Abstract
Effects of susceptibility variants may depend on from which parent they are inherited. While many associations between sequence variants and human traits have been discovered through genome-wide associations, the impact of parental origin has largely been ignored. Combining genealogy with long range phasing, we demonstrate that for 38,167 Icelanders genotyped using SNP chips, the parental origin of most alleles can be determined. We then focused on SNPs that associate with diseases and are within 500kb of known imprinted genes. Seven independent SNP associations were examined. Five, one each with breast cancer and basal cell carcinoma, and three with type 2 diabetes (T2D), exhibit parental-origin specific associations. These variants are located in two genomic regions, 11p15 and 7q32, each harbouring a cluster of imprinted genes. Furthermore, a novel variant rs2334499 at 11p15 was seen to associate with T2D where the allele that confers risk when paternally inherited is protective when maternally transmitted. We identified a differentially methylated CTCF binding site at 11p15 and demonstrated correlation of rs2334499 with decreased methylation of that site.
The effect of sequence variants on phenotypes may depend on parental origin. The most obvious scenario, although not the only one1, is imprinting where the effect is limited to the allele inherited from a parent of specific sex. Despite this, most reports of genome-wide association studies (GWAS) treated the paternal and maternal alleles as exchangeable. While this is understandable as the information required is often unavailable, it reduces the power to discover some susceptibility variants and underestimates the effects of others, contributing to unexplained heritability. Here we describe a new method that allows us to systematically determine parental origin of haplotypes even when parents of probands are not genotyped. The results are used to discover associations that exhibit parental-origin specific effects.
DETERMINING PARENTAL ORIGIN
Previously2, we introduced the method of long range phasing that allows for accurate phasing of Icelandic samples typed with Illumina bead chips for regions up to10 cMs in length. Two advances have been made since then, stitching and parental origin determination. Genome-wide, long range phasing was applied to overlapping tiles, each 6 cM in length, with 3 cM overlap between consecutive tiles. For each tile, we attempted to determine the parental origins of the two phased haplotypes regardless of whether the parents of the proband were chip-typed. Using the Icelandic genealogy database, for each of the two haplotypes of a proband, a search was performed to identify, among those individuals also known to carry the same haplotype, the closest relative on each of the paternal and maternal sides (Figure 1). Results for the two haplotypes were combined into a robust single-tile score reflecting the relative likelihood of the two possible parental origin assignments (score > 0 supporting one assignment and score < 0 supporting the other assignment, see Methods for details). We then tried to stitch the haplotypes from consecutive tiles together based on sharing at the overlapping region. Stitching and parental origin determination are complementary tasks. Specifically, if parental origin is determined with high confidence for one tile the information can be propagated to other tiles through stitching (Supplementary Figure 1A). Conversely, in cases where the overlap between two adjacent tiles is homozygous for all SNPs, stitching can still be accomplished if parental origins can be determined for both tiles independently (Supplementary Figure 1B). For haplotypes derived by stitching, a contig-score for parental origin is computed by summing the individual single-tile scores.
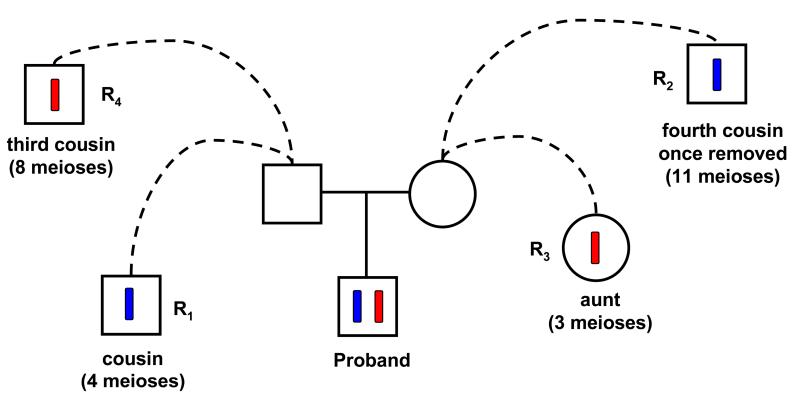
Figure 1. An example of parental origin determination.
