Abstract
Developmentally programmed genome rearrangement has been observed in a variety of eukaryotes from vertebrates to worms to protists, and it provides an interesting exception to the general rule of the constancy of the genome. DNA elimination in the ciliated protozoan Tetrahymena is one of the most well-characterized programmed genome rearrangement events. DNA elimination in the newly formed macronucleus of Tetrahymena is epigenetically regulated by the DNA sequence of the parental macronucleus. Dicer-produced, Piwi-associated small RNAs mediate this epigenetic regulation, probably through a whole-genome comparison of the germline micronucleus to the somatic macronucleus. However, a correlation between small RNAs and programmed genome rearrangement could not be detected in the worm Ascaris suum. Therefore, different types of eukaryotes may have developed unique solutions to perform genome rearrangement.
Introduction
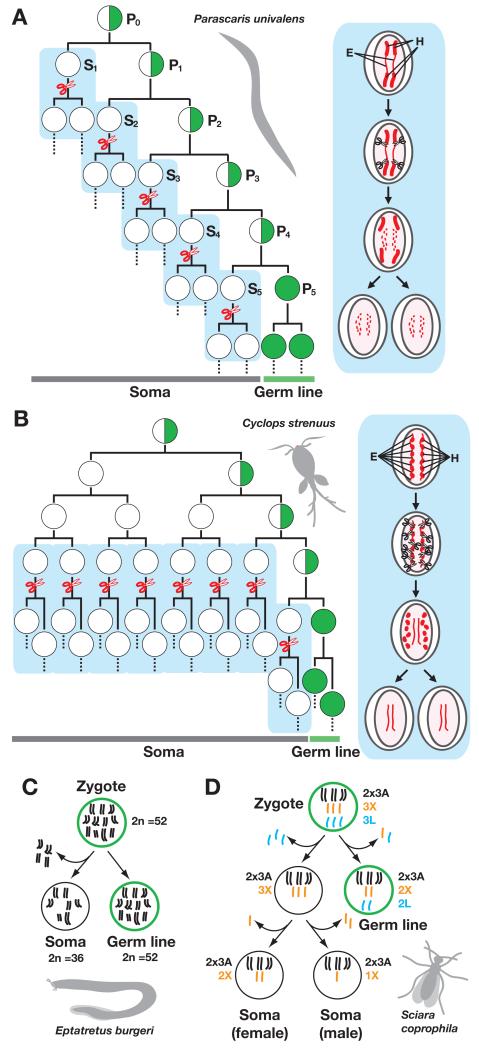
The human genome contains potentially harmful genetic elements that constantly require silencing. However, rather than constantly silencing these elements, why not simply delete them altogether? In fact, certain eukaryotic organisms delete these elements through the process of developmentally programmed genome rearrangements. Theodor Boveri was one of the first biologists to propose that chromosomes carry factors regulating Mendelian inheritance. In 1899, which was still prior to the general acceptance of chromosomes as the basis for genetic inheritance, he reported the first example of developmentally programmed genome rearrangement through observations in the early embryogenesis of Parascaris equorum, which is a parasitic nematode found in horses (reviewed in (Satzinger, 2008)). In many parasitic nematodes, the elimination of heterochromatic regions occurs in presomatic blastomeres. This process, designated chromatin diminution, is strictly developmentally regulated, as it only occurs at defined stages soon after the somatic linage is separated from the germline (Fig. 1A). The timing of chromatin diminution and the amount of DNA eliminated from the presumptive somatic cells differs among species. In Ascaris suum, a parasite found in pigs, about 25% of the genome is eliminated; however, more than 80% of the germline genome is deleted in the horse parasite Parascaris univalens. These deleted DNA segments are highly enriched for repeated satellite DNA elements (reviewed in (Muller & Tobler, 2000)).
Figure 1. Programmed genome rearrangements in different types of eukaryotes.
(A) Chromatin diminution in the parasitic nematode Parascaris univalens. (B) Chromatin diminution in the copepod crustacean Cyclops strenuus. In both (A) and (B), a schematic representation of the cell lineages in early embryogenesis is shown at left, and a schematic representation of a presomatic cell undergoing chromatin diminution is shown at right. The presumptive primordial germ cells are indicated by half-green circles. The primordial germ cells give rise to all germ cells, which are represented by all-green circles. Blank circles represent presomatic cells. The mitotic processes in which chromatin diminutions occur are marked with scissors. Heterochromatin (H) and euchromatin (E) are represented by thick and thin red lines, respectively. (C) Chromosome elimination in a hagfish. A schematic representation of the segregation of germline and somatic cells and chromosome elimination in Eptatreus burgeri is shown. The zygote and germline cells have 52 chromosomes per diploid cell (2n), whereas somatic cells have only 36. (D) Chromosome elimination in a sciarid fly. A schematic representation of the segregation of germline and somatic cells and chromosome elimination in Sciara coprophila is shown. Autosomes (black), X chromosomes (orange), and germline specific chromosomes (blue) are marked by A, X, and L, respectively.
A similar chromatin diminution process has been observed in the somatic cell lineages of different Cyclops species (copepod crustaceans) (Fig. 1B) (Beermann, 1977). Additionally, the deletion of entire chromosomes (i.e., chromosome elimination) has been observed in the somatic cells of early hagfish embryos (Fig. 1C) (Kohno et al., 1986) and sciarid flies (Fig. 1D) (Goday & Esteban, 2001). Interestingly, chromosome elimination is linked to germ-soma differentiation and sex determination in sciarid flies. Additionally, mating-type switching in yeasts and VDJ recombination in mammalian immune cells can be considered as types of developmentally programmed genome rearrangements. As Gilbert (2010) suggested, developmentally programmed genome rearrangement “is a fascinating exception to the general rule of the constancy of the genome and it provides an interesting alternative to differential gene expression. (Gilbert, 2010)” However, the molecular mechanisms regulating developmentally programmed genome rearrangements are still poorly understood. One of the most extreme examples of programmed genome rearrangement was discovered to occur during the differentiation of the somatic macronucleus in ciliated protozoa, and the molecular mechanisms regulating this process have been delineated. Here, I review the current understanding of how programmed genome rearrangements are regulated by small non-coding RNAs in ciliates, mainly Tetrahymena thermophila. The role of DNA elimination, the eliminated sequences from Tetrahymena thermophila, and the evolutionary relationship between Tetrahymena DNA elimination and programmed genome rearrangements in other eukaryotes will also be discussed.
