Abstract
To gain insight into the structural basis of DNA bending by adenine–thymine tracts (A-tracts) and their role in DNA recognition by gene-regulatory proteins, we have determined the crystal structure of the high-affinity DNA target of the cancer-associated human papillomavirus E2 protein. The three independent B-DNA molecules of the crystal structure determined at 2.2-Å resolution are examples of A-tract-containing helices where the global direction and magnitude of curvature are in accord with solution data, thereby providing insights, at the base pair level, into the mechanism of DNA bending by such sequence motifs. A comparative analysis of E2–DNA conformations with respect to other structural and biochemical studies demonstrates that (i) the A-tract structure of the core region, which is not contacted by the protein, is critical for the formation of the high-affinity sequence-specific protein–DNA complex, and (ii) differential binding affinity is regulated by the intrinsic structure and deformability encoded in the base sequence of the DNA target.
Keywords: A-tract, papillomavirus E2–DNA target, transcriptional control
The relationship between DNA sequence, structure, and function has been studied and discussed extensively for the last 20 years. A particular effort has been directed toward the structural elucidation of short runs of four to six adenine–thymine residues, known as A-tracts, in an attempt to reveal the structural basis of DNA curvature induced by such sequence motifs when they are inserted in phase with the helical periodicity (1, 2). Despite numerous efforts, including x-ray crystallographic and solution studies, the structural basis of A-tract-induced bending has remained enigmatic (3). Because no single structure could explain the whole phenomenon, it was necessary to rely on models, several of which had been proposed (4). They generally conform to the gel migration data, which suggest that the center of curvature is toward the minor groove of the A-tracts and toward the major groove of the intervening general sequences (5). However, they differ substantially in the details of the stereochemical origin of curvature. This issue is of particular biological significance, as sequence-dependent DNA curvature or bending is an important determinant of DNA recognition by proteins (6).
The E2 regulatory system of human papillomaviruses provides an example where sequence-specific binding of proteins to A-tracts is crucial to the organism's life cycle. The E2 proteins from all viral strains activate or repress transcription in a context-dependent manner and are required for the initiation of replication in vivo. Their function depends on sequence-specific binding to a highly conserved 12-bp sequence of the general form ACCGNNNNCGGT, where N4 is variable (7, 8). However, the E2 binding sites in the human papillomavirus (HPV) genomes, including the cancer-related strains HPV-16 and HPV-18 (9), exhibit a further level of specificity in the interaction with their cognate E2 proteins. This specificity is manifested by the identity of the central 4-bp region (e.g., AATT, AAAA/TTTT), which is of the kind known to induce DNA curvature in solution (1, 10), and by the corresponding binding affinities (11–15). Unlike HPV, in other papillomaviruses such as the well-studied bovine papillomavirus type 1 (BPV-1), the central region is highly variable (16).
The DNA-binding domains of several E2 proteins and their complexes with DNA have been studied in terms of their structure and DNA-binding affinity (13, 14, 17). It was found that the DNA is largely deformed by the protein and that high-affinity and low-affinity DNA–protein complexes exhibit similar DNA conformations and protein–DNA contacts (14, 17). Hence, the structural basis for sequence-specific recognition and differential DNA binding affinity in this system should be sought in the structural alterations between free and protein-bound DNA targets.
Here we present the crystal structure of the high-affinity E2–DNA target of human papillomaviruses, ACCGAATTCGGT, determined at 2.2-Å resolution. We show that the three independent B-DNA molecules of the crystal structure are intrinsically bent in the free state in a direction that positions the minor groove of the A-tract region at the concave side of the helix, as required for the E2–DNA complex and in agreement with solution studies of A-tract-containing DNA. The implications of these findings for A-tract-dependent DNA bending and its role in sequence-specific protein–DNA interactions are discussed.
Materials and Methods
DNA Preparation and Crystallization.
The DNA dodecamer ACCGAATTCGGT was synthesized and purified by a described procedure (18). Crystals of ACCGAATTCGGT were grown at 19°C from 5-μl hanging drops containing 2 mg/ml DNA, 76 mM MgCl2, 1 mM spermine⋅4HCl, 20 mM Na cacodylate buffer (pH 7.0), and 15% (wt/vol) 2-methyl-2,4-pentanediol equilibrated against a reservoir of 37% (wt/vol) 2-methyl-2,4-pentanediol in 20 mM Na cacodylate buffer (pH 7.0). Crystals appeared after several days but diffracted poorly and were highly mosaic; however, occasionally the crystals dissolved over a period of several months and recrystallized with a similar morphology and an average size of 0.1 × 0.1 × 0.2 mm3. These crystals belong to space group P1 with three duplexes in the unit cell and diffract to a resolution of 2.2 Å.
