Abstract
Localization of mRNAs to specific destinations within a cell or an embryo is important for local control of protein expression. mRNA localization is well-known to function in very large and polarized cells such as neurons, and to facilitate embryonic patterning during early development. However, recent genome-wide studies have revealed that mRNA localization is more widely utilized than previously thought to control gene expression. Not only can transcripts be localized asymmetrically within the cytoplasm, they are often also localized to symmetrically-distributed organelles. Recent genetic, cytological, and biochemical studies have begun to provide molecular insight into how cells select RNAs for transport, move them to specific destinations, and control their translation. This review will summarize recent insights into the mechanisms and function of RNA localization with a specific emphasis on molecular insights into each step in the mRNA localization process.
1. Introduction
RNA localization is the process by which a cell or organism transports and anchors specific RNAs at predetermined locations. Studies in a variety of systems have shown that proper RNA localization is an important feature of early embryonic patterning, neuronal cell function and various aspects of cell motility and polarization (Martin and Ephrussi, 2009; St Johnston, 2005). Localization of mRNA serves several functions, all of which are used by various cells and organisms. First, localization of an mRNA, which can produce many copies of protein through repeated rounds of translation, can be more efficient than translating a protein at a random location and relying of transport or diffusion to localize it to the site of action. This rationale for localizing mRNAs is especially important in very large cells, such as neurons, oocytes, and embryos. Second, mRNA localization can be used to prevent the accumulation of protein in a place that would be harmful to the cell or organism because mRNA localization is tightly coupled to translational repression, such that mRNAs in transit are translationally repressed. Third, mRNA localization can allow a cell to respond quickly to a stimulus by activating translation of a localized transcript. This mode of cellular response is more rapid than relying on transducing a signal to the nucleus to mount a transcriptional response. Finally, an underappreciated function of localized RNAs is that they can serve as a scaffold for assembly of protein complexes and function in a manner independent of simple protein translation.
Classic studies of mRNA localization have highlighted the importance of localized mRNAs in oocytes/early embryos and highly polarized cells (such as neurons)(Du et al., 2007). In these large cell types, it is hypothesized that diffusion alone would not be sufficient to localize a protein to the site of action in a reasonable amount of time. These studies provide us with the greatest understanding of the function of mRNA localization and the mechanisms that act to target and transport mRNAs to their destinations. However, many recent studies have shown that mRNA localization is not restricted to large, highly specialized cells, but that it is a common feature of nearly all cell types. Furthermore, many genome-wide studies have begun to suggest that the majority of all mRNAs will have specific destinations (Blower et al., 2007; Du et al., 2008; Lecuyer et al., 2007; Sharp et al., 2011; Shepard et al., 2003) so that localization of mRNAs is the rule rather than a specialized process used by a few mRNAs in a few specialized cell types.
In general, all localized mRNAs require a common set of processes to occur. First, the mRNA must contain a cis acting sequence that targets the mRNA to a specific destination (commonly referred to as a ‘zipcode’(Jambhekar and Derisi, 2007)). Second, the zipcode must be recognized by RNA-binding proteins that serve to link the mRNA to the appropriate transport machinery. Third, the complex of RNA and RNA-binding proteins must be localized to the appropriate destination, either by diffusion or more commonly through active transport along the cytoskeleton. Fourth, the RNA must be anchored at the final destination. Finally, an interconnected theme is that localized mRNAs are most often translationally repressed during the process of transport and anchoring, so there must be a mechanism to release the mRNAs from the transport/anchoring complex to allow the mRNA to be translated at the final destination(Oleynikov and Singer, 1998). This review will briefly summarize the current state of the field for each of these areas, highlight recent studies that provide exciting new information about each of these topics and point the reader to more comprehensive reviews on each of the individual topics. One large class of specifically localized RNAs that will not be addressed in this review is noncoding RNAs that are retained in the nucleus. I point the reader to an excellent recent review on this topic(Wang and Chang, 2011).