Life of Ciliates
Ciliates are primarily single-celled eukaryotes that are found in almost every body of water. They are characterized by the presence of hair-like organelles called cilia, which are used for swimming, feeding, attachment and/or sensation (Lynn, 2008). Ciliates, from the group Alveolata, are evolutionarily distant from Unikonts, which include animals, fungi and amoebas, and Plantae, which include algae and land plants (Baldauf et al., 2000). Ciliates tend to have large cellular dimensions with some reaching 2 mm in length and are one of the most structurally complex groups of protozoans (Lynn, 2008). Tetrahymena thermophila (Fig. 2A, hereafter referred to as Tetrahymena) is a free-living freshwater ciliate that is one of the most commonly used protozoa in laboratories.
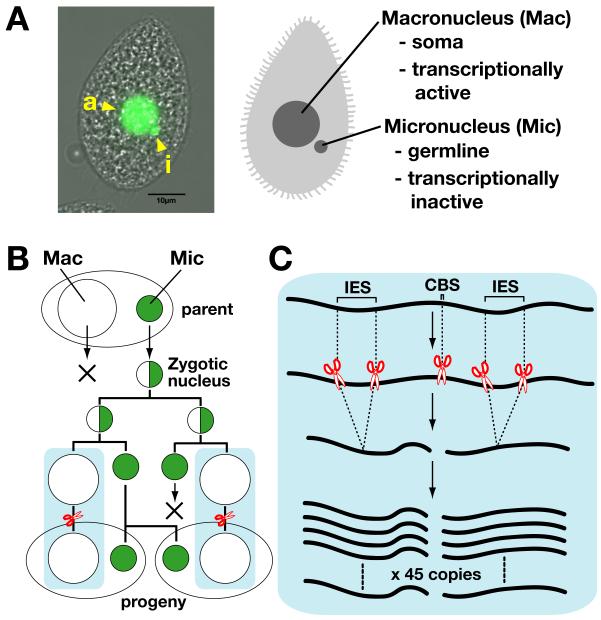
Figure 2. Programmed DNA elimination in Tetrahymena.
(A) Nuclear dimorphism of Tetrahymena thermophilla. In the left picture, the DNA of a Tetrahymena cell is stained by DAPI (green). Tetrahymena has one somatic macronucleus, which is marked by an arrowhead with ”a”, and one germline micronucleus, which is marked by an arrowhead with ”i” in the interphase stage. (B) Fates of the nuclei during sexual reproduction. The micronucleus (green circle) undergoes meiosis and produces the zygotic nucleus (half green circle) by fusion with a meiotic product received from its mating partner (not shown). The zygotic nucleus divides twice mitotically; two of the new nuclei become micronuclei (green circle), and the other two differentiate to become macronuclei (open circle). Two new macronuclei are distributed one to each daughter cell. One of the two micronuclei is destroyed, whereas the other divides mitotically and segregates into two daughter cells. The parental macronucleus is destroyed at the end of sexual reproduction. (C) Programmed genome rearrangements. In the newly formed macronucleus, internal eliminated sequences (IESs) are eliminated by DNA elimination, and chromosome breaks occur at chromosome breakage sequences (CBSs). Each macronuclear chromosome is endoreplicated to ~45 copies.
Most ciliates demonstrate nuclear dimorphism through the presence of a diploid germline micronucleus and a somatic polyploid macronucleus (Fig. 2A). The micronucleus is transcriptionally inert, although an exception occurs during early sexual reproduction and will be explained later; thus, all gene expression occurs in the macronucleus (Fig. 2A) (reviewed in (Karrer, 2000)). Only the micronucleus has the ability to undergo meiosis, which is followed by fertilization to form the zygotic nucleus. In Tetrahymena, the zygotic nucleus produces four nuclei, two of which differentiate into macronuclei, while the other two become the new micronuclei (Fig. 2B). The parental macronucleus is destroyed by a macroautophagy-like process at the end of sexual reproduction (Akematsu et al., 2010).
Programmed Genome Rearrangement in Tetrahymena
During the differentiation of the zygotic nucleus into the new macronucleus, two types of programmed genome rearrangements occur in Tetrahymena. The first is chromosome breakage at the chromosome breakage sequence (CBS) site, accompanied by short DNA trimming and de novo telomere formation (Fig. 2C). There are ~200 CBSs per haploid micronuclear genome, and 5 micronuclear chromosomes are fragmented to ~200 pieces of the macronuclear chromosomes. CBSs share a non-palindromic 15-bp element containing a 10 bp invariant core (5′-AAACCAACCYC-3′, Y = C or T) (Hamilton et al., 2006). The mechanism regulating these chromosomal breaks is currently unclear.
The second type of genome rearrangement process in Tetrahymena is the elimination of internal DNA segments followed by the ligation of the two flanking macronuclear-destined sequences (Fig. 2C). Although this process can be called chromatin diminution, the conventional term DNA elimination will be used in this review. A stretch of DNA that is destined to be eliminated is called the internal eliminated sequence (IES). In Tetrahymena, it has been estimated that there are approximately 6,000 different IESs that range from ~0.5–20 kb in length. The most recent data from the genome sequencing project indicate that the sizes of the micronuclear and macronuclear genomes are ~154 Mb and ~103 Mb, respectively (Eisen et al., 2006) (Coyne et al., 2008), (Coyne, et al. http://www.broadinstitute.org/annotation/genome/Tetrahymena/MultiHome.html). Therefore, more than 30% of the micronuclear DNA is eliminated from the macronucleus in Tetrahymena. In other ciliates, such as Euplotes, Stylonychia and Oxytricha, more than 95% of the genome is eliminated. All known IESs of Tetrahymena are located in intergenic regions or in introns (Heinonen & Pearlman, 1994). Many of the known IESs are transposon-like repeats (Wuitschick et al., 2002) (Fillingham et al., 2004), and the rest are comprised of other types of repeated or single-copy sequences (Yao et al., 1984).
DNA elimination in Tetrahymena is epigenetically regulated
DNA elimination in Tetrahymena occurs reproducibly at a specific site or at a limited number of alternative sites. The boundaries of DNA eliminations are relatively precise (i.e., within several nucleotides) (Austerberry et al., 1989). However, no consensus sequence motif has been identified in or around the IESs of Tetrahymena. The only common identified elements that are associated with DNA elimination are short (1–8 bp) direct repeats of varied sequences at both ends of the IESs (Austerberry & Yao, 1988). However, these sequences have no known function in IES removal. The flanking regions of some IESs have cis-acting sequences that are required for IES removal (Godiska et al., 1993), but no sequence homology has been observed across different elements.