Data Collection, Structure Determination, and Refinement.
X-ray diffraction data from a single crystal coated with Exxon Paraton oil and flash cooled to 100 K were measured on a Rigaku R-AXIS IIC image plate detector mounted on a Rigaku rotating anode generator (FRC model), with Ni-filtered CuKα radiation focused by Yale-type mirrors. Data were processed with denzo-scalepack (19). Crystal data and intensity statistics are given in Table 1.
Table 1.
Crystallographic data and refinement statistics
| Space group | P1 |
| Unit cell dimensions, Å and ° | a = 39.8, b = 40.2, c = 39.5 |
| α = 74.4, β = 76.4, γ = 59.9 | |
| No. of independent DNA duplexes | 3 |
| Volume per base pair, Å3 | 1,450 |
| Resolution limits, Å | 19.0–2.2 |
| No. of measured reflections with I > 0 | 36,389 |
| No. of unique reflections/used in refinement* | 9,810/8,474 |
| Completeness of data, % | 95.7 (95.0) |
| Rsym(I), % | 5.1 (56.4) |
| R-factor/R-free, %† | 22.4/29.2 |
| No. of DNA atoms per no. of water molecules | 1,458/114 |
| rms deviations | |
| Bond length, Å | 0.01 |
| Bond angle, ° | 1.6 |
| Bonded B-factor, Å2 | 3.8 |
Values in parentheses refer to the upper resolution shell (2.24–2.20 Å).
Number of observed reflections after correcting for anisotropic scaling (see text).
R-factor = Σ|FO − |FC∥/|FO|. R-free was calculated with 5% of the data excluded from the refinement.
The structure was determined by the molecular Fourier transform method as implemented in the program mft (20), combined with a modified version of ultima (21), with the use of a search model based on standard B-DNA fiber coordinates (22). The search parameters and resolution range used here were similar to that used for another triclinic dodecamer (18, 20).
The structure was subjected to a rigid-body refinement by X-PLOR, Version 3.1 (23), and the duplexes were divided into groups as described for other DNA structures (18). The high-resolution data (beyond 3 Å) suffered from significant anisotropy, and therefore the refinement was continued with SHELXL-97, with the use of the options of individual atom refinement, together with HOPE (for anisotropic scaling) and SWAT (to model the solvent as a diffuse continuum) (24). The observed structure factors were corrected for the anisotropy in the data and were used for further refinement by X-PLOR. This refinement used cycles of simulated annealing followed by energy minimization and restrained individual temperature-factor refinement, with bulk solvent correction and overall anisotropic temperature factor optimization. DNA geometrical parameters from Parkinson et al. (25) were used. The refinement cycles were alternated with examination of the electron density maps for manual correction of the model and location of ordered solvent molecules by O (26). Solvent molecules were accepted if they had an acceptable geometry of contact with the DNA or other solvent molecules. The refinement results are in Table 1.
Results and Discussions
Intrinsically Bent DNA Molecules.
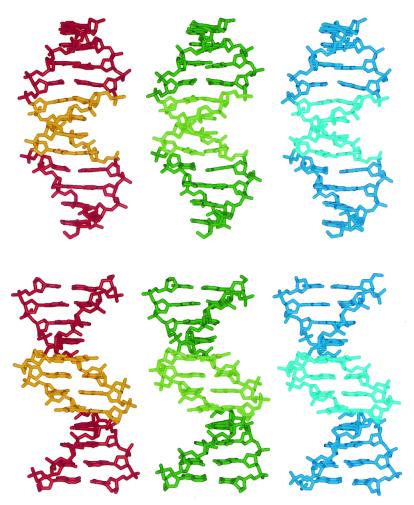
The crystal structure of the E2–dodecamer represents a packing arrangement of B-DNA dodecamers with three independent molecules (see Materials and Methods). The interduplex contacts are head-to-tail base pair stacking and backbone/major-groove interactions that are commonly found in B-DNA crystals (27). The three duplexes of ACCGAATTCGGT are illustrated in Fig. 1. The rms pairwise differences among the three molecules range from 0.5 to 0.7 Å, including all atoms, reflecting their high similarity. This similarity is not induced by crystal packing interactions, as the three molecules are embedded in different crystalline environments and are “loosely” packed in a manner similar to that of the E2 binding sites with non-A-tract core regions (18).