2. Extent and locations of mRNA localization
Classic studies of mRNAs localization during oogenesis and embryogenesis have shown that mRNAs localize to specific places in oocytes and early embryos and that the localization of these mRNAs is important for establishing pattern formation during early development (Bashirullah et al., 1998). Another very prominent cell type that exhibits dramatic RNA localization are neurons, where several well-characterized mRNAs are known to localize to axons and function in neuronal signaling (Holt and Bullock, 2009; Martin and Ephrussi, 2009). These studies have been reviewed is detail elsewhere and will not be discussed in detail here. However, many of these important early studies lead to the view that localization of mRNAs was a niche process used only in very specialized cells. Over the past decade the application of genome-wide technologies has changed this view dramatically and suggests that the majority of the transcriptome is localized to a nonrandom position within the cell. Below I highlight some of the novel destinations that serve as sites of mRNA localization.
2.1 Endoplasmic reticulum
2.1.1 Co-translational Signal Sequence-dependent mRNA localization
The ER is a large, membrane-bound organelle that regulates protein secretion, aspects of calcium handling and lipid biosynthesis. The ER is also one of the major cellular sites of protein translation as most transmembrane and secreted proteins are translated on the surface of the ER. One of the best-understood mechanisms of mRNA localization involves the co-translational localization of mRNAs encoding N-terminal signal sequences (SS) to the endoplasmic reticulum (Walter and Blobel, 1981a, b; Walter et al., 1981). Many mRNAs encoding transmembrane, lumenal, and secreted proteins contain a N-terminal signal sequence. When these mRNAs are exported from the nucleus to the cytoplasm, translation initiates on cytoplasmic ribosomes. Translation of the signal sequence triggers binding of the Signal Recognition Particle (SRP) to the SS, which stalls translation and triggers localization of the mRNA:nascent peptide:ribosome complex to the signal recognition receptor on the ER surface (Gilmore et al., 1982). Translation then restarts and the newly synthesized peptide is co-translationally inserted into the lumen of the ER through the Sec61 channel. This is a classic case of mRNA localization to the site of action of the coded protein, and provides the paradigm for co-translational mRNA localization.
2.1.2 Translation-independent mRNA localization
The signal sequence hypothesis suggested that all mRNAs that are bound to the surface of the ER will contain an N-terminal signal sequence, will be bound to actively translating ribosomes, and localize to the ER in a translation-dependent manner. However, cotranslational localization of mRNA to the ER does not account for all mRNA localization to the ER(Adesnik et al., 1976). Additionally, many mRNAs that do contain signal sequences can localize to the ER in a translation-independent manner (de Jong et al., 2006; Diehn et al., 2000). Furthermore, mutation or RNAi of components of the SRP does not affect the viability or growth rate of any organism where it has been studied, suggesting that there are alternative methods to localize mRNAs to the ER membrane(Hann et al., 1989; Ren et al., 2004). Taken together, these studies have suggested that alternative methods exist to localize mRNAs to the ER. A recent genome-wide study used biochemical fractionation and sequential detergent extraction to demonstrate that there is a wide range of mRNA association with the ER, from mRNAs exclusively localized to the ER to mRNAs equally present in the ER and cytosol to mRNAs exclusively present in the cytosol (Chen et al., 2011). First, many mRNAs encoding cytosolic or nuclear proteins were associated with the ER, although the overall distribution of these types of mRNAs was biased towards the cytoplasmic fraction. Second, only mRNAs encoding secreted proteins showed the expected translation-dependent mRNA localization to the ER, while mRNAs encoding resident proteins of the endomembrane system showed a stable and translation-independent association with the ER(Chen et al., 2011), consistent with a FISH-based assay of mRNA localization to the ER (Cui et al., 2012). This study demonstrated that many mRNA-binding proteins are present on the surface of the ER, and that the p180 protein plays a role in translation-independent mRNA localization to the ER (Cui et al., 2012). Taken together, these studies demonstrate that there are multiple modes of mRNA association with the ER. mRNAs encoding selected protein components of the endomembrane system can localize independently of translation(possibly through the p180 protein or other unidentified receptors). mRNAs encoding proteins encoding secreted proteins localize in an SRP-dependent manner, and mRNAs encoding cytosolic proteins can localize in a currently undefined manner.