In blue and red are two phased haplotypes of a proband. Among other typed individuals, the closest paternal relative known to also carry the blue haplotype is R1, a cousin, while the corresponding maternal relative is R2. For the red haplotype, a maternal aunt (R3) carries the haplotype, while the closest known carrier on the father side is R4. Since R1 is a closer than R2 and R3 is a closer than R4, the blue and red haplotypes are likely paternally and maternally inherited respectively. The single-tile score (see Methods) supporting this assignment is 0.194.
After filtering based on various quality and yield criteria, 289,658 autosomal markers and 8,411 markers on chromosome X were used. Excluding those with no parent listed in the genealogy database or with less than 98% genotyping yield, 38,167 individuals, a majority typed with Illumia HumanHap300/CNV370 bead chips (Supplementary Information), were processed. For these individuals, 97.8% of the heterozygous genotypes were long range phased, 99.8% of these had parental origin determined. Overall, 3,841,331,873 heterozygous genotypes, 97.7% of those called, had parental origin assigned. The data includes 2,879 typed trios. To empirically evaluate the accuracy of our method, a run was performed with the data of parents in these trios removed when determining parental origin. For 231,585,437 heterozygous genotypes in the probands/offspring, parental origin was determined both by our method and by the trio data directly, with 500,330 discrepancies, an error rate of 500,300/231,585,437 = 0.22%. Since the trios tested have passed heritability checks in pre-processing, the error rate for individuals with fewer than two parents genotyped is probably higher. Still, the overall error rate is likely below 0.4% (Supplementary Information).
IMPRINTING AND DISEASE ASSOCIATION
While many mechanisms can lead to parental origin specific association with a phenotype, a priori sequence variants located close to imprinted genes are more likely to exhibit such behaviour. Through two sources, Luedi et al.3 and the Imprinted Gene Catalogue4, 5, we found forty eight genes known to be imprinted in humans (Supplementary Table 1). Selecting regions that fall within 500 kb of any of these genes (Build 36 of the Human Genome Assembly) amounts to approximately 1% of the genome. The 500 kb threshold was chosen because imprinted genes often occur in clusters and the imprinting status of genes close to known imprinted genes is often undetermined. It is also known that a sequence variant can directly affect the function of a gene located some distance away. Among the 298,069 SNPs we processed, 3840 fall within these selected regions.
Consulting the Catalogue of Published Genome-Wide Association Studies6, we intersected reported SNP-disease associations with P < 5 × 10−8 with the selected regions (Supplementary Table 2). Further restricting to diseases for which we have published genome scans, 4 associations remained. Three other SNP associations we were aware of that fall within the imprinted regions, one recently published by us for basal cell carcinoma7 and two established by the Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium, were also examined.
ASSOCIATION ANALYSIS
For each disease-SNP association, five tests were performed (Table 1). A standard case-control test without taking parental origin into account was performed to provide a baseline. Then a case-control analysis was performed separately for the paternally and maternally inherited alleles. A two-degree of freedom test was applied to evaluate the joint effect. A multiplicative model was assumed for the two alleles, but the magnitude and direction of the effect were allowed to differ. Finally, the difference between the effects of the paternally and maternally inherited alleles was directly tested by comparing their allele frequencies within cases. The information for this test mainly comes from the counts of the two types of heterozygotes within cases. (see Supplementary Information for details).
Table 1.