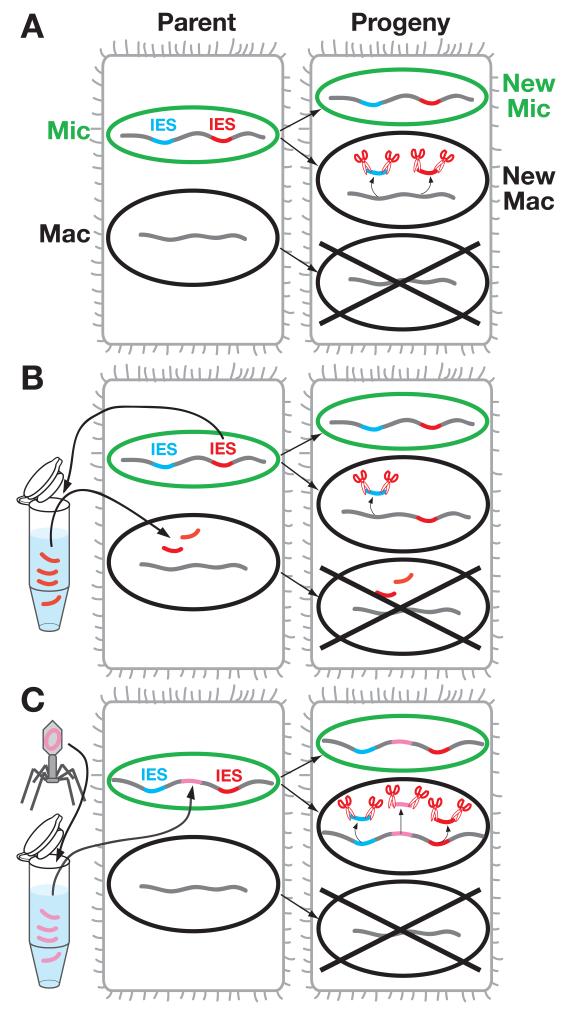
DNA elimination in the newly formed macronucleus of Tetrahymena is epigenetically regulated by the DNA sequence of the parental macronucleus. This property has been demonstrated by the introduction of a DNA fragment containing an IES into the parental macronucleus of a Tetrahymena, which, in turn, causes retention of the complementary sequence of this IES in the newly formed macronucleus (Fig. 3A, B) (Chalker & Yao, 1996). Similar epigenetic regulation of DNA elimination by parental macronuclear DNA has been observed in Paramecium (Epstein & Forney, 1984) (You et al., 1994) (Duharcourt et al., 1995). It has also been observed that a bacteriophage-derived neomycin resistance gene integrated into the parental micronuclear chromosome is often eliminated from the new macronucleus (Fig. 3C) (Yao et al., 2003) (Liu et al., 2005), (Howard-Till & Yao, 2007). These phenomena indicate that DNA elimination is not regulated by the primary sequence of the micronucleus or new macronucleus; instead, it is regulated by the DNA sequence of the parental macronucleus such that the new macronucleus copies the sequence pattern of the parental macronucleus. To achieve these maternal regulations, it has been suggested that some sequence-specific information must be transferred from the parental macronucleus to the new macronucleus (Meyer & Duharcourt, 1996) (Chalker & Yao, 1996). Several lines of evidence indicate that this trans-nuclear information is carried by ~29-nt small RNAs that are made by an RNAi-related pathway in Tetrahymena.
Figure 3. DNA elimination in Tetrahymena is epigenetically regulated.
Cells before conjugation (parent) are on the left and cells post-conjugation (progeny) are on the right. (A) DNA elimination in a wild-type Tetrahymena cell. IESs are eliminated during new macronuclear development. (B) Introduction of DNA containing the red IES into the parental macronucleus results in the retention of the complementary sequence of this IES in the new macronucleus. (C) The bacteriophage-derived neomycin resistance gene (pink) integrated into the parental micronucleus is often eliminated from the new macronucleus.
An RNAi-related mechanism is required for DNA elimination
RNAi-related pathways commonly utilize Argonaute family proteins, which hold small (~20–30 nt) RNAs. Base-pairing interactions between small RNAs and target sequences induce various types of gene regulation, including RNA degradation, translational repression, transcriptional silencing and, occasionally, transcriptional activation (reviewed in (Ender & Meister, 2010)). Argonaute proteins consist of two subfamilies: Ago and Piwi proteins. The Ago proteins associate with small RNAs (small interfering (si) and micro (mi) RNA’s) produced by Dicer family proteins from various double-stranded RNAs. In contrast, Piwi proteins in metazoans interact with small RNAs (Piwi-associated (pi) RNAs) produced by a Dicer-independent mechanism from single-stranded RNAs. piRNAs, as well as some si- and miRNAs, are 2′-O-methylated at their 3′ termini by conserved homologs of the RNA methyltransferase Hen1 (Yu et al., 2005) (Horwich et al., 2007) (Kirino & Mourelatos, 2007) (Saito et al., 2007). Metazoans have both Ago and Piwi proteins, whereas plants and fungi have only Ago-subfamily proteins (Seto et al., 2007).
DNA elimination of Tetrahymena is regulated by small RNAs which are produced by a Dicer protein, are associated with a Piwi protein, and are modified by a Hen1 protein. The small RNAs are called scan (scn) RNAs and are expressed exclusively during sexual reproduction (Mochizuki et al., 2002). Although the length of scnRNAs was originally suggested to be ~28 nt (Mochizuki et al., 2002), recent sequencing analyses by our group (Kurth, HM and KM, unpublished) indicate that most of them range from 28-30 nt with a peak at 29 nt. scnRNAs interact with the Argonaute protein Twi1p (Mochizuki & Gorovsky, 2004a), are produced by the Dicer protein Dcl1p (Malone et al., 2005) (Mochizuki & Gorovsky, 2005) and are stabilized by 2′-O-methylation at their 3′-termini by the RNA methyltransferase Hen1p (Kurth & Mochizuki, 2009). Twi1p and Dcl1p are essential for DNA elimination, whereas Hen1p is required for efficient DNA elimination (Mochizuki et al., 2002) (Malone et al., 2005) (Kurth & Mochizuki, 2009). Similar requirements of scnRNA and RNAi machineries for DNA elimination have been reported in Paramecium tetraurelia. The accumulation of scnRNAs in Paramecium is dependent on the Piwi proteins Ptiwi01 and Ptiwi09 and on the Dicer proteins Dcl2 and Dcl3; RNAi knockdown of Ptiwi01/09 or Dcl2/3 causes defective DNA elimination (Bouhouche et al., 2011) (Lepere et al., 2008).