Figure 1.
Two views of the three unique helices of the E2-dodecamer, ACCGAATTCGGT. The top view is perpendicular to the dyad axis through the center. The bottom view is along that dyad. The AATT regions of the three molecules are shown in light colors.
All three molecules have a global curvature whose center points toward the minor groove of the A-tract and toward the major groove of the flanking regions. The angles between the end base pairs of the central decamers in the three molecules are 8°, 10°, and 9°, respectively. The angles between the end base pairs of the dodecamers are close to 0°, presumably to optimize end-to-end stacking interactions in the crystal. Similar bend angles (8°, 11°, 8°) were obtained by fitting global curved axes with curves (28). Both the A-tract and the non-A-tract regions contribute to the global curvature, as discussed below.
All helices are slightly underwound compared with standard B-DNA, with an average helical periodicity of 10.3 bp, as also observed in the other E2 binding sites (18). The groove dimensions of the three helices deviate significantly from canonical B-DNA (22) and from the E2 binding sites with general-sequence core regions (18). In particular, the present three helices have a very narrow minor groove at the A-tract region with gradual widening toward the helical ends. Narrowing of the minor groove of the A/T-rich zone is associated with large propeller twisting of the base pairs (av. −16° at the center compared with −7° at the flanking base pairs). These features are common to all A-tracts studied (3).
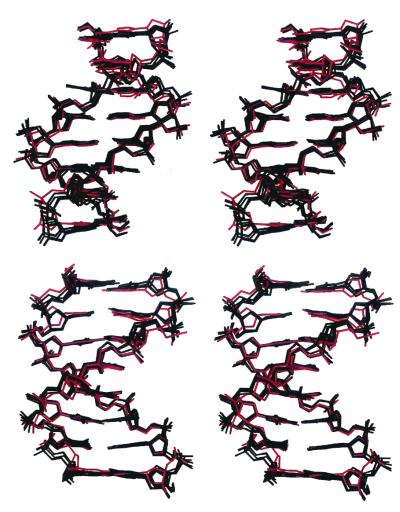
The specific conformational features of the central base pair region (CGAATTCG) of the present helices are common to all other DNA oligomers containing the same sequence. Crystal structures of this sequence incorporated in 9- to 12-bp duplexes and crystallized under a variety of conditions and in the presence of different cations (Na+, K+, Mg2+, Ca2+) were determined at high resolution (29–35), and several of them were isomorphous to the Dickerson–Drew dodecamer (36). The superpositioning of such structures with the present one is shown in Fig. 2. The rms differences between the central octamers of the current molecules and those of the others range from 0.9 to 1.2 Å when all of the atoms are examined and from 0.4 to 0.8Å when only base atoms are used. These findings demonstrate that the conformation of the double-helical octamer CGAATTCG observed in the crystal structures is an intrinsic property of the particular base sequence. Local conformational alterations as a result of environmental effects are confined mainly to the flexible sugar-phosphate backbone. The global bend directed toward the A-tract minor groove in the present E2 helices is also apparent in all of the other structures. It should be noted, however, that in most of the reported AATT structures, the minor-groove bend at the central region is masked by another bend induced by crystal packing interactions. As a result, the overall curvature in these molecules is asymmetric and 90° away from the direction found in solution studies as discussed (10).
Figure 2.
Superpositioning of the central base pair region (CGAATTCG) of the present dodecamer (red) with identical regions from six different crystal structures determined at high resolution, ranging from 0.9 Å to 1.7 Å (the Nucleic Acid Database access codes are bd0014, bd0016, bd0018, bd0019, bd0029, and bd1084). The top view is along the dyad of the central minor groove. The bottom view is perpendicular to that dyad.
Insights into the Structural Basis of A-Tract-Induced Bending.
The key question is, what kind of mechanism, at the atomic level, is causing DNA bending by an adenine–thymine tract? The three independent molecules of the present crystal structure are examples of A-tract-containing helices with an overall curvature in accord with the solution data. The structural data provide insights into the stereochemical basis of A-tract-induced bending as outlined below.