Parental Origin Specific Analyses of Disease Susceptibility Variants
| Disease, SNP[Alleles]a | Standard case-control test |
Tests of association with parental origins |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position B36 | (M)b | Paternal Alleled | Maternal Alleled | 2-df teste | P. vs M. (case only) | |||||
|
|
||||||||||
| N (Case Sample Size) | Con. Frq. | OR | P † | OR | P † | OR | P † | P † | n12 : n21f | P † |
| Breast Cancer | ||||||||||
| rs3817198 c [C/T] | (34909) | |||||||||
| C11 1,865,582 | 0.303 | 1.04 | 0.36 | 1.17 | 0.0038 | 0.91 | 0.11 | 0.0040 | 437 : 339 | 6.2 × 10−4 |
| (1803) | ||||||||||
| Basal Cell Carcinoma | ||||||||||
| rs157935 [T/G] | (37041) | |||||||||
| C7 130,236,093 | 0.676 | 1.23 | 1.8×10−5 | 1.40 | 1.5×10−6 | 1.09 | 0.19 | 3.8 × 10−6 | 237 : 182 | 0.010 |
| (1118) | ||||||||||
| T2D, rs2237892 [C/T] | ||||||||||
| C11 2,796,327 | (34706) | |||||||||
| Discovery (1468) | 0.925 | 1.19 | 0.044 | 1.14 | 0.24 | 1.24 | 0.071 | 0.095 | 81 : 90 | 0.51 |
| Replication (783) | 1.08 | 0.43 | 0.87 | 0.30 | 1.43 | 0.024 | 0.050 | 35 : 59 | 0.014 | |
| Combined (2251) | 1.15 | 0.043 | 1.03 | 0.71 | 1.30 | 0.0084 | 0.027 | 116 : 149 | 0.054 | |
| T2D, rs231362 c [C/T] | ||||||||||
| C11 2,648,047 | (33377) | |||||||||
| Discovery (1423) | 0.551 | 1.09 | 0.051 | 0.97 | 0.67 | 1.23 | 0.0010 | 0.0037 | 329 : 401 | 0.014 |
| Replication (750) | 1.10 | 0.073 | 1.00 | 0.99 | 1.22 | 0.011 | 0.037 | 158 : 191 | 0.098 | |
| Combined (2173) | 1.10 | 0.013 | 0.98 | 0.73 | 1.23 | 6.2×10−5 | 2.6 × 10−4 | 487 : 592 | 0.0032 | |
| T2D, rs4731702 [C/T] | ||||||||||
| C7 130,083,924 | (34706) | |||||||||
| Discovery (1468) | 0.439 | 1.15 | 0.0018 | 1.07 | 0.24 | 1.23 | 6 .4×10−4 | 0.0013 | 335 : 374 | 0.17 |
| Replication (783) | 0.95 | 0.38 | 0.84 | 0.024 | 1.08 | 0.31 | 0.048 | 163 : 204 | 0.037 | |
| Combined(2251) | 1.08 | 0.039 | 0.99 | 0.79 | 1.17 | 0.0010 | 0.0041 | 498 : 578 | 0.022 | |
| T2D, rs2334499 [T/C] | ||||||||||
| C11 1,653,425 | (34706) | |||||||||
| Discovery (1468) | 0.412 | 1.11 | 0.017 | 1.41 | 4.3×10−9 | 0.87 | 0.020 | 3.5 × 10−9 | 437 :276 | 7.0×10−9 |
| Replication (783) | 1.02 | 0.71 | 1.23 | 0.0055 | 0.84 | 0.023 | 0.0018 | 222 : 157 | 8.0×10−4 | |
| Combined (2251) | 1.08 | 0.034 | 1.35 | 4.7×10−10 | 0.86 | 0.0020 | 5.7 × 10−11 | 659 : 433 | 4.1×10−11 | |
The first allele is the risk allele based on analyses that do not take into account parent of origin.
Size of the control set.
Imputed allele probabilities were used.
Effect of the paternally inherited allele is tested by comparing the corresponding alleles in cases to those in controls. Effect of the maternally inherited allele is similarly tested.
Test assumes a multiplicative effect for the paternally and maternally inherited alleles, but allows the effects to be different under the alternative hypothesis when the null hypothesis of no effect is tested.
To directly test whether the paternally and maternally inherited alleles have different effects, their allele frequencies were compared within the cases. Information for this test is mainly captured by the counts of the two types of heterozygotes: n12 denotes the number of cases who have inherited allele 1 from the father and allele 2 from the mother, and n21 is the reverse.
Genomic control was applied.
Among the 7 associations examined, one with prostate cancer (PrC) and another with coronary artery disease (CAD), did not exhibit parental-origin specific effects (Supplementary Information and Supplementary Table 3). The 5 associations that did are presented here.
