin before they are eliminated, and the formation of this structure is essential for DNA elimination. Because histone modifications and the chromodomain protein Pdd1p, which binds to these modifications, accumulate specifically on IESs in a Dcl1p- and Twi1p-dependent manner [42] [40,41], scnRNAs selected for IES specificity must identify their complementary sequence and recruit the histone methyltransferase Ezl1p. It has been suggested that nascent non-coding transcripts mediate the interaction between chromatin and scnRNA-Twi1p complexes in the new macronucleus as in the parental macronucleus (Fig. 6F) [37]. However, it is not yet clear how the RNAi-related mechanism and Ezl1p are linked.
A study in which an IES element was extensively mutagenized revealed that the rearrangement efficiency of an IES is correlated with the overall length of the IES and that the lower size threshold is 300 bp [45]. It has been suggested that either a length of more than 300 bp is required for proper targeting by small RNAs or a structure larger than that of a di-nucleosome is necessary to induce efficient DNA elimination.
Because artificial tethering of the heterochromatin component Pdd1p to a locus is sufficient to induce its ectopic DNA elimination [42], Pdd1p can recruit all proteins required for DNA elimination. Recently it has been reported that a PiggyBac transposase-like protein is required for DNA elimination in the oligohymenophorean ciliates Paramecium and Tetrahymena (Fig. 6I) [46] [47]. In both ciliates, the PiggyBac transposase-like proteins are encoded in the macronuclear genome, suggesting that a transposase invaded an ancestor of these ciliates and has now been domesticated in the host genome. Because the Tetrahymena PiggyBac-like protein Tpb2p localizes to heterochromatin foci on the nuclear periphery in the developing macronucleus [47], this protein may directly recognize heterochromatin structures and excise IESs. Tpb2p shows some sequence preference for a DNA-cutting site in vitro [47]. This sequence preference may also contribute to the precise elimination of IESs in addition to homology-dependent IES recognition by scnRNAs.
RNA-directed DNA descrambling in spirotrich ciliates
In spirotrich ciliates such as Euplotes, Stylonychia, and Oxytricha, more than 95% of the genome is eliminated during macronuclear development. The lest of their genome is extensively fragmented to approximately 20,000 chromosomes in the macronucleus (per haploid genome), with an average size of around 2 kb. These chromosomes often contain only single genes and are therefore referred to as “gene-sized” chromosomes [7] [48]. Some spirotrich ciliates, such as Stylonychia and Oxytricha, not only eliminate IESs but also perform “descrambling” of protein-encoding sequences during DNA rearrangement (Fig. 3C). In this process, “scrambled” protein-encoding sequences in the micronucleus are joined and assembled (descrambled) into the proper order in the macronucleus. Studies of DNA rearrangement in a developmental time course revealed that the removal of conventional IESs tends to precede the complex events of inversion and translocation [49]. It has been estimated that 20-30% of genes in these spirotrichs are scrambled in the micronucleus.
Short-sequence repeats, called “pointers,” found at MDS-IES junctions are involved in DNA rearrangement in spirotrich ciliates [48]. A pointer at the junction between an MDS, n, and the downstream IES is generally identical to a pointer between MDS n+1 and its upstream IES. However, pointer repeats are very short. The average repeat length is 4 bp between non-scrambled segments and 11 bp between scrambled ones. Therefore, these pointers are probably not sufficient for the identification of sites of DNA rearrangement.