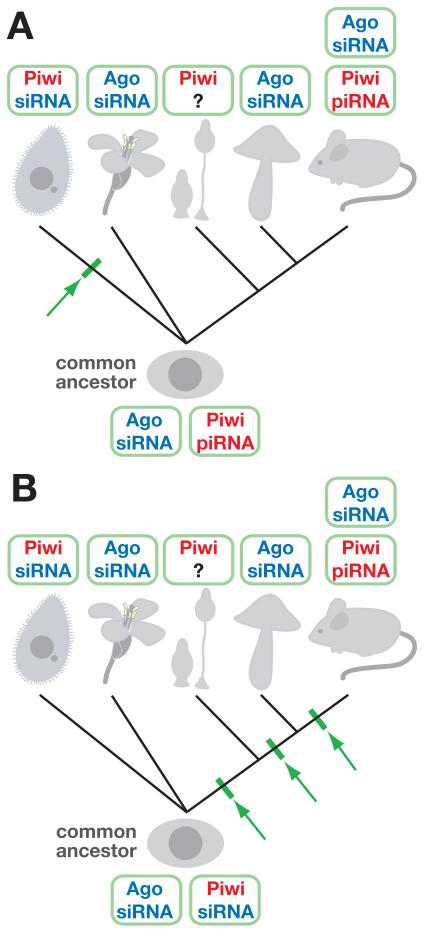
Although the production of Piwi-associated small RNAs in metazoans (piRNAs) appears to be Dicer-independent, this is clearly not in case at least in some Piwi-associated small RNAs in Tetrahymena. Because scnRNAs are associated with the Piwi-protein Twi1p in Tetrahymena, they can be called piRNAs. The presence of 2′-O-methylation on scnRNAs further demonstrates the similarity between piRNAs in metazoans and scnRNAs in Tetrahymena. However, in contrast to piRNAs, scnRNAs are produced from long double-stranded RNA substrates through a Dicer-dependent mechanism. In this context, scnRNAs clearly constitute a class of siRNAs. In addition to Twi1p and Dcl1p, the Tetrahymena genome encodes eleven other Argonaute proteins (Couvillion et al., 2009) and three Dicer proteins (Malone et al., 2005) (Mochizuki & Gorovsky, 2005) (Lee & Collins, 2006). Interestingly, all twelve Argonaute proteins identified in Tetrahymena are more closely related to the Piwi proteins of metazoans than to Ago proteins (Seto et al., 2007). Among them, Twi2p, 3p, 4p 5p and 6p are very similar, and Twi2p is known to associate with 23-24 nt siRNAs produced by the Dicer homologue Dcr2p (Lee & Collins, 2007) (Couvillion et al., 2009). The association of Piwi proteins with Dicer-produced small RNAs in Tetrahymena might indicate that the common ancestor of ciliates and other eukaryotes had both Ago-siRNA and Piwi-piRNA systems and that Piwi proteins were co-opted to use siRNAs specifically through ciliate evolution (Fig. 4A). Alternatively, Piwi proteins associating with siRNAs might be the ancestral form of a Piwi-small RNA complex, and Piwi-piRNA relationships might have evolved after ciliates and other eukaryote lineages were separated (Fig. 4B).
Figure 4. Evolution of Argonaute proteins in eukaryotes.
The presence of different types of Argonaute-small RNA complexes (i.e., Ago-siRNA, Piwi-piRNA and/or Piwi-siRNA) in different types of eukaryotes (left to right: ciliates, plants, slime molds, fungi and metazoans) is shown. It is not known which type of small RNA is complexed with Piwi in slime molds. (A) If the ancestor of these eukaryotes had Ago-siRNA and Piwi-piRNA complexes, the Piwi-siRNA complex found in ciliates must have emerged during the evolution of the ciliate lineage (indicated by arrow). (B) If the ancestor of these eukaryotes had Ago-siRNA and Piwi-siRNA complexes, then the Piwi-piRNA complex in metazoans could have emerged at one of the three time points during their evolution as indicated by arrows.
Similarly to metazoans and ciliates, the slime mold Dictyostelium discodium also utilizes Piwi proteins (Seto et al., 2007). To understand Piwi-piRNA evolution more clearly, it would be interesting to know whether the slime mold uses Dicer in the production of Piwi-associated small RNAs. Also, because the molecular mechanisms underlying the production of small RNAs which are associated to the Piwi proteins Twi7p, 8p, 9p, 10p 11p and 12p in Tetrahymena are not clear, it is important to study their biogenesis as well.
The Ascaris genome, like as other metazoan genomes, contains both Ago and Piwi encoding genes. However, it is not known whether some of these Argonaute proteins have any role in chromatin diminution process.
DNA elimination is regulated by scnRNAs
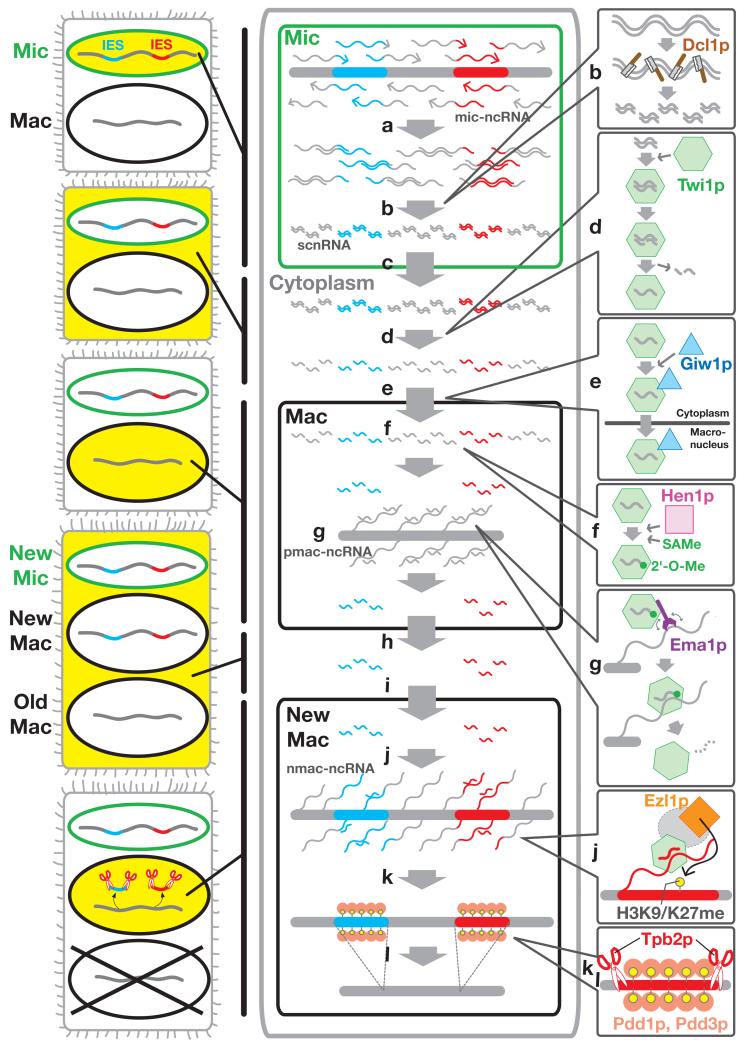
As described above, DNA elimination in the newly formed macronucleus of Tetrahymena is epigenetically regulated by the DNA sequence of the parental macronucleus and is dependent on scnRNAs. The biogenesis and actions of scnRNAs are temporally and spatially regulated. In this review, a current model for an scnRNA-mediated DNA elimination process will be explained by dividing it into five steps (Fig. 5): 1) scnRNA production in the micronucleus; 2) scnRNA-Twi1p complex formation in the cytoplasm and its transport into the parental macronucleus; 3) selective scnRNA degradation in the parental macronucleus; 4) scnRNA-mediated heterochromatin formation in the new macronucleus and 5) DNA elimination in the new macronucleus.