Breast Cancer
Allele C of rs3817198 in the 11p15 region (Figure 2) was reported8 to be associated with breast cancer with an allelic OR of 1.07 (P = 3 × 10−9 ). This study included about 21,860 cases and 22,578 controls allowing for such a modest effect to achieve genome-wide significance. A study9 of CGEMS (The Cancer Genetic Markers of Susceptibility Project) with 9,770 cases and 10,799 controls reported ORs of 1.02 and 1.12 for heterozygous and homozygous carriers of the same variant respectively. Using information in their supplementary material, we deduced a P of 0.06. Rs3817198 is not on the Illumina chips used to type the majority of the Icelandic samples, but is included on the 1M Illumina chips for which we have data on 124 trios. Single track assay was used to type another 90 trios, giving a total of 214 trios with genotypes for rs3817198 that translates to a training set of 856 haplotypes. Adapting the statistical model employed by IMPUTE10, allele probabilities of rs3817198 were calculated for individuals with phased and parental origin determined haplotypes for this region (see Supplementary Information for details). With the imputation results for 1,803 cases and 34,909 controls (Table 1), the standard case-control test gave a non-significant OR of 1.04 (P = 0.36). However, when parental origin was taken into account, the paternally inherited allele showed a significant association (OR = 1.17, P = 0.0038). The direct test of parental-origin specific effects that used only the case data was even more significant (P = 6.2 × 10−4). This is because the estimated effect of the C allele when maternally inherited, while not significant (P = 0.11), is protective (OR = 0.91).
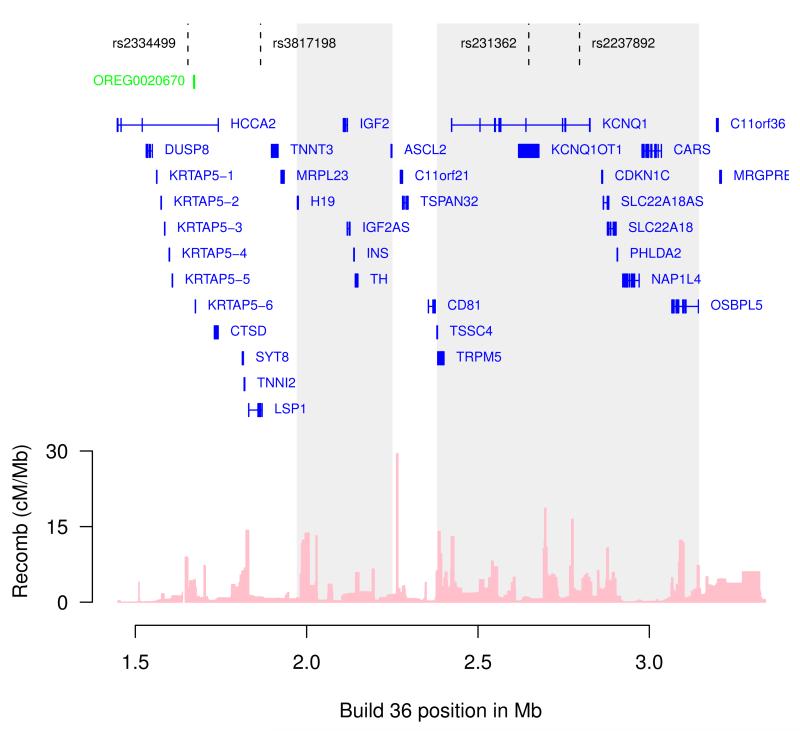
Figure 2. Chromosome 11p15 locus.
Markers associated with T2D (rs2334499, rs231362, rs2237892) as well as breast cancer (rs3817198), are indicated. The two regions containing clusters of imprinted genes are shaded. Location of the CTCF binding region studied (OREG0020670) and gene annotations were taken from the University of California Santa Cruz genome browser. Estimated recombination rates (from HapMap) are plotted to reflect the linkage disequilibrium structure in the region.
Basal Cell Carcinoma
We recently identified association of allele T of rs157935, located at 7q32 (Figure 3), with basal cell carcinoma (OR = 1.23, P = 5.7 × 10−10)7. Limiting the analysis to samples for which parental origin could be determined, the paternally inherited allele was significantly associated with the disease (OR = 1.40, P = 1.5 × 10−6), but the effect of maternally inherited allele, while in the same direction, was not significant (OR = 1.09, P = 0.19) (Table 1). Tested directly, the effects of the paternally and maternally inherited alleles were significantly different (P = 0.010).