Two types of non-coding RNAs have been suggested to be involved in DNA rearrangement in spirotrich ciliates. The first consists of 25- to 30-nt small RNAs [50]. These small RNAs are homologous to micronucleus-specific DNA sequences, like scnRNAs during the development of new macronuclei in Tetrahymena. Moreover, RNAi-mediated knock-down of Stylonychia PIWI, which is a potential binding partner of the small RNAs, caused loss of histone H3 methylation at lysine 9 [50] and an arrest in the development of new macronuclei [51] in Stylonychia. Therefore, it has been proposed that spirotrichs have a small RNA-directed DNA rearrangement mechanism like that of oligohymenophoreans, in which small RNAs specify the IESs to be removed and recruit chromatin-modifying enzymes to induce heterochromatin formation at these sequences [50]. However, as over 95% of the IESs in Stylonychia are shorter than 100 bp, that is, shorter than the length of DNA in a nucleosome, it is unlikely that small RNA-directed chromatin modification can direct precise DNA elimination of these short IESs. Moreover, many IESs in spirotrich ciliates are even smaller than 10 bp and therefore could hardly be targeted precisely by small RNAs 25-30 nt in length. As it has not been demonstrated that PIWI interacts with small RNAs or that PIWI and small RNAs have a direct role in DNA rearrangement, further studies are necessary to conclude that small RNAs play a role in DNA rearrangement in spirotrichs.
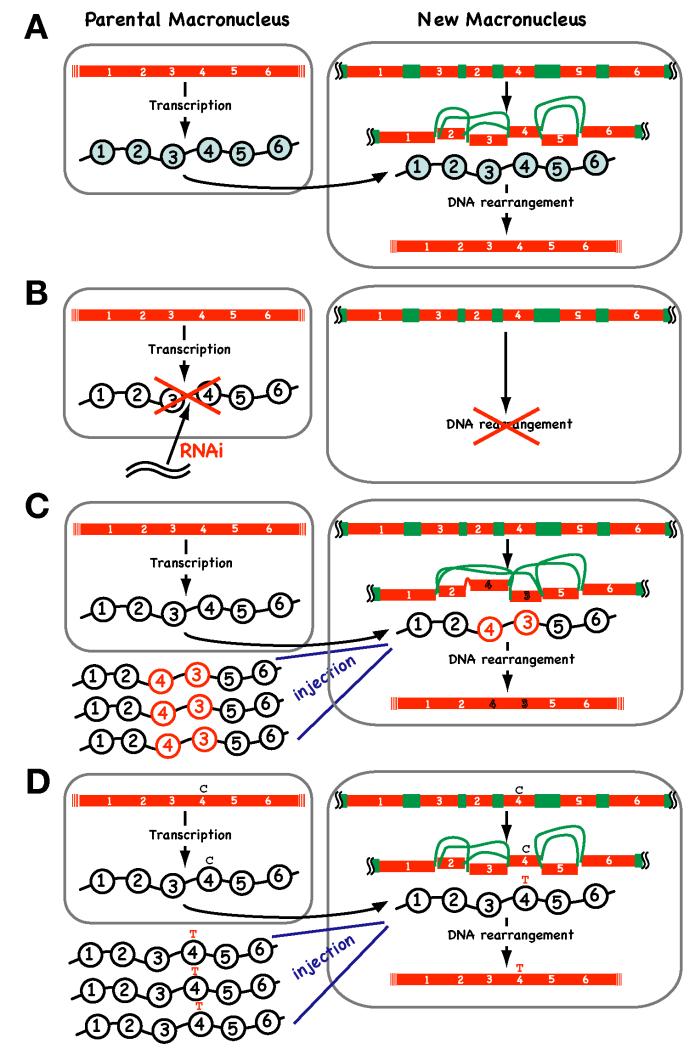
Although the involvement of small RNAs in DNA rearrangement in spirotrichs is still under debate, it is certain that they are not able to support the complex DNA descrambling process (Fig. 3C). Recent studies in Oxytricha strongly suggest that another type of non-coding RNAs, long macronuclear RNAs, is predominantly utilized to guide the descrambling event (Fig. 7A). Long RNA-guided DNA descrambling was first proposed as a theoretical model [52] [53] and has been demonstrated experimentally recently [54]. Bi-directional transcription of the parental macronuclear genome occurs during early conjugation in Oxytricha. Macronuclear long RNAs are likely produced by “telomere to telomere” transcription of short “gene-sized” macronuclear chromosomes (Fig. 7A). RNAi knock-down of specific parental macronuclear long RNAs inhibits descrambling in addition to eliminating non-scrambled IESs in the corresponding loci in the new macronucleus (Fig. 7B). Moreover, the injection of artificial RNAs reprograms the descrambling order (Fig. 7C). Therefore, long macronuclear RNAs act as templates in DNA rearrangement in Oxytricha.