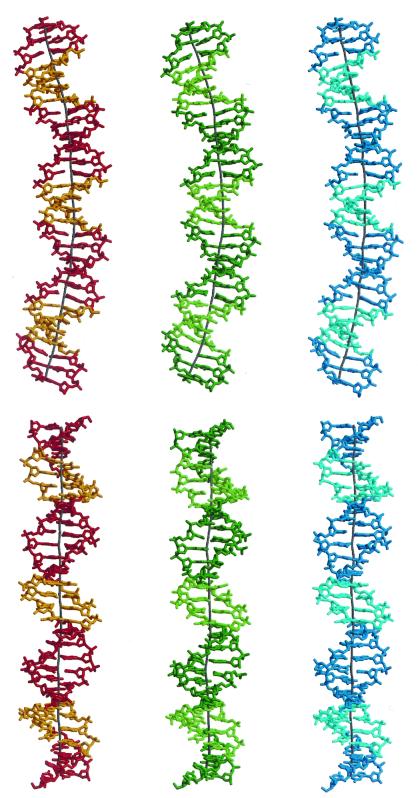
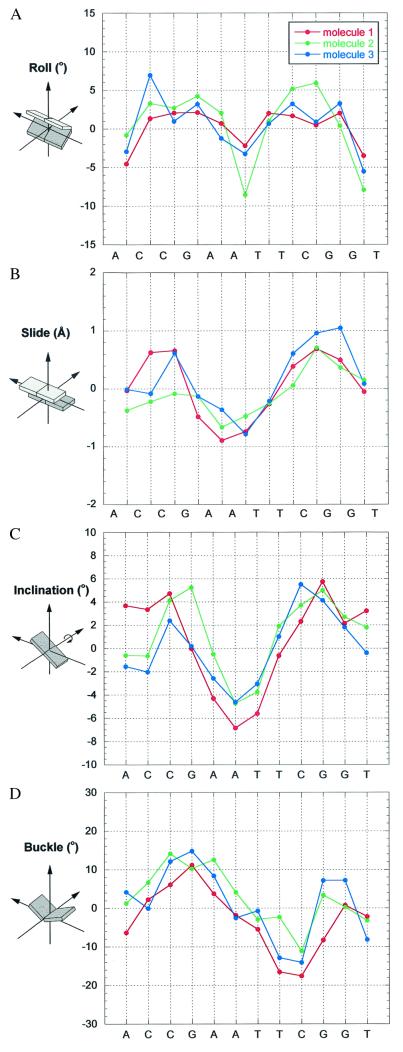
Continuous 30-bp helices based on the central 10 bp of each of the three molecules are shown together with their global helix axes in Fig. 3. These helices clearly demonstrate that curvature is achieved by both bending toward the minor groove at the A-tract sequences and toward the major groove at the intervening G/C-rich sequences. The magnitude of curvature in each of the three helices is close to 10° per decameric repeat and partitioned evenly along the double helix. This value compares well with data obtained from solution studies on similar sequence motifs (10). Analysis of the helical parameters at the base pair level shows that the major contribution to the global curvature comes from negative roll angles (closing the base pairs toward the minor groove) in the A-tract region and positive roll angles (closing toward the major groove) in the intervening segments (Fig. 4A). The roll angles are positively correlated with the corresponding slide values that measure the relative translation of the base pairs along their long axes (Fig. 4B).
Figure 3.
Thirty-base pair-long helices of CCGAATTCGG/CCGAATTCGG/CCGAATTCGG based on the central 10 base pairs of each molecule of ACCGAATTCGGT. The geometry of the G/C junction was obtained by best molecular fitting, where the first two base pairs of each dodecameric duplex were fitted onto the last two base pairs of an identical duplex. The base pairs at the G/C junctions are nearly parallel to each other, and hence the overall curvature is determined by the decameric helices. The global helix axis was derived with the program CURVES (28). The top view is perpendicular to the dyad through the central A-T base pair step. The bottom view is along the central dyad. The color code is as in Fig. 1.
Figure 4.
Local parameter plots of the three E2-dodecamers. Roll (A), slide (B), and buckle (D) were calculated with FREEHELIX (37). Inclination angles (C) were calculated with respect to the fitted curved axis by CURVES (28). See definitions in the text and in ref. 38. As shown by Lu and Olson (38), the values of certain local parameters are sensitive to the choice of reference frame but are only slightly affected by the algorithm used. FREEHELIX, which employs the consensus reference frame, was chosen here to allow direct comparisons with related DNA structures (14, 18).