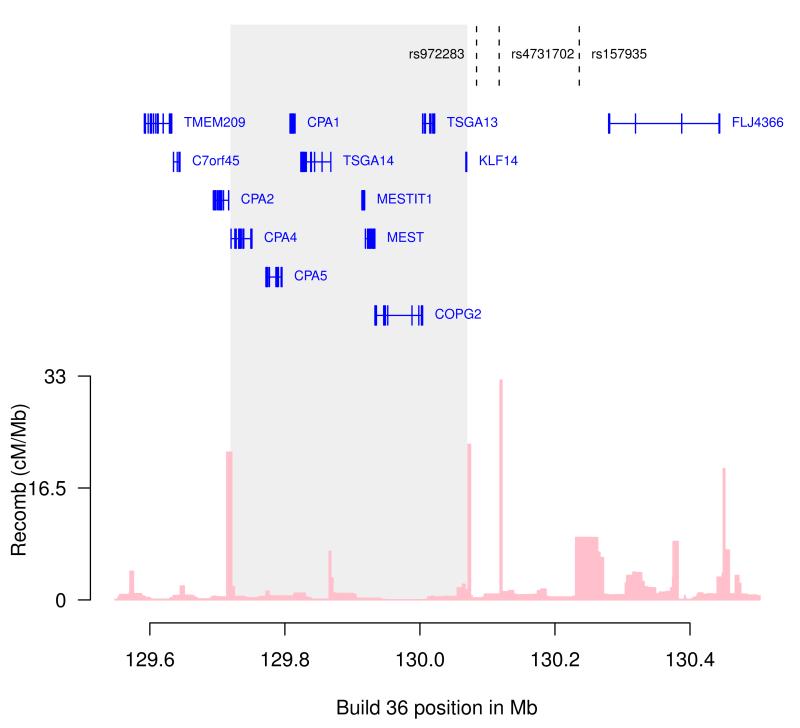
Figure 3. Chromosome 7q32 locus.
Markers associated with T2D (rs4731702, rs972283) as well as basal cell carcinoma (rs157935), are indicated. Rs972283, reported in the DIAGRAM study is not on the Illumina chip. Data on the correlated marker rs4731702 (r2 = 1, HapMap Ceu) is reported here. The region containing the known imprinted genes is shaded. Gene annotations were taken from the University of California Santa Cruz genome browser. Estimated recombination rates (from HapMap) are plotted to reflect the linkage disequilibrium structure in the region.
Type 2 Diabetes
Allele C of rs2237892 in the maternally expressed gene KCNQ1 was first observed to associated with T2D in Asian populations11, 12. Power to detect association in populations of European ancestry is low due to the high frequency of the variant there (~93% compared to ~61% in Asians), but the association has nonetheless been replicated11, 12. In the T2D samples we have previously employed in genome-scans (Table 1) that include 1,468 cases, none of the tests involving parental origin were significant for rs2237892. But when a new T2D list (Supplementary Information) allowed us to add 783 patients, giving a total of 2,251 cases, allele C was significantly associated with the disease (OR = 1.30, P = 0.0084) when maternally transmitted, while the results for the paternally inherited allele were flat (OR = 1.03, P = 0.71).
Through a meta-analysis of large numbers of T2D genome-wide scans with additional follow-up within the DIAGRAM consortium (Supplementary Information), allele C of rs231362 was shown to associate with the disease (OR = 1.08, P = 3 × 10−13). Rs231362 is also located in KCNQ1 (Figure 2), but it is not substantially correlated with rs2237892 (r2 = 0.002). Rs231362 is not on any of the Illumina chips used. A training set of 912 haplotypes, created through single track assay genotyping of 228 trios, was used for imputation of rs231362 into the Icelandic samples. Employing the imputed results, the standard case-control test gave an OR of 1.10 (P = 0.013). The effect, however, appears to be limited to the maternally inherited allele (OR = 1.23, P = 6.2 × 10−5).
Another association with T2D established by the DIAGRAM consortium is allele C of rs4731702 at 7q32 (OR = 1.07, P = 2 × 10−10) (Figure 3). In our combined samples, the association was again restricted to the maternally inherited allele (OR = 1.17, P = 0.0010) (OR = 0.99, P = 0.79 for the paternally inherited allele).
Evaluating the 7 known susceptibility variants jointly, the five highlighted above plus the two variants for PrC and CAD mentioned earlier, the test of no parental-specific effect for all gave a P < 5 × 10−6. Also, false-discovery rate13 analysis indicates that it is likely that at least four out of five highlighted variants have true parental-origin specific effect (Supplementary Information).