Figure 7. Long non-coding RNA-guided DNA descrambling in Oxytricha.
(A) A model for RNA-guided DNA descrambling in Oxytricha. Telomere-to-telomere transcription of parental macronuclear “gene-sized” chromosomes produces guide RNAs (wavy lines with circles), which are then transported to the newly developed macronucleus where they act as scaffolds to guide DNA rearrangement. (B) Disruption of long non-coding RNAs by RNAi causes a defect in DNA rearrangement. (C) Microinjection of artificial templates (in this example, RNA having a 1-2-4-3-5-6 sequence) alters the order of the DNA descrambling pattern in the new macronucleus. (D) Microinjection of artificial templates that have base substitutions (C to T in this example) alters the DNA sequence of the new macronucleus.
Transposases encoded in a group of abundant micronucleus-limited DNA transposons are expressed in a developmentally programmed fashion and mediate both DNA descrambling and IES elimination in Oxytricha [55]. Further research is needed to understand how long non-coding RNAs and transposases are physically and functionally linked.
RNA-directed DNA proofreading
An even more surprising function for long non-coding RNAs has been identified in Oxytricha. Nowacki et al. [54] found that base substitutions introduced into injected RNAs were occasionally transferred to new macronuclear DNA (Fig. 7D). This indicates that there is an RNA-directed DNA proofreading mechanism that allows acquired somatic mutations to be transmitted to the next generation. Similar non-Mendelian inheritance phenomena have been described in the plant Arabidopsis [56]. In loss-of-function HOTHEAD mutants of Arabidopsis, DNA polymorphisms absent in the parent but present in previous generations reappear at a high frequency in the progeny of homozygous mutant plants. It ha s been postulated that heritable RNAs covering the genome guide template-directed changes in DNA sequence. Therefore a DNA proofreading mechanism mediated by an “RNA cache” may be widely distributed among eukaryotes.
Evolution of non-coding RNA-directed DNA rearrangements
In Tetrahymena, an RNAi-related mechanism is required for the formation of the heterochromatin state that precedes IES elimination. Because it is known that RNAi machinery is also required for heterochromatin formation and subsequent transcriptional gene silencing in other eukaryotes [57], the molecular mechanism regulating IES elimination in Tetrahymena (and probably in Paramecium) is evolutionarily related to RNAi-directed heterochromatin formation. Many IES sequences in ciliates are related to transposable elements. Therefore, it is believed that one of the main roles of DNA elimination is eliminating transposable elements from the transcriptionally active macronucleus. Because RNAi machinery is also required for transposon silencing in other eukaryotes [58], DNA elimination in ciliated protozoa probably evolved from the RNAi-directed and heterochromatin-mediated transposon silencing pathway.
On the other hand, the evolutionary advantage of DNA descrambling is less evident. Analogous to mRNA splicing, DNA descrambling may increase the chance of inventing genes with novel functions by changing the rearrangement patterns of DNA segments. Alternatively, there may be neutral evolutionary selective pressure on micronuclear DNA scrambling, as long as the RNA-directed descrambling mechanism exists.
Conclusion
DNA elimination in the oligohymenophorean ciliate Tetrahymena is mediated by small non-coding RNAs that are produced by an RNAi-related mechanism. The small RNA-mediated comparison of the complete micronuclear and macronuclear genomes is proposed to identify the DNA sequences to be eliminated. The macronucleus-encoded domesticated PiggyBac-like transposase is probably directly involved in the DNA excision process. In contrast, DNA descrambling in the spirotrich ciliate Oxytricha is regulated by long non-coding RNAs, and transposases encoded by a group of abundant micronuclear transposons are required for this DNA rearrangement. These long RNAs most likely act as templates to guide the descrambling process. How these seemingly distinct mechanisms have evolved in different classes of ciliates is unclear. Future studies on other ciliate classes may help address this question. Additionally, the detailed molecular mechanisms regulating the biogenesis and selection of scnRNAs in Tetrahymena, the transcription of long-template RNAs in Oxytricha, and the induction of DNA rearrangements by non-coding RNAs are still emerging. I expect that established genetic tools in these model ciliates, such as gene deletion [59] and RNAi techniques [60] [61] [62], and fully sequenced genomes [63] [64] will help to answer these unsolved questions in the near future.