The concerted variations in the relative position of the base pairs lead to bending into the minor groove at the A-tract region and into the major groove at the flanking G/C sequences. As a result of this geometry and the helix twist, the A⋅T and G⋅C base pairs are inclined to the global helix axis in opposite directions with respect to their long dimension, as shown by the corresponding inclination angles of Fig. 4C and illustrated in Fig. 5. An abrupt change in inclination of the base pairs would impair the base-stacking geometry of the concerned base pairs. To minimize such unfavorable effects, the change in the direction of inclination is accompanied by “buckling” of the base pairs at the junctions. Instead of being coplanar, the base pairs are buckled; that is, the two bases are tilted toward each other by 10–20° about the short axis of the base pair. Whereas base pair buckling does not appear to significantly affect the Watson–Crick hydrogen bond energy, it allows for continuous partial base pair stacking interactions (involving only one of the bases of a pair) between the A-tract base pairs and the non-A-tract base pairs (Fig. 5). The buckle angles change from positive values at one junction of the helix to negative ones at the other junction, as expected for a nearly symmetric conformation (Fig. 4D). Buckling of the base pairs is characteristic of all of the other CGAATTCG-containing oligomers (from 9 to 12 base pairs) shown in Fig. 2 and therefore represents an intrinsic feature of the base sequence. Buckle-mediated bending is also shown by the protein-bound E2–DNA targets (14) and appears to be a common determinant of DNA bending by proteins (e.g., 39, 40).
Figure 5.
Schematic representation of an unrolled helix viewed at the minor groove. A⋅T base pairs are in light blue, and G⋅C base pairs are in dark blue. The global helix axis is along the vertical direction. The figure illustrates the mediating effect of base pair buckling between the A-tract region (negative inclination of the base pairs) and the flanking segments (positive inclination of the base pairs).
Homo-oligomeric A-tracts (e.g., An/Tn, n = 4–6), embedded in a G/C-rich environment and repeated in phase with the helical periodicity, were shown to induce larger curvature in solution than their symmetric counterparts (3, 10). The mechanism of A-tract-dependent bending is likely to be different in such nonsymmetric sequences. Other AT-rich motifs such as ATAT and TTAA do not confer curvature on the DNA helix, a phenomenon that has been attributed to the conformational polymorphism of such sequences because of the dynamical nature of the T-A sites (10).
DNA Recognition by the E2 Proteins and the Structural Basis of Differential Binding Affinity.
Based on the high-resolution structural data on the E2–DNA target ACCGACGTCGGT, in its free state and in its complex with the DNA binding domain of the BPV-1 E2 protein (referred to as BPV-E2) (17, 18), we have shown that the conformational features of the conserved regions (ACCG/CGGT) contacted directly by the BPV-E2 protein are only slightly modified by the protein, whereas a considerable deformation is imposed on the noncontacted core region (ACGT). This finding led to the conclusion that the inherent structure of the conserved regions, as well as the deformability (deformation at low energy cost) of the nonconserved core region, is critical for the recognition process. Such conformational features are encoded in the base sequence and constitute a structural code for its recognition by the protein (18).
The recent structural and biochemical data on the DNA-binding domains of the E2 proteins from BPV and HPV complexed with different DNA targets have shown as in the BPV-E2 system, in the HPV-E2 complex the DNA is bent toward the dimeric protein by the interaction of a pair of symmetrically disposed α-helices with the two conserved ACC/GGT sites in symmetrically related regions of the major groove, whereas the central region facing the protein from the minor-groove side has no contacts with the protein (14). The crystal structures have also shown that high-affinity and low-affinity targets of HPV-E2 (represented by ACCGAATTCGGT and ACCGACGTCGGT, referred to as AATT and ACGT), which differ in the identity of the central noncontacted region, exhibit very similar protein-bound conformations (14). The combined structural and biochemical data on the different E2–DNA targets in their free state and in complexes with the BPV-E2 and HPV-E2 proteins further substantiate the predominance of the DNA structural code in sequence-specific recognition and provide insights into the structural basis of DNA recognition and differential binding affinity as discussed below.