These studies have revealed an unexpected complexity in mRNA localization to the ER, but many important questions remain unanswered. First, what features in an mRNA direct translation-independent localization? Analysis of an ER-localized mRNA that contained both a signal sequence and transmembrane domain revealed that mutation of the reading frame preserves localization potential (Cui et al., 2012). These data suggest that mRNA localization information may be encoded in protein coding sequences. This principle would be similar to that found for the RNA sequence coding or the signal peptide, which directed mRNA export from the nucleus independently of the translated protein (Palazzo et al., 2007). Using the mRNA sequence that codes for a specific protein domain to localize the mRNA to the destination of the protein would be an efficient manner for the cell to encode two functionally coupled pieces of information into the same RNA sequence. While this is an attractive hypothesis with some experimental support, it will need to be tested directly. Another outstanding question is what proteins are involved in targeting mRNAs to the ER in a translation-independent manner. The p180 protein has been implicated in localization of a subset of mRNAs to the ER, but is not required for all mRNA localization. Furthermore, the RNA-binding domain in p180 shows no sequence preference (Cui et al., 2012), so it is unclear how specific mRNAs would be targeted to the ER. Elucidation of both the cis-acting RNA sequences and trans-acting protein factors will allow a greater understanding of translation-independent mRNA localization to the ER.
2.1.3 Localization of cytosolic mRNAs to the ER
As stated above, several studies have shown that mRNAs encoding cytoplasmic proteins copurify with the ER. There is essentially nothing known about the mechanism that drives these mRNAs to the ER, but a recent study examining the subcellular location of protein translation has provided a rationale for mRNA localization to the ER (Reid and Nicchitta, 2012). The recent development of ‘ribosome profiling’ to measure ribosome protected fragments on mRNAs as a proxy for translation rate has enabled the rapid, genome-wide analysis of translation in a variety of systems (Ingolia, 2010; Ingolia et al., 2009; Ingolia et al., 2011). A recent study using ribosome profiling combined with cell fractionation examined the rates of translation in the cytosol to that on the surface of the ER.(Reid and Nicchitta, 2012). Consistent with the studies discussed above, this study found that both signal sequence-containing and cytosolic mRNAs were actively translated on the ER. Interestingly, this study found that the density of ribosomes per mRNA was approximately twice as high on the ER as it was in the cytosol, suggesting that the surface of the ER is a more efficient environment for protein translation (Reid and Nicchitta, 2012). More efficient translation could provide a rationale for localization of mRNAs encoding cytoplasmic proteins to the ER. Although this study has provided an intriguing rationale for mRNA localization to the ER, the functional consequences or mechanism of localization are currently unknown. Important future questions will involve defining how mRNAs are recruited to the ER for translation, and the how translation on ER-bound ribosomes impacts protein function.
2.1.4 Links to asymmetric cytoplasmic mRNA localization pathways
In addition to coupling localization of secreted and endomembrane protein mRNAs to ER-bound ribosomes, localization of mRNAs to the ER has many links to asymmetric cytoplasmic mRNA localization pathways. In budding yeast, localization of the ASH1 mRNA to the bud tip on actin cables is coincident with movement of the ER (Schmid et al., 2006). In Xenopus oocytes, the mRNA binding protein Vera cofractionates with the ER and is important for the localization of the Vg1 mRNA to the vegetal cortex of the oocyte (Deshler et al., 1997). Furthermore, the Balbiani Body, which is the major site of mRNA localization in Xenopus, is rich in mitochondria and ER (Kloc et al., 2004). Finally, localization of seed storage protein mRNAs in rice endosperm cells is coupled to the ER (Choi et al., 2000). Taken together, these studies suggest that the ER may be one of the main sites of intracellular mRNA localization and that this organelle is likely to control many different processes in addition to the production of proteins of the endomembrane and secretory systems. Furthermore, the fact that many asymmetric cytoplasmic mRNA localization pathways are coupled to the ER suggests that understanding how and why particular mRNAs are localized to the ER will be key to understanding the many aspects of mRNA localization in a cell.