A novel diabetes susceptibility variant
Properly evaluating the statistical significance of the susceptibility variants described above requires adjusting for relatedness of the participants using the method of genomic control14. This required performing genome scans for these diseases (Supplementary Table 4 gives parental-origin test results for established susceptibility variants located outside the selected regions). The T2D scan performed with the initial sample set (Supplementary Information and Supplementary Figure 2) gave a striking result (Table 1). Allele T of rs2334499, at 11p15 (Figure 2), showed such a weak association (OR = 1.11, P = 0.017) in the standard case-control test that it does not stand out in a genome-wide scan. However, taking into account parental origin, both the paternally inherited allele (OR = 1.41, P = 4.3 × 10−9) and the 2-df of freedom test (P = 3.5 × 10−9) were genome-wide significant. Most intriguing, the maternally inherited allele also showed nominally significant association, but the effect of allele T was protective (OR = 0.87, P = 0.020). Tested directly, the difference between the effects of the paternally and maternally inherited alleles was also genome-wide significant (P = 7.0 × 10−9). This SNP falls within 350 Kb of a large cluster of imprinted genes, making the results even more compelling. Still, the observation that allele T is protective when maternally inherited called for replication. For this, we used an additional set of 783 chip-typed T2D cases. All tests involving parental origin were significantly replicated. For the combined analysis of the two sample sets (Supplementary Information and Supplementary Figure 3), the paternally inherited allele had an OR of 1.35 (P = 4.7 × 10−10) and the maternally inherited allele has an OR of 0.86 (P = 0.0020). The 2-df test and the paternal versus maternal tests gave P of 5.7 × 10−11 and 4.1 × 10−11 respectively.
As there are known examples in an imprinted setting where the paternal and maternal alleles interact15, we tested rs2334499 for an interactive effect. This test was not significant (P > 0.4, Supplementary Information) indicating that the multiplicative model provides an adequate fit. Specifically, compared to CT (first allele paternal, second allele maternal), CC, TT and TC have relative risks of 1.17, 1.35 and 1.57 respectively.
The transmitted maternal allele has an effect in all four T2D variants in Table 1. Since prenatal maternal conditions may be a factor in conferring risk on the offspring, we examined the role of the non-transmitted maternal allele. No significant effect was observed (Supplementary Information).
IMPRINTED REGIONS ON 11p15 AND 7q32
Imprinted genes at 11p15.5 fall into two clusters, H19/IGF2 and KCNQ1 (Figure 2), regulated through separate imprinting control regions (ICR) that each controls expression of a number of genes within the cluster16. The H19/IGF2 ICR is regulated through a differentially methylated region (DMR) that is normally methylated only on the paternal chromosome. Binding of the insulator protein CTCF in the ICR is permitted only on the unmethylated maternal chromosome, resulting in expression of IGF2 only from the paternal methylated chromosome and expression of H19 from the choromosome17. The breast cancer paternally associated marker rs3817198 resides within LSP1, 100 kb downstream of H19 and within the same LD block. The effect of this marker on breast cancer could thus be through the H19/IGF2 imprinted locus. Loss of imprinting (LOI) at the H19/IGF2 locus, resulting in activation of IGF2 expression, has been reported in a number of different tumor types18. Furthermore, LOI at the H19/IGF2 locus in normal tissue has also been shown to predispose to colorectal cancer18.
The KCNQ1 cluster is regulated through an ICR located in the promoter region of KCNQ1OT1, a paternally expressed non-coding antisense RNA. Hypermethylation of the maternal allele results in monoallelic activity of the neighboring maternally expressed protein coding genes. The two T2D associated markers at this locus rs231362 and rs2237892 are both located within the maternally expressed KCNQ1, consistent with the risk associations. Rs231362 also residing within the KCNQ1OT1 antisense transcript (Figure 2).
While both the T2D marker rs2334499 and the breast cancer marker rs3817198 fall within 350 kb of imprinted genes, the region harboring both rs2334499 and rs3817198 has not been reported to be imprinted (Figure 2)19 The T2D associated marker rs2334499 resides within the first intron of HCCA2, a gene spanning 300 kb including several other genes (Figure 2) including KRTAP5-1 to -5 genes, DUSP8 and CTSD20. To determine if genes in this region showed signs of imprinting we performed allele specific expression analysis of the HCCA2, CTSD and DUSP8 genes (Figure 2) as well as three genes known to be imprinted in the 11p15.5 region (IGF2, KCNQ1 and KCNQ1OT1, in RNA isolated from peripheral blood and adipose. While allele specific expression of IGF2, KCNQ1 and KCNQ1OT1 was confirmed in this dataset clear biallelic expression was seen for HCCA2 and DUSP8. However, excess paternal expression could not be ruled out for CTSD (Supplementary Information, Supplementary Table 6).