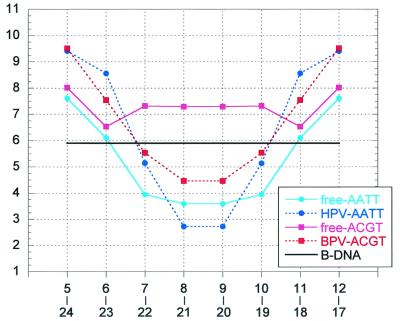
Inspection of the structures of the high-affinity HPV-E2 target of the present study, ACCGAATTCGGT, and that reported for a 16-mer DNA target incorporating the same central dodecamer complexed to the DNA binding domain of the HPV-18 E2 protein (14) shows that many features of the free DNA target are preserved in the protein-bound conformation. These include the unusually narrow minor groove of the A-tract core and the direction of curvature where the A-tract region bends toward the minor groove and the flanking regions toward the major groove. The predisposition of the free DNA target to adoption of a conformation compatible with the one required to form the complex is critical to its high affinity for the HPV-E2 protein. The global curvature of the free DNA is further enhanced in the protein–DNA complex to optimize the interactions between the dimeric protein and the two symmetrically disposed conserved sites. This enhancement is achieved by the cumulative effect of small changes in the relative orientation of the base pairs. As a result, the minor groove is narrower at the A-tract zone and wider at the flanking regions with respect to the free DNA (Fig. 6).
Figure 6.
Minor-groove width plots of the free and protein-bound DNA targets, ACCGAATTCGGT and ACCGACGTCGGT (referred to as AATT and ACGT). All values (Å) were end-to-end averaged, following the symmetry of the base sequence. The values of free-AATT represent an average of the three molecules of the present study. The values of HPV-AATT were derived from the coordinates of the HPV-E2/AATT complex (14). The values of free-ACGT and BPV-ACGT were derived from the coordinates of the corresponding high-resolution structures (18, 17). Because the conformations of the AATT and ACGT DNA targets bound to the same protein are similar to each other (14), only one set of values is shown for each type of protein–DNA complex. The groove width is defined as the shortest P–P distance across the groove less 5.8 Å (the sum of the van der Waals radii of the two phosphate groups). The numbering of the P atoms is 2 to 12 and 14 to 24 (5′ to 3′ direction) for the two strands, respectively.
The binding affinities of the AATT and other A-tract-containing targets for the BPV-E2 protein are one to two orders of magnitude less than that for the HPV-E2 proteins, whereas the affinities of the ACGT target and other non-A-tract-binding sites for the two proteins are comparable (13, 14). This observation can be rationalized on the basis of the deformations induced in the two DNA targets on interaction with the two proteins. Whereas the conformations of different DNA targets bound to the same protein are highly similar, significant conformational differences are observed for the same DNA target between the two different proteins BPV-E2 and HPV-E2 (14), as also indicated by solution studies (11, 12, 15). The most prominent conformational change is in the dimensions of the central minor groove, where the corresponding groove widths are smaller than 3 Å and greater than 4 Å for the HPV– and BPV–DNA complexes, respectively (14), as illustrated in Fig. 6. The minor-groove width of the free AATT target at the core region is midway between those of the two bound conformations. However, unlike the HPV-E2/AATT complex, where DNA bending into the minor groove is associated with an expected compression of the groove relative to that of the free DNA, the bending of AATT in the BPV-E2 complex is associated with a significant widening of the central minor groove compared with its free form, which is opposed to the natural tendency of the double helix. This widening of the groove would lead to a considerable loss in the enthalpic contribution to the stability of the BPV-E2/AATT complex compared with its HPV-E2 counterpart. Such observations explain why the HPV-E2 protein shows a preference for A-tract core regions that have unusually narrow minor grooves, whereas the BPV-E2 protein does not show such a tendency (16).
The conformational changes induced by the two different proteins in the conserved sequences (ACCG/CGGT) that are contacted by the proteins are small and similar for the two DNA targets AATT and ACGT, emphasizing the role of the intrinsic structure of these regions in the specificity of interaction in the E2 system. In contrast, the distortions imposed by the two proteins on the central noncontacted region of the ACGT target are much larger compared with that for the AATT target, particularly for the HPV-E2 complex, where the change is from a wide minor groove (wider than 7 Å) in the free state to an extremely narrow minor groove (narrower than 3 Å) in the bound state (Fig. 6). The binding affinities of the ACGT target for the two proteins are similar (13, 14), because of the deformable nature of the noncontacted core region as discussed (18). However, the binding affinity of the ACGT target for the HPV-E2 protein is two orders of magnitude less than that of the AATT target (14), suggesting that in this system the energetic advantage rendered by deformability of the noncontacted DNA region of the former is much lesser than that obtained by an intrinsically compatible A-tract conformation of the same region in the latter. Hence, differential binding affinity in the E2 system appears to be determined to a large extent by the energy required to deform different DNA targets from their free conformations to the protein-bound conformations, a finding that may be relevant to other systems as well. For example, the intrinsic structure and deformability of the 434 λ bacteriophage operator sites are likely to play a similar role in differential binding affinity observed in their complexes with the corresponding repressor and Cro proteins (refs. 41 and 42 and references therein).