2.2 Mitochondria
Mitochondria are the cellular sites of respiration, energy production and some aspects of calcium handling. Mitochondria are unique organelles as they contain a fraction of the genetic information required for their maintenance, and must also import many proteins that are encoded by nuclear DNA (Ernster and Schatz, 1981). Import of proteins into the mitochondria was thought to occur posttranslationally, as proteins translated in vitro are capable of import into the mitochondria (Neupert and Herrmann, 2007). However, ribosomes are present on the surface of yeast mitochondria, suggesting that there may be cotranslational import of some proteins into the mitochondria (Kellems et al., 1974, 1975). Genome-wide studies of purified yeast mitochondria have demonstrated that several hundred mRNAs copurify with mitochondria-bound ribosomes (Gadir et al., 2011; Saint-Georges et al., 2008). Interestingly, the majority of the mitochondria-localized mRNAs code for proteins that function in the mitochondria, consistent with a role for co-translational protein import. Localization of a subset of these mitochondria-localized mRNAs has revealed that translation of the N-terminal mitochondrial targeting sequence (MTS), as well as specific RNA elements both in the coding regions and 3′UTRs are important for mRNA targeting to mitochondria (Corral-Debrinski et al., 2000).
Globally, the majority of mitochondria-localized transcripts depend on translation to target to the mitochondria, but as is the case with the ER, there is a class of mRNAs that localize independently of translation (Saint-Georges et al., 2008). Two studies have shown the Puf3 RNA-binding protein is important for localization of a subset of transcripts to the mitochondria: Puf3 associates primarily with transcripts that function at the mitochondria(Gerber et al., 2004) and is required for the localization of these transcripts to the organelle (Gadir et al., 2011; Saint-Georges et al., 2008). Interestingly, deletion of mitochondria targeting elements within the ATP2 mRNA resulted in defects in mitochondrial respiration and defects in protein import into the mitochondria, demonstrating that mRNA localization to mitochondria is critical for the proper function of the organelle (Margeot et al., 2002). In addition, a recent study of in Xenopus cultured neurons identified a nuclear protein, Lamin B2, as being translated in the axon. Surprisingly, the axonal pool of translated LB2 protein localized to mitochondria where it was required for proper mitochondrial function (Yoon et al., 2012). It will be interesting to determine how some messages localize independently of translation and what role those messages play in mitochondrial function.
2.3 Peroxisomes
Peroxisimes are membrane-bound organelles involved in oxygen scavenging and some aspects of lipid metabolism. Peroxisomes derive from budding of ER-derived vesicles and import of peroxisosomal proteins (Ma et al., 2011). A recent study examined the localization of many mRNAs encoding peroxisomal proteins in budding yeast and found that a subset of mRNAs are localized to the peroxisome (Zipor et al., 2009). The peroxisome-localized mRNAs included both structural peroxisomal proteins and proteins that undergo post-translational import into peroxisomes. Interestingly, a subset of peroxisomal mRNAs also localized to the ER, suggesting that the complex biogenesis of peroxisomes may also be reflected in complex patterns of mRNA localization. Similar to the localization of mRNAs to mitochondria, there is a contribution of a cis-acting zipcode that functions independently of protein translation to localize mRNAs to the peroxisome. For example, localization of the PEX14 mRNA was shown to be partially dependent on the RNA-binding protein Puf5. Surprisingly, localization of the PEX14 mRNA to peroxisomes was not required for protein function, as a partially localization-deficient construct was able to complement a null mutation of PEX14 (Zipor et al., 2009). Localization of mRNAs to peroxisomes appears to have many features in common with mRNA localization to mitochondria. Future work will be required to determine if there are functional consequences to disruption of peroxisimal mRNA localization and if this phenomenon is conserved in other species.
2.4 Endo/Lysosomes
Endosomes are formed by the invagination of the plasma membrane and are triggered by the activation of cell surface receptors (Hurley, 2008). Endosomes control the sorting of activated cell surface receptors either to the plasma membrane for further use or to the lysosome for degradation. Several studies have found an unexpected link between lysosomes and RNA regulation. For example, two recent studies have linked components of the MultiVesicular Body (MVB) biogenesis pathway and RNAi-mediated gene repression (Gibbings et al., 2009; Lee et al., 2009). Both studies found that components of the RNAi machinery associate with endosomes/MVBs and that depleting components of the endosome machinery impair the ability of the cell to silence transcripts containing miRNA target sequences. Although it was not examined directly, these results strongly suggest that mRNAs targeted for repression will be localized to endosomes/MVBs. Another study found that components of endosomes colocalize with markers of cytoplasmic P-bodies (Gibbings et al., 2009), which are thought to be sites of mRNA storage or degradation. Taken together, these studies demonstrate that endosomal membranes are sites of mRNA storage and processing for degradation.