The imprinted region on 7q32 consists of maternally expressed genes (CPA4 and KLF14) flanking paternally expressed genes (MEST, MESTIT1) (Figure 3). The T2D associated marker rs4731702 is located 14 kb from the maternally expressed KLF14 transcription factor21 and only increases risk of T2D when carried on the maternal chromosome. The basal cell carcinoma variant rs157935, conferring risk through the paternal allele, is located 170 kb telomeric to the imprinted region.
We previously22 correlated SNP genotypes from the Illumina 300K chip to gene expression using RNA samples from adipose tissue (N=603) and peripheral blood (N=745). Here, taking parental origin into account, we re-evaluated the correlation between the six variants in Table 1 and expression of genes at the 7q32 and 11p15.5 loci. Interestingly, the T2D risk allele of rs4731702 at 7q32 correlated with lower expression of KLF14 in adipose tissue (P = 3 × 10−21) when inherited maternally, but there was no effect when inherited paternally (Supplementary Table 7). Similar correlation was not seen in blood. Conversely, no strong correlation with parent of origin specific gene expression was seen for the other disease associated variants on 7q32 or 11p15.5 (Supplementary Table 7).
METHYLATION OF NOVEL CTCF BINDING SITE
Recent studies have mapped regions of CTCF binding genome-wide for identification of insulator elements23, 24. One of the sites identified (OREG0020670) is a 2 kb region located 17 kb centromeric to our new T2D marker rs2334499 (Figure 2, Supplementary Figure 4). We assessed the methylation status of this CTCF binding region in DNA samples derived from peripheral blood, using bisulfite sequencing. We identified a differentially methylated region of 180 bp including seven CpG dinucleotides (Supplementary Figure 4) where the ratio of 5-methyl cytosine (Cp) varied from around 0.1-0.6. Methylation at five of the seven CpGs (CpG-1 through CpG-5) (Supplementary Figure 4), was highly correlated (Supplementary Table 9). The estimated Cp ratio was tested for correlation with SNPs in a 2 Mb surrounding region. The most significant correlation was observed between methylation status at CpG-4 and rs2334499, P = 2.6 × 10−13 (Table 2). Furthermore, correlation between rs2334499 and methylation of CpG-1 through CpG-5 was significant. For all five CpGs the T2D risk allele correlated with decreased methylation and this effect was observed regardless of whether the allele was inherited from the father or the mother. By contrast, neither the breast cancer variant nor the two other T2D markers at 11p15.5 showed any correlation with the methylation status of this CTCF site.
Table 2.
Methylation of a CTCF binding region is correlated with the T2D risk variant rs2334499
| % Methylation Mean (SE)a |
Effectb | P-valuec | |
|---|---|---|---|
| CpG-1 | 22.9 (0.9) | −5.7 | 6.5×10−7 |
| CpG-2 | 15.3 (0.7) | −3.1 | 0.00055 |
| CpG-3 | 13.3 (0.8) | −2.5 | 0.017 |
| CpG-4 | 56.7 (0.9) | −8.4 | 2.6×10−13 |
| CpG-5 | 34.9 (0.9) | −6.7 | 6.8×10−8 |
| CpG-6 | 22.2 (0.6) | −1.0 | 0.24 |
| CpG-7 | 52.9 (1.1) | −1.5 | 0.30 |
% Methylation at each site shown is the mean and the standard error (SE) of the C/T ratio estimated by bisulfite sequencing of 168 individuals.
Change in percentage methylation per allele T of rs2334499 carried.
Significance of the correlation between methylation and rs2334499.
DISCUSSION
Being able to determine parental origin of alleles and haplotypes in large samples opens up new avenues to study associations between sequence variants and human traits. Standard association analysis provides suboptimal power to discover disease susceptibility variants that exhibit parental-origin specific effects. Even when association can be established, the true effect is underestimated. Rs2334499 did not capture serious attention even with the large collaborative effort of the DIAGRAM consortium. However, its contribution to T2D, measured by the recurrent risks of siblings generated, is second only to that of the TCF7L2 variant among the known susceptibility variants (Supplementary Information and Supplementary Figure 2). Sequence variants such as rs2334499 that can confer both risk and protection depending on parental origin can lead to balanced selection and in that promote diversity.