Figure 5. A model for small RNA-directed DNA elimination in Tetrahymena.
(a) Bi-directional transcription of the micronuclear (Mic) genome produces double-stranded micronuclear non-coding RNA (mic-ncRNA). (b) The Dicer-like protein Dcl1p digests double-stranded mic-ncRNAs to short (~29 nt) RNAs, named scnRNAs, in the micronucleus. (c) scnRNA is exported to the cytoplasm where (d) it complexes with the Argonaute protein Twi1p and its passenger strand is cut and removed by the endoribonuclease (Slicer) activity of Twi1p. (e) Giw1p binds to Twi1p complexed with single-stranded scnRNA and transports the complex to the parental macronucleus (Mac). (f) The RNA methyltransferase Hen1p transfers a methyl group from S-adenosylmethionine (SAMe) to 2′-OH of the last nucleotides of single-stranded scnRNAs. This modification (2′-O-Me) stabilizes the scnRNA. (g) The RNA helicase Ema1p facilitates the interaction between the Twi1p–scnRNA complex and nascent parental macronuclear ncRNA (pmac-ncRNA). This interaction induces scnRNA degradation. (h) The selected Twi1p–scnRNA complexes are exported to the cytoplasm and (i) are imported into the newly formed macronucleus (New Mac). The mechanisms underlying this transport are not clear. (j) Similar to the parental macronucleus, the RNA helicase Ema1p facilitates the interaction between the Twi1p–scnRNA complex and the nascent macronuclear ncRNA (nmac-ncRNA). This interaction recruits the histone methyltransferase Ezl1p through an unknown mechanism. Ezl1p catalyzes the methylation of histone H3 at lysine 9 (H3K9me) and lysine 27 (H3K27me). (k) The chromodomain proteins Pdd1p and Pdd3p bind to the methylated histone H3 and establish a heterochromatin structure. (l) The PiggyBac transposase-like protein Tpb2p recognizes the heterochromatin and catalyzes the final DNA excision process.
1) scnRNA production
Because Tetrahymena scnRNAs can hybridize to micronuclear-specific DNA sequences in vitro (Mochizuki et al., 2002) (Mochizuki & Gorovsky, 2004a) (Chalker et al., 2005) (Aronica et al., 2008), they are, at least in part, derived from the micronucleus. Although the micronucleus is transcriptionally inert during most stages of the Tetrahymena life cycle, it becomes transcriptionally active during the prophase of meiosis (Sugai & Hiwatashi, 1974) (Martindale et al., 1985). During this stage, all regions of the micronuclear genome studied produce detectable transcripts, and certain regions were confirmed to be transcribed bi-directionally (Chalker & Yao, 2001) (Aronica et al., 2008). Therefore, it is plausible that the entire micronuclear genome is transcribed bi-directionally at this stage (Fig. 5a). We call the non-coding transcripts micronuclear-non-coding (nc) RNAs (mic-ncRNAs). The localization of a subunit of RNA polymerase II (RNAPII) but not RNAPI or III to the micronucleus during the prophase of meiosis (Mochizuki & Gorovsky, 2004b) indicates that mic-ncRNAs are transcribed by RNAPII. The Dicer protein Dcl1p also localizes to the micronucleus during meiosis. Disruption of the DCL1 gene results in a loss of scnRNA production and overaccumulation of mic-ncRNAs (Malone et al., 2005) (Mochizuki & Gorovsky, 2005). These results indicate that Dcl1p processes mic-ncRNAs into scnRNAs in the micronucleus during meiosis (Fig. 5b).
2) scnRNA-Twi1p complex formation and nuclear import
The Piwi protein Twi1p localizes to the cytoplasm during meiosis in Tetrahymena (Mochizuki et al. 2002). Thus, scnRNAs produced in the micronucleus are probably transported to the cytoplasm (Fig. 5c) where they complex with Twi1p (Fig. 5d). How scnRNAs are exported from the micronucleus and how they complex with Twi1p are currently not clear. In the mid-conjugation stages, Twi1p localizes to the parental macronucleus (Mochizuki et al., 2002). Dcl1p, which is required for the production of scnRNAs, is also required for the macronuclear localization of Twi1p (Noto et al., 2010). Therefore, Twi1p must be complexed with scnRNA to be imported into the macronucleus. Twi1p has an endoribonuclease activity that is vital for removing one of the two strands of scnRNA from the Twi1p–scnRNA complex (Fig. 5d) and is essential for the macronuclear localization of Twi1p (Noto et al., 2010). Giw1p interacts to Twi1p in a Twi1p endoribonuclease-dependent manner and is required for the macronuclear localization of Twi1p (Noto et al., 2010). Therefore, Giw1p probably senses the state of Twi1p-associated scnRNAs and selectively transports mature Twi1p–scnRNA complexes to the macronucleus (Fig. 5e).
Recombinantly expressed Hen1p of Tetrahymena only methylates single-stranded RNAs in vitro (Kurth & Mochizuki, 2009). In addition, TWI1 mutants with defective endoribonuclease activity fail to remove one of the two strands of scnRNAs and subsequently fail to 2′-O-methylate scnRNAs (Kurth & Mochizuki, 2009). Therefore, scnRNAs are probabably 2′-O-methylated in vivo by Hen1p only after the Twi1p–scnRNA complex releases one of the two scnRNA strands. This methylation step most likely occurs in the macronucleus because Hen1p is localized there (Kurth and Mochizuki 2009) (Fig. 5f). The Hen1p-mediated 2′-O-methylation of scnRNA is required for stable accumulation of scnRNA and for proper DNA eliminations (Kurth & Mochizuki, 2009).