The link of RNAs to endosomes has several exciting implications. First, mRNAs and miRNAs have recently been found to be excreted from cells in microvesicles/exosomes (Skog et al., 2008; Valadi et al., 2007), and it was recently demonstrated that the core components of the machinery required for MVB formation are also required for the formation of exosomes (Baietti et al., 2012). Therefore, RNA-binding proteins present on endosomes/MVBs could play a role in determining which RNAs are selected for inclusion into exosomes and secretion outside of the cell. Another exciting possibility is that mRNAs linked to endosomes could couple activation of a cell surface receptor to a translational response. One study found that stimulation of neuronal cells with nerve growth factor triggered axonal translation of the transcriptional-activator CREB. Newly synthesized CREB was then transported to the nucleus on endosomes and was important for the transcriptional response to the stimulus (Cox et al., 2008). This suggests that localization of the CREB mRNA to distal axons, perhaps in the context of endosomes, could serve to poise the mRNA to increase axon-localized translation in response to activation of an extracellular receptor.
Another interesting link of endosomes to mRNA transport has come from studies of two more established systems of mRNA localization, localization of the bicoid mRNA in Drosophila oocytes, and transport of mRNAs in the hyphae of Ustilago maydis (Baumann et al., 2012; Irion and St Johnston, 2007). A visual genetic screen for mutants that influence the localization of the bicoid mRNA identified a component of the ESCRT-II complex, a complex that is part of the machinery required for MVB formation, as being required for bicoid mRNA localization (Irion and St Johnston, 2007). The authors further demonstrated that all members of ESCRT-II are required for bicoid localization, while components of ESCRT-I and ESCRT-III are not required for this process. This suggests that ESCRT-II has a role in RNA localization that is independent of its role in MVB formation. The filamentous fungus U. maydis represents a very different system of RNA transport, where mRNAs are transported bidirectionally in the hyphae through the action of both dynein and kinesin motors, yet endosomes were similarly required for mRNA localization. The majority of mRNA transport is mediated by the Rrm4 protein (Becht et al., 2006). A recent study demonstrated that motile mRNAs colocalize with shuttling endosomes and that endosome biogenesis is required for efficient mRNA movement (Baumann et al., 2012). These results are similar to the observation that the Drosophila Rab11 protein, which is a marker for recycling endosomes, colocalizes with the oskar RNA at the posterior pole of oocytes (Dollar et al., 2002). Furthermore, the genomic RNAs of retroviruses are localized to endosomes en route to the plasma membrane for viral budding (Basyuk et al., 2003).
Taken together, these studies suggest that endosomes are likely to be major sites of mRNA localization and control. Linkage of endosomes to the RNAi pathway suggests that endosomes are likely to be sites of storage for translationally-repressed mRNAs. Since endosomes have many well-documented connections to cytoskeletal transport pathways (Scita and Di Fiore, 2010), the cell may use the well-established endosome transport pathways to move mRNAs along with endosomes. The fact that endosomes are the site of activated cell surface receptors provides the cell with a mechanism to link the translation of localized mRNAs to extracellular signals. Finally, the link of endosomes/MVBs with exosomes suggests that the cell may also use endosomes to compartmentalize specific mRNAs into exosomes for the purpose of cell-to cell communication.
2.5 Actin cytoskeleton
Although the actin cytoskeleton is not a membrane-bound organelle such as those previously discussed, it is a prominent site of mRNA localization. Indeed, the mRNA encoding chicken β-actin mRNA was shown to localize to cell protrusions and to the leading edge of motile fibroblasts (Lawrence and Singer, 1986). Furthermore, impairment of β-actin mRNA localization was shown to interfere with the speed and directionality of cell motility (Kislauskis et al., 1994, 1997), which are actin-dependent processes. Several recent studies have augmented our understanding of mRNA localization to the actin cytoskeleton. One hypothesis for the importance of mRNA localization is that co-translation of all members of a multiprotein complex could facilitate the assembly of that complex. This hypothesis was confirmed for the actin-nucleating Arp2/3 complex, as the mRNAs for all members of the complex localize to cell protrusions (Mingle et al., 2005). Interestingly, knockdown of one member of the complex resulted in changes in the speed and direction of cell migration. These changes in motility could be rescued by a properly localized mRNA, but not completely restored by an mRNA that was targeted to another cellular compartment, demonstrating that the localization of one of the mRNAs of the Arp2/3 complex is important for the function of the complex (Liao et al., 2011). In addition, a recent genome-wide study of mRNAs associated with actin-rich cell protrusions demonstrated that ~50 mRNAs were enriched in these protrusions and that a subset of these mRNAs were localized to the cell protrusions in a microtubule-dependent manner that required the APC protein (Mili et al., 2008). Altogether, studies of actin-localized mRNAs suggest that mRNAs present on the actin cytoskeleton are likely to be involved in control of actin dynamics and other processes related to cell motility.