References
- 1.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 3.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 4.Collins K. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech TR, Rio DC. Localization of transcribed regions on extrachromosomal ribosomal RNA genes of Tetrahymena thermophila by R-loop mapping. Proc Natl Acad Sci U S A. 1979;76:5051–5055. doi: 10.1073/pnas.76.10.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 7.Jahn CL, Klobutcher LA. Genome remodeling in ciliated protozoa. Annu Rev Microbiol. 2002;56:489–520. doi: 10.1146/annurev.micro.56.012302.160916. [DOI] [PubMed] [Google Scholar]
- 8.Yao MC, Duharcourt S, Chalker DL. Genome-wide rearrangements of DNA in ciliates. In: Craig N, Craigie R, Gellert M, Lambowiz A, editors. Mobile DNA II. Academic Press; 2002. pp. 730–758. [Google Scholar]
- 9.Heinonen TY, Pearlman RE. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J Biol Chem. 1994;269:17428–17433. [PubMed] [Google Scholar]
- 10.Yao MC, Gorovsky MA. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48:1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- 11.Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 2002;30:2524–2537. doi: 10.1093/nar/30.11.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fillingham JS, Thing TA, Vythilingum N, Keuroghlian A, Bruno D, Golding GB, Pearlman RE. A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila. Eukaryot Cell. 2004;3:157–169. doi: 10.1128/EC.3.1.157-169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao MC, Choi J, Yokoyama S, Austerberry CF, Yao CH. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell. 1984;36:433–440. doi: 10.1016/0092-8674(84)90236-8. [DOI] [PubMed] [Google Scholar]
- 14.Austerberry CF, Snyder RO, Yao MC. Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic Acids Res. 1989;17:7263–7272. doi: 10.1093/nar/17.18.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer E, Chalker DL. Epigenetics in Ciliates. In: Allis CD, Jenuwein T, Reinberg D, Caparros ML, editors. Epigenetics. Cold Spring Habor Laboratory Press; 2006. pp. 127–150. [Google Scholar]
- 16.Mochizuki K: RNA-directed epigenetic regulations of DNA rearrangements. Essays Biochem. 2010 doi: 10.1042/bse0480089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu CY, Rana TM. Small RNAs: regulators and guardians of the genome. J Cell Physiol. 2007;213:412–419. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. Rna. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. Rna. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- 30.Lepere G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2009;37:903–915. doi: 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugai T, Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J Protozool. 1974;21:542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- 32.Martindale DW, Allis CD, Bruns PJ. RNA and protein synthesis during meiotic prophase in Tetrahymena thermophila. J Protozool. 1985;32:644–649. doi: 10.1111/j.1550-7408.1985.tb03094.x. [DOI] [PubMed] [Google Scholar]
- 33.Chalker DL, Yao MC. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stargell LA, Gorovsky MA. TATA-binding protein and nuclear differentiation in Tetrahymena thermophila. Mol Cell Biol. 1994;14:723–734. doi: 10.1128/mcb.14.1.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mochizuki K, Gorovsky MA. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot Cell. 2004;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noto T, Kurth HM, Kataoka K, Aronica L, Desouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K. The Tetrahymena Argonaute-Binding Protein Giw1p Directs a Mature Argonaute-siRNA Complex to the Nucleus. Cell. 2010;140:692–703. doi: 10.1016/j.cell.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aronica L, Bednenko J, Noto T, Desouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bednenko J, Noto T, Desouza LV, Siu KW, Pearlman RE, Mochizuki K, Gorovsky MA. Two GW Repeat Proteins Interact with the Tetrahymena Argonaute and Promote Genome Rearrangement. Mol Cell Biol. 2009 doi: 10.1128/MCB.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci U S A. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 43.Madireddi MT, Coyne RS, Smothers JF, Mickey KM, Yao MC, Allis CD. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- 44.Coyne RS, Nikiforov MA, Smothers JF, Allis CD, Yao MC. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol Cell. 1999;4:865–872. doi: 10.1016/s1097-2765(00)80396-2. [DOI] [PubMed] [Google Scholar]