3) Selective scnRNA degradation
Although scnRNAs complementary to both macronuclear-destined sequences (MDSs) and IESs are produced in the early conjugation stage, only the second class of scnRNAs becomes enriched during the mid-conjugation stages (Mochizuki & Gorovsky, 2004a) (Aronica et al., 2008). During these stages, Twi1p-scnRNA complexes are localized to the parental macronucleus. Therefore, a specific mechanism must selectively degrade scnRNAs that are complementary to the parental macronuclear genome. The three Twi1p-associated proteins Ema1p (putative RNA helicase), CnjBp (GW repeat and Zinc-finger protein), and Wag1p (GW repeat protein) play important roles in the selective degradation of scnRNAs (Aronica et al., 2008) (Bednenko et al., 2009). Ema1p is also required for the interaction between parental macronuclear ncRNAs (pmac-ncRNAs) and scnRNA-Twi1p complexes. Therefore, it has been proposed that scnRNA degradation is induced by a base-pairing interaction between scnRNA and pmac-ncRNA in the parental macronucleus (Fig. 5g) (Aronica et al., 2008). The requirement of pmac-ncRNAs for proper DNA elimination has been directly demonstrated in Paramecium, whereby downregulation of pmac-ncRNAs by RNAi blocks the scnRNA selection process in targeted regions and induces ectopic DNA elimination (Lepere et al., 2008). The GW repeat proteins CnjBp and Wag1p have redundant roles in the selective degradation of scnRNAs (Bednenko et al., 2009). In addition, the GW repeat proteins Nowa1p and Nowa2p in Paramecium are essential for DNA elimination (Nowacki et al., 2005). However, the molecular mechanisms underlying the involvement of these GW repeat proteins in DNA elimination remain unclear.
4) scnRNA-directed heterochromatin formation
During the development of the new macronucleus, the localization of Twi1p changes from the parental macronucleus to the new macronucleus (Fig. 5h, i). Because scnRNAs complementary to MDSs are degraded in the parental macronucleus, only scnRNAs complementary to IESs move with Twi1p to the new macronucleus. Twi1p is required for the methylation of histone H3 at lysine 9 (H3K9me) and lysine 27 (H3K27me) in the new macronucleus (Liu et al., 2004) (Liu et al., 2007). The enhancer-of-zeste homolog Ezl1p is required for the accumulation of these histone methylations (Liu et al., 2007). As in the parental macronucleus, Ema1p is required for the interaction between nascent transcripts (new macronuclear ncRNAs = nmac-ncRNAs) and Twi1p-scnRNA complexes in the new macronucleus. H3K9/K27me is also greatly reduced in the absence of Ema1p (Aronica et al., 2008). Therefore, Twi1p-scnRNA complexes are likely to be recruited to chromatin sites by a base-pairing interaction between scnRNAs and nmac-ncRNAs, and this interaction eventually recruits Ezl1p (Fig. 5j). However, it is not clear how Ezl1p is recruited by the RNAi-related mechanism.
The modified histones H3K9/K27me are bound to the chromodomain-containing proteins Pdd1p and Pdd3p (Taverna et al., 2002) (Liu et al., 2007). During the process of DNA elimination, IESs localize to dense heterochromatic regions at the nuclear periphery (Madireddi et al., 1996). Twi1p, Ezl1p, and Pdd1p are required for the formation of this heterochromatic structure and for DNA elimination (Coyne et al., 1999) (Mochizuki et al., 2002) (Liu et al., 2007). Therefore, IESs form heterochromatin before they are eliminated, and the formation of the nuclear peripheral heterochromatic structure probably plays an important role in DNA elimination (Fig. 5k). As H3K9/K27me and Pdd1p accumulate specifically on IESs in a Dcl1p- and Twi1p-dependent manner (Taverna et al., 2002) (Liu et al., 2004) (Malone et al., 2005), scnRNAs selected for IES specificity must identify their complementary sequence and recruit the histone methyltransferase Ezl1p.
Because heterochromatic modifications seem to trigger DNA elimination, it is interesting to note that the rearrangement efficiency of an IES is correlated with the overall length of the IES and that the lower size threshold is 300 bp (Kowalczyk et al., 2006). As Kowalczyk et al. (2006) suggested a dinucleosome may be necessary to induce efficient DNA elimination.
5) A domesticated transposase is involved in DNA elimination
It has recently been reported that the PiggyBac transposase-like proteins Pgm and Tpb2p are required for DNA elimination in Paramecium (Baudry et al., 2009) and Tetrahymena (Cheng et al., 2010), respectively. Recombinantly expressed Tpb2p produces double-strand DNA breaks possessing 4 nt 5′-overhangs (Cheng et al., 2010), which were also detected at the end of IESs in vivo during DNA elimination in Tetrahymena (Saveliev & Cox, 1996). Tpb2p shows some sequence preference for a DNA-cutting site in vitro, which weakly fits to the observed boundary sequences of IESs in vivo (Vogt A and KM, unpublished data). This sequence preference may contribute to the precise elimination of IESs along with the homology-dependent IES recognition by scnRNAs.
It is known that artificially tethering of Pdd1p to an ectopic locus is sufficient to induce its DNA elimination (Taverna et al., 2002). Therefore, it appears Pdd1p is capable of recruiting all of the proteins necessary to induce DNA elimination. One such protein is probably Tpb2p because Tpb2p co-localizes to heterochromatin foci containing Pdd1p and H3K9/27me in the developing macronucleus (Cheng et al., 2010). Tpb2p may directly recognize Pdd1p or one of its associated heterochromatin proteins and mediate elimination of IES (Fig. 5l). It is known that some families of transposons are preferentially integrated into heterochromatic regions (e.g., (Zou et al., 1996)). Tpb2p may be derived from such a heterochromatin-preferring transposon; however, the PiggyBac transposon in insects does not have such preference (Wang & Fraser, 1993). Further studies are required to determine the domestication process of Tpb2p as well as whether and how Tpb2p mechanistically recognizes heterochromatin components to perform the final DNA excision process.
Potential Roles of DNA elimination and IESs
Because many IES sequences in ciliates are related to transposable elements, DNA elimination may be a mechanism to remove transposons from a transcriptionally active macronucleus. In a wide variety of eukaryotes, transposons are silenced by RNAi-related mechanisms (reviewed in (Girard & Hannon, 2008)). For example, piRNAs mediate transcriptional and post-transcriptional silencing of transposons in animals, and siRNAs induce RNA-directed DNA methylation (RdDM) to repress transcription from transposons in plants. DNA elimination in ciliated protozoans might have a common evolutionary origin with RNAi-directed transposon silencing pathways in other eukaryotes.
If IESs can be eliminated from a transcriptionally active somatic macronucleus, why do ciliates keep these DNAs in their germline micronucleus? One reason is that IESs may have important functions for the germline micronucleus.