Summary and Conclusions
In the present study, the crystal structure of the high-affinity DNA target of the E2 protein from HPV is analyzed and compared with several related DNA structures. The comparative analysis demonstrates that unique conformational features, such as an unusually narrow minor groove at the A-tract region and an overall curvature formed by minor-groove compression of the A-tract region and major-groove compression of the flanking G + C-rich segments, are inherent to the specific base sequence. The direction and magnitude of curvature are in agreement with solution studies. The detailed conformational analysis at the base-pair level shows that global curvature is brought about mainly by correlated variations in the roll angles between the base pairs in all helical regions and in the buckling of the base pairs at the junctions between the A-tract and non-A-tract segments.
Comparisons between the structure of the A-tract-containing E2 dodecamer and those of other E2-DNA targets, both in the free state and in their complexes with proteins, demonstrate that the intrinsic structure of the conserved regions that are in direct contact with the protein is critical for sequence-specific protein–DNA interactions in all E2 systems. The high specificity of interaction observed in the HPV E2 system is achieved by the intrinsic A-tract structure of the core region, which is not contacted by the protein. Differential binding affinity in the E2 system is finely tuned by the intrinsic structure and deformability encoded in the base sequence of the DNA target. Such studies provide a basis for concepts of structural codes for DNA recognition by proteins as well as structural insights into the relationship between structure and function in protein–DNA interactions.
Acknowledgments
This article is dedicated to the memory of Paul B. Sigler. We thank Linda Shimon, Joseph Kalb (Gilboa), Richard Lavery, and Aaron Klug for their helpful comments on the manuscript. The work was supported by grants from the Israel Science Foundation administered by the Israel Academy of Sciences and Humanities and from the United States–Israel Binational Science Foundation. Z.S. holds the Helena Rubinstein Professorial Chair of Structural Biology at the Weizmann Institute of Science.
Abbreviations
- A-tracts
adenine–thymine tracts
- HPV
human papillomavirus
- BPV-1
bovine papillomavirus type 1
Footnotes
Data deposition: The atomic coordinates have been deposited in the Nucleic Acid Database, Department of Chemistry, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (NDB structure ID code BD0052).
References
- 1.Crothers D M, Haran T E, Nadeau J G J. J Biol Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 2.Hagerman P J. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 3.Crothers D M, Shakked Z. In: Oxford Handbook of Nucleic Acid Structures. Neidle S, editor. London: Oxford Univ. Press; 1999. pp. 455–468. [Google Scholar]
- 4.Goodsell D S, Dickerson R E. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zinkel S S, Crothers D M. Nature (London) 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 6.Travers A A. In: DNA-Protein: Structural Interactions. Lilley D M J, editor. London: Oxford Univ. Press; 1995. pp. 49–75. [Google Scholar]
- 7.Steger G, Ham J, Yaniv M. Methods Enzymol. 1996;274:173–185. doi: 10.1016/s0076-6879(96)74016-8. [DOI] [PubMed] [Google Scholar]
- 8.McBride A A, Romanczuk H, Howley P M. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 9.zur Hausen H. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 10.Shatzky-Schwartz M, Arbuckle N D, Eisenstein M, Rabinovich D, Bareket-Samish A, Haran T E, Luisi B F, Shakked Z. J Mol Biol. 1997;267:595–623. doi: 10.1006/jmbi.1996.0878. [DOI] [PubMed] [Google Scholar]
- 11.Bedrosian C, Bastia D. Virology. 1990;174:557–575. doi: 10.1016/0042-6822(90)90109-5. [DOI] [PubMed] [Google Scholar]