- 45.Kowalczyk CA, Anderson AM, Arce-Larreta M, Chalker DL. The germ line limited M element of Tetrahymena is targeted for elimination from the somatic genome by a homology-dependent mechanism. Nucleic Acids Res. 2006;34:5778–5789. doi: 10.1093/nar/gkl699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Betermier M. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng CY, Vogt A, Mochizuki K, Yao MC. A Domesticated piggyBac Transposase Plays a Key Role in Heterochromatin Dynamics and DNA Cleavage during Programmed DNA Deletion in Tetrahymena thermophila. Mol Biol Cell. 2010 doi: 10.1091/mbc.E09-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prescott DM. Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nat Rev Genet. 2000;1:191–198. doi: 10.1038/35042057. [DOI] [PubMed] [Google Scholar]
- 49.Mollenbeck M, Zhou Y, Cavalcanti AR, Jonsson F, Higgins BP, Chang WJ, Juranek S, Doak TG, Rozenberg G, Lipps HJ, et al. The pathway to detangle a scrambled gene. PLoS One. 2008;3:e2330. doi: 10.1371/journal.pone.0002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juranek SA, Rupprecht S, Postberg J, Lipps HJ. snRNA and heterochromatin formation are involved in DNA excision during macronuclear development in stichotrichous ciliates. Eukaryot Cell. 2005;4:1934–1941. doi: 10.1128/EC.4.11.1934-1941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paschka AG, Jönsson F, Maier V, Möllenbeck M, Paeschke K, Postberg J, Rupprecht R, Lipps HJ. The use of RNAi to analyze gene function in spirotichous ciliates. Eur J Protistol. 2003;39:449–454. [Google Scholar]
- 52.Prescott DM, Ehrenfeucht A, Rozenberg G. Template-guided recombination for IES elimination and unscrambling of genes in stichotrichous ciliates. J Theor Biol. 2003;222:323–330. doi: 10.1016/s0022-5193(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 53.Angeleska A, Jonoska N, Saito M, Landweber LF. RNA-guided DNA assembly. J Theor Biol. 2007;248:706–720. doi: 10.1016/j.jtbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 54.Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lolle SJ, Victor JL, Young JM, Pruitt RE. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature. 2005;434:505–509. doi: 10.1038/nature03380. [DOI] [PubMed] [Google Scholar]
- 57.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hai B, Gaertig J, Gorovsky MA. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 2000;62:513–531. doi: 10.1016/s0091-679x(08)61554-x. [DOI] [PubMed] [Google Scholar]
- 60.Mollenbeck M, Postberg J, Paeschke K, Rossbach M, Jonsson F, Lipps HJ. The telomerase-associated protein p43 is involved in anchoring telomerase in the nucleus. J Cell Sci. 2003;116:1757–1761. doi: 10.1242/jcs.00351. [DOI] [PubMed] [Google Scholar]
- 61.Howard-Till RA, Yao MC. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol Cell Biol. 2006;26:8731–8742. doi: 10.1128/MCB.01430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beisson J, Betermier M, Bre MH, Cohen J, Duharcourt S, Duret L, Kung C, Malinsky S, Meyer E, Preer JR, Jr., et al. Silencing specific Paramecium tetraurelia genes by feeding double-stranded RNA. CSH Protoc. 2010 doi: 10.1101/pdb.prot5363. pdb prot5363. [DOI] [PubMed] [Google Scholar]
- 63.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
Further reading list
- Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nature medicine. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- Chalker DL. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim Biophys Acta. 2008;1783:2130–2136. doi: 10.1016/j.bbamcr.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Lepere G, Meyer E. Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 2009;25:344–350. doi: 10.1016/j.tig.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Landweber LF. Epigenetic inheritance in ciliates. Current opinion in microbiology. 2009;12:638–643. doi: 10.1016/j.mib.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]