Because a micronucleus divides by mitosis, whereas the macronucleus divides by amitosis, it is hypothesized that only the micronucleus has centromeric DNAs. The exclusive localization of the centromeric histone H3 variant Cna1p to the micronucleus supports this view (Cervantes et al., 2006) (Cui & Gorovsky, 2006). Thus, it is reasonable to conclude that centromeric DNAs probably reside within IESs. However, only a limited number of IESs could have this function because the micronuclear chromosomes are probably unicentric and there are only five micronuclear chromosomes per haploid genome.
The other important requirement for the germline chromosome is the ability to undergo meiosis. Because recombination is potentially mutagenic, it is logical to induce recombination at sequences possessing no function or an unnecessary function. Indeed, it is known that some junk DNAs are recombination hot spots for meiotic homologous recombination in certain eukaryotes (Majewski & Ott, 2000). Based on this logic, IESs could be hypothesized to act as hot spots for meiotic recombination, although no experimental evidence exists to support such a claim.
In addition to their putative function in germline genome integrity, IESs may play a role in gene expression regulation. The micronucleus is transcriptionally inert. Therefore, if an IES includes a cis-regulatory module, such as an enhancer or promoter, it could only be active in a newly developed macronucleus that has not yet undergone DNA elimination. Once the IES is removed, however, its regulatory capacity would be eliminated. This narrow window of time for gene activation would provide an ideal temporal regulation for genes that are required only to be expressed during the development of the new macronucleus, including genes involved in DNA elimination. Although links between transposons and cis-regulatory elements have been reported in many eukaryotes (reviewed in (van de Lagemaat et al., 2003) (Biemont & Vieira, 2006)), IESs containing promoter or enhancer activity have not yet been reported in Tetrahymena. Conversely, if an IES is interrupting a regulatory region or the coding sequence of a gene, the gene will only be expressed after the IES is removed by DNA elimination. An example of gene regulation by DNA elimination has been reported in the ciliate Euplotes; EcTERT-2, which is required for de novo telomere synthesis in a new macronucleus, is expressed only after the removal of an IES in the EcTERT-2 locus (Karamysheva et al., 2003).
Are DNA eliminations in other eukaryotes regulated by small RNAs?
Heterochromatins are targeted for elimination in most programmed genome rearrangement events (DNA elimination, chromatin diminution and chromosome elimination), which may imply that heterochromatin is mechanistically linked to genome rearrangement processes; however, it is also possible that heterochromatin is simply associated with the general repeat-rich nature of eliminated DNAs. Because heterochromatin formation is directed by an RNAi-related mechanism in many eukaryotes (Grewal, 2010), it seems reasonable to expect that programmed genome rearrangements, which are often associated with heterochromatin, may also be regulated by small RNAs.
However, despite the involvement of heterochromatin in the chromatin diminution of the parasitic nematode Ascaris suum, a recent analysis of small RNAs from this worm detected only a tiny amount of small RNAs corresponding to the eliminated satellite DNA (Wang et al., 2011). Moreover, these small RNAs were not upregulated at the stage of chromatin diminution. Therefore, no apparent correlation exists between small RNAs and the chromatin diminution of satellite DNA in this worm.
There is a fundamental difference between chromatin diminution in parasitic nematodes and DNA elimination in ciliates. In parasitic nematodes, the elimination of DNA is coupled with mitosis, and the eliminated DNA segments seem to be discarded because they do not attach to mitotic spindles (Goday et al., 1992). On the other hand, DNA elimination in Tetrahymena occurs independently of mitosis or DNA replication (Nikiforov et al., 1999) and probably removes centromeres because the macronuclear chromosomes segregate by amitosis. Because the Argonaute protein CSR-1 and small RNAs (22G-RNAs) are required for centromere function in segregation of holocentric chromosome in C. elegans, it has been suggested that DNAs in parasitic nematodes may be eliminated because they are not targeted for chromosome segregation by small RNAs (Claycomb et al., 2009). This is in contrast with the small RNA targeted DNA elimination in Tetrahymena.
Because programmed genome rearrangements in different types of eukaryotes probably evolved independently in different lineages, it is reasonable to conclude that unique species may use distinct mechanisms to achieve chromatin elimination. Thus, it would be interesting to compare the mechanisms of programmed genome rearrangements across species to understand how evolution has used many different mechanisms to perform the singular task of DNA elimination.
Acknowledgments
Research in our laboratory is supported by the European Research Council (ERC) Starting Grant (204986) under the European Community’s Seventh Framework Programme, by the Austrian Science Fund (FWF) Doktoratskolleg RNA Biology and SFB “RNA regulation of the transcriptome” and by the Austrian Academy of Sciences.
References
- Akematsu T, Pearlman RE, Endoh H. Gigantic macroautophagy in programmed nuclear death of Tetrahymena thermophila. Autophagy. 2010;6:901–911. doi: 10.4161/auto.6.7.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica L, Bednenko J, Noto T, et al. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes & development. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry CF, Snyder RO, Yao MC. Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic acids research. 1989;17:7263–7272. doi: 10.1093/nar/17.18.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry CF, Yao MC. Sequence structures of two developmentally regulated, alternative DNA deletion junctions in Tetrahymena thermophila. Molecular and cellular biology. 1988;8:3947–3950. doi: 10.1128/mcb.8.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science (New York, N.Y. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Baudry C, Malinsky S, Restituito M, et al. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes & development. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednenko J, Noto T, Desouza LV, et al. Two GW Repeat Proteins Interact with the Tetrahymena Argonaute and Promote Genome Rearrangement. Molecular and cellular biology. 2009;29:5020–5030. doi: 10.1128/MCB.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann S. The diminution of Heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda) Chromosoma. 1977;60:297–344. doi: 10.1007/BF00292858. [DOI] [PubMed] [Google Scholar]
- Biemont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Bouhouche K, Gout JF, Kapusta A, Betermier M, Meyer E. Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic acids research. 2011 doi: 10.1093/nar/gkq1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Xi X, Vermaak D, Yao MC, Malik HS. The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus. Molecular biology of the cell. 2006;17:485–497. doi: 10.1091/mbc.E05-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Fuller P, Yao MC. Communication between parental and developing genomes during tetrahymena nuclear differentiation is likely mediated by homologous RNAs. Genetics. 2005;169:149–160. doi: 10.1534/genetics.104.032300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Molecular and cellular biology. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes & development. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Vogt A, Mochizuki K, Yao MC. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Molecular biology of the cell. 2010;21:1753–1762. doi: 10.1091/mbc.E09-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Lee SR, Hogstad B, et al. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes & development. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Nikiforov MA, Smothers JF, Allis CD, Yao MC. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Molecular cell. 1999;4:865–872. doi: 10.1016/s1097-2765(00)80396-2. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Thiagarajan M, Jones KM, et al. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics. 2008;9:562. doi: 10.1186/1471-2164-9-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Gorovsky MA. Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila. Molecular and cellular biology. 2006;26:4499–4510. doi: 10.1128/MCB.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes & development. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS biology. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Meister G. Argonaute proteins at a glance. Journal of cell science. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- Epstein LM, Forney JD. Mendelian and non-mendelian mutations affecting surface antigen expression in Paramecium tetraurelia. Molecular and cellular biology. 1984;4:1583–1590. doi: 10.1128/mcb.4.8.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham JS, Thing TA, Vythilingum N, et al. A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila. Eukaryotic cell. 2004;3:157–169. doi: 10.1128/EC.3.1.157-169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. Sinauer Associates Inc; Sunderland, MA: 2010. [Google Scholar]