Functional imprinting is extremely tissue and stage-specific and while some genes retain their imprinted status throughout life the main role of imprinting is believed to be during prenatal growth and development. However, the associations of rs4731702 C with T2D and KLF14 expression in adult adipose tissue, in both cases only when maternally inherited, strongly implicates this transcription factor as the disease gene.
We searched for evidence of epigenetic marks around the novel T2D risk variant rs2334499, as it is located some distance away from the established 11p15.5 imprinted genes. A CTCF binding site in the region was found to be differentially mehtylated and the rs2334499 risk allele was shown to be correlated with decreased methylation. Given the well established role of CTCF in imprinting, this novel site could differentially influence the dosage of imprinted genes on the two parental chromosomes.
Despite the successes, GWAS have so far yielded sequence variants that only explain a small fraction of the estimated heritability of most of the human traits studied. Obvious contributors to the unexplained heritability, or Dark Matter, include rare variants not well tagged by common SNPs and common variants that have very small effects individually. Results presented here demonstrate that a portion of the heritability of some common/complex traits is hidden in more complex relations between sequence variants and the risks of these variants.
METHODS
Parental Origin Assignment
Let H be a haplotype for a tile T. For a particular proband, f(T,H) and m(T,H) were calculated as respectively the meiotic distance to the closest relative on the father side and the mother side known to carry H. Descendants of the parents of the proband, e.g. siblings of the proband, were excluded for this calculation. Also, a value of 10,000 was assigned when no relatives carrying the haplotype was found. Let A and B be the two phased haplotypes of the proband. The single-tile score for parental origin was calculated as
A score that is > 0 supports the assignment of A as the paternally inherited haplotype and B as the maternally inherited haplotype, while a score < 0 supports the reverse. While not meant to be optimal in any formal sense, this scoring was chosen to have two properties. Firstly, for the same absolute difference between m(T,H) and f(T,H), the absolute value of score(T,H) is higher when the minimum of m(T,H) and f(T,H) is smaller, thus giving more weight to situations when a close relative who shared a haplotype is found. Secondly, the scoring was designed to be robust so that the result from one haplotype in one tile could not completely dominate the contributions from other haplotypes and adjacent tiles when results were combined (see below). When haplotypes for n consecutive tiles, T1,…,Tn, could be stitched together to form A = (A1,…,An) and B = (B1,…,Bn), then the contig score for parental origin assignment was calculated as
Parental original were assigned based on whether the contig-score was greater or smaller than zero. Most importantly, the accuracy of this procedure was evaluated using the trio test.
Acknowledgments
This project was funded in part by FP7-MC-IAPP Grant agreement no. 218071 (CancerGene) to deCODE Genetics.
Footnotes
Supplementary information accompanies this paper
Author Information The authors declare competing financial interest.
References
- 1.Rampersaud E, Mitchell BD, Naj AC, Pollin TI. Investigating parent of origin effects in studies of type 2 diabetes and obesity. Curr Diabetes Rev. 2008;4:329–39. doi: 10.2174/157339908786241179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008 doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luedi PP, et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–30. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001;29:275–6. doi: 10.1093/nar/29.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–65. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. [Accessed April 25th 2009];A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies.
- 7.Stacey SN, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–14. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 11.Yasuda K, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–7. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 12.Unoki H, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 13.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 15.Georges M, Charlier C, Cockett N. The callipyge locus: evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 2003;19:248–52. doi: 10.1016/S0168-9525(03)00082-9. [DOI] [PubMed] [Google Scholar]
- 16.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg M, Wei M, Yuan L, Murty VV, Tycko B. Biallelic expression of HRAS and MUCDHL in human and mouse. Hum Genet. 2003;112:334–42. doi: 10.1007/s00439-003-0907-7. [DOI] [PubMed] [Google Scholar]
- 20.Authier F, Metioui M, Fabrega S, Kouach M, Briand G. Endosomal proteolysis of internalized insulin at the C-terminal region of the B chain by cathepsin D. J Biol Chem. 2002;277:9437–46. doi: 10.1074/jbc.M110188200. [DOI] [PubMed] [Google Scholar]
- 21.Parker-Katiraee L, et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3:e65. doi: 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 23.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]