- 12.Thain A, Webster K, Emery D, Clarke A R, Gaston K. J Biol Chem. 1997;272:8236–8242. doi: 10.1074/jbc.272.13.8236. [DOI] [PubMed] [Google Scholar]
- 13.Hines C S, Meghoo C, Shetty S, Biburger M, Brenowitz M, Hegde R S. J Mol Biol. 1998;276:809–818. doi: 10.1006/jmbi.1997.1578. [DOI] [PubMed] [Google Scholar]
- 14.Kim S-S, Tam J K, Wang A-F, Hegde R S. J Biol Chem. 2000;275:31245–31254. doi: 10.1074/jbc.M004541200. [DOI] [PubMed] [Google Scholar]
- 15.Ferreiro D U, Mauricio L, Lima T R, Nadra A D, Alonso L G, Goldbaum F A, de Prat-Gay G. Biochemistry. 2000;39:14692–14701. doi: 10.1021/bi001694r. [DOI] [PubMed] [Google Scholar]
- 16.Li R, Knight J, Bream G, Stenlund A, Botchan M. Genes Dev. 1989;3:510–526. doi: 10.1101/gad.3.4.510. [DOI] [PubMed] [Google Scholar]
- 17.Hegde R S, Grossman S R, Laimins L A, Sigler P B. Nature (London) 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 18.Rozenberg H, Rabinovich D, Frolow F, Hegde R S, Shakked Z. Proc Natl Acad Sci USA. 1998;95:15194–15199. doi: 10.1073/pnas.95.26.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Rabinovich D, Rozenberg H, Shakked Z. Acta Crystallogr D. 1998;54:1336–1342. doi: 10.1107/s090744499800465x. [DOI] [PubMed] [Google Scholar]
- 21.Rabinovich D, Shakked Z. Acta Crystallogr A. 1984;40:195–200. [Google Scholar]
- 22.Chandrasekaran R, Arnott S. J Biomol Struct Dyn. 1996;13:1015–1027. doi: 10.1080/07391102.1996.10508916. [DOI] [PubMed] [Google Scholar]
- 23.Brünger A T, editor. X-PLOR. A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 24.Sheldrick G M, Schneider T R. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 25.Parkinson G, Vojtechovsky J, Clowney L, Brüunger A T, Berman H M. Acta Crystallogr D. 1996;52:57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- 26.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Berman H M. Biopolymers. 1997;44:23–44. doi: 10.1002/(SICI)1097-0282(1997)44:1<23::AID-BIP3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 28.Lavery R, Sklenar H. J Biomol Struct Dyn. 1992;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 29.Shui X, Sines C C, McFail-Isom L, VanDerveer D, Williams L D. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 30.Shui X, McFail-Isom L, Hu G H, Williams L D. Biochemistry. 1998;37:16877–16887. doi: 10.1021/bi982063o. [DOI] [PubMed] [Google Scholar]
- 31.Minasov G, Tereshko V, Egli M. J Mol Biol. 1999;291:83–99. doi: 10.1006/jmbi.1999.2934. [DOI] [PubMed] [Google Scholar]
- 32.Soler-Lopez M, Malinina L, Liu J, Huynh-Dinh T, Subirana J A. J Biol Chem. 1999;274:23683–23686. doi: 10.1074/jbc.274.34.23683. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Subirana J A. J Biol Chem. 1999;274:24749–24752. doi: 10.1074/jbc.274.35.24749. [DOI] [PubMed] [Google Scholar]
- 34.Johansson E, Parkinson G, Neidle S. J Mol Biol. 2000;300:551–561. doi: 10.1006/jmbi.2000.3907. [DOI] [PubMed] [Google Scholar]
- 35.Sines C C, McFail-Isom L, Howerton S B, VanDerveer D, Williams L D. J Am Chem Soc. 2000;122:11048–11056. [Google Scholar]
- 36.Dickerson R E, Drew H R. J Mol Biol. 1981;149:761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- 37.Dickerson R E. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X-J, Olson W K. J Mol Biol. 1999;285:1563–1575. doi: 10.1006/jmbi.1998.2390. [DOI] [PubMed] [Google Scholar]
- 39.Nikolov D B, Chen H, Halay E D, Hoffmann A, Roeder R G, Burley S K. Proc Natl Acad Sci USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice P A, Yang S-W, Mizuuchi K, Nash H A. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 41.Rodgers D W, Harrison S C. Structure (London) 1993;1:227–240. doi: 10.1016/0969-2126(93)90012-6. [DOI] [PubMed] [Google Scholar]
- 42.Albright R A, Matthews B A. Proc Natl Acad Sci USA. 1998;95:3431–3436. doi: 10.1073/pnas.95.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]