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends in cell biology. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goday C, Esteban MR. Chromosome elimination in sciarid flies. Bioessays. 2001;23:242–250. doi: 10.1002/1521-1878(200103)23:3<242::AID-BIES1034>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Goday C, Gonzalez-Garcia JM, Esteban MR, Giovinazzo G, Pimpinelli S. Kinetochores and chromatin diminution in early embryos of Parascaris univalens. The Journal of cell biology. 1992;118:23–32. doi: 10.1083/jcb.118.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiska R, James C, Yao MC. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes & development. 1993;7:2357–2365. doi: 10.1101/gad.7.12a.2357. [DOI] [PubMed] [Google Scholar]
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Current opinion in genetics & development. 2010 doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Williamson S, Dunn S, et al. The highly conserved family of Tetrahymena thermophila chromosome breakage elements contains an invariant 10-base-pair core. Eukaryotic cell. 2006;5:771–780. doi: 10.1128/EC.5.4.771-780.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen TY, Pearlman RE. A germ line-specific sequence element in an intron in Tetrahymena thermophila. The Journal of biological chemistry. 1994;269:17428–17433. [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Howard-Till RA, Yao MC. Tudor nuclease genes and programmed DNA rearrangements in Tetrahymena thermophila. Eukaryotic cell. 2007;6:1795–1804. doi: 10.1128/EC.00192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva Z, Wang L, Shrode T, Bednenko J, Hurley LA, Shippen DE. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell. 2003;113:565–576. doi: 10.1016/s0092-8674(03)00363-5. [DOI] [PubMed] [Google Scholar]
- Karrer KM. Tetrahymena genetics: two nuclei are better than one. Methods in cell biology. 2000;62:127–186. doi: 10.1016/s0091-679x(08)61529-0. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA (New York, N.Y. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno S, Nakai Y, Satoh S, Yoshida M, Kobayashi H. Chromosome elimination in the Japanese hagfish, Eptatretus burgeri (Agnatha, Cyclostomata) Cytogenetics and cell genetics. 1986;41:209–214. doi: 10.1159/000132231. [DOI] [PubMed] [Google Scholar]
- Kowalczyk CA, Anderson AM, Arce-Larreta M, Chalker DL. The germ line limited M element of Tetrahymena is targeted for elimination from the somatic genome by a homology-dependent mechanism. Nucleic acids research. 2006;34:5778–5789. doi: 10.1093/nar/gkl699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA (New York, N.Y. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes & development. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nature structural & molecular biology. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes & development. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Song X, Gorovsky MA, Karrer KM. Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryotic cell. 2005;4:421–431. doi: 10.1128/EC.4.2.421-431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes & development. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DH. The Ciliated Protozoa. Springer; 2008. [Google Scholar]
- Madireddi MT, Coyne RS, Smothers JF, Mickey KM, Yao MC, Allis CD. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J. GT repeats are associated with recombination on human chromosome 22. Genome research. 2000;10:1108–1114. doi: 10.1101/gr.10.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Molecular and cellular biology. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale DW, Allis CD, Bruns PJ. RNA and protein synthesis during meiotic prophase in Tetrahymena thermophila. The Journal of protozoology. 1985;32:644–649. doi: 10.1111/j.1550-7408.1985.tb03094.x. [DOI] [PubMed] [Google Scholar]
- Meyer E, Duharcourt S. Epigenetic regulation of programmed genomic rearrangements in Paramecium aurelia. The Journal of eukaryotic microbiology. 1996;43:453–461. doi: 10.1111/j.1550-7408.1996.tb04504.x. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes & development. 2004a;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryotic cell. 2004b;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes & development. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Tobler H. Chromatin diminution in the parasitic nematodes ascaris suum and parascaris univalens. International journal for parasitology. 2000;30:391–399. doi: 10.1016/s0020-7519(99)00199-x. [DOI] [PubMed] [Google Scholar]
- Nikiforov MA, Smothers JF, Gorovsky MA, Allis CD. Excision of micronuclear-specific DNA requires parental expression of pdd2p and occurs independently from DNA replication in Tetrahymena thermophila. Genes & development. 1999;13:2852–2862. doi: 10.1101/gad.13.21.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Kurth HM, Kataoka K, et al. The Tetrahymena Argonaute-Binding Protein Giw1p Directs a Mature Argonaute-siRNA Complex to the Nucleus. Cell. 2010;140:692–703. doi: 10.1016/j.cell.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Zagorski-Ostoja W, Meyer E. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr Biol. 2005;15:1616–1628. doi: 10.1016/j.cub.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes & development. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satzinger H. Theodor and Marcella Boveri: chromosomes and cytoplasm in heredity and development. Nature reviews. 2008;9:231–238. doi: 10.1038/nrg2311. [DOI] [PubMed] [Google Scholar]
- Saveliev SV, Cox MM. Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. The EMBO journal. 1996;15:2858–2869. [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Molecular cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Sugai T, Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. The Journal of protozoology. 1974;21:542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Van De Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Wang HG, Fraser MJ. TTAA serves as the target site for TFP3 lepidopteran transposon insertions in both nuclear polyhedrosis virus and Trichoplusia ni genomes. Insect molecular biology. 1993;1:109–116. doi: 10.1111/j.1365-2583.1993.tb00111.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Czech B, Crunk A, et al. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome research. 2011 doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic acids research. 2002;30:2524–2537. doi: 10.1093/nar/30.11.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Choi J, Yokoyama S, Austerberry CF, Yao CH. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell. 1984;36:433–440. doi: 10.1016/0092-8674(84)90236-8. [DOI] [PubMed] [Google Scholar]
- Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science (New York, N.Y. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- You Y, Scott J, Forney J. The role of macronuclear DNA sequences in the permanent rescue of a non-mendelian mutation in Paramecium tetraurelia. Genetics. 1994;136:1319–1324. doi: 10.1093/genetics/136.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, et al. Methylation as a crucial step in plant microRNA biogenesis. Science (New York, N.Y. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Ke N, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes & development. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]