2.6 Microtubule cytoskeleton
Microtubules are used for the majority of long-distance transport within cells. Microtubules are important for the localization of the majority of known localized mRNAs (with the notable exception of localized mRNAs in Saccharomyces cerevisiae, which require actin-based transport) through the action of various motor proteins. However, several studies suggest that microtubules are not just a highway for transporting mRNAs, but can also serve a destination for some localized transcripts (Martin and Ephrussi, 2009). Purification of microtubules from a variety of organisms (sea urchin, frog, human) has demonstrated that hundreds of mRNAs co-purify with microtubules (Blower et al., 2007; Hamill et al., 1994; Rodriguez et al., 2005; Sharp et al., 2011; Suprenant, 1993; Suprenant et al., 1993; Suprenant et al., 1989). The majority of microtubule-localized mRNAs code for proteins that appear to function in cell cycle and microtubule-related processes. Our group recently tested this idea directly by knocking down the mRNAs for 10 uncharacterized microtubule-localized transcripts in HeLa cells. We found that the majority of these mRNAs coded for proteins required for proper microtubule organization during both interphase and mitosis (Sharp et al., 2011). Work in Xenopus oocytes has also found that both spindle localization and translational control of a subset of mRNAs are important for completion of both meiotic divisions, highlighting the link between mRNA localization and translational control (Eliscovich et al., 2008).
In addition to containing mRNAs that control the microtubule cytoskeleton, microtubules also contain a number of transcripts that appear to be translationally repressed and are likely to be passive cargo on the spindle rather than active participants in controlling microtubule-related events (Blower et al., 2007). This idea is consistent with recent work looking at RNA localization in early cell divisions of the snail Ilyanassa obsoleta (Lambert and Nagy, 2002; Rabinowitz and Lambert, 2010). In this work, the authors used RNA FISH to examine several mRNAs important for early development. Some of these transcripts show an unequal distribution during cell division by selectively associating with one of the two centrosomes of a dividing blastomere. This suggests that unequal segregation of important developmental mRNAs by the mitotic spindle could be an important mechanism for the establishment of early patterning asymmetries. While many mRNAs are known to localize to meiotic and mitotic microtubules, little is known about the sequences or proteins involved in this process. There is also nothing known about the functional consequences of disrupting mRNA localization to microtubules. These will all be important future areas of research.
2.7 Summary
Taken together, there is an enormous diversity in the destinations of localized mRNAs. However, several common themes are evident. First, mRNAs tend to localize to the site of action of the coded protein. Second, many types of mRNA localization appear to require a combination of cis-acting RNA sequences and co-translational mRNA targeting. Finally, intracellular membranes are one of the most common destinations for localized mRNAs, suggesting that the cell may use well-established membrane sorting machinery to shuttle mRNAs to different destinations within the cell.
3. Mechanisms of mRNA localization
3.1 Cis-acting mRNA localization signals
A basic principle of mRNA localization is that some mRNAs contain sequence elements, termed ‘zipcodes’, that are recognized by specific protein factors that link them to the transport machinery. Zipcodes can range from a few nucleotides (5–6) to hundreds of base pairs. With a few notable exceptions, there is no consensus sequence for an RNA zipcode. Further, localization activity of an mRNA can be conferred by many low specificity sequence elements acting in concert. There are several excellent reviews covering the state of the field of cis-acting sequence elements (Jambhekar and Derisi, 2007) and trans-acting proteins (Martin and Ephrussi, 2009; St Johnston, 2005), so I will not go into any detail on these topics here, but will highlight some recent studies that have revealed unexpected complexity in types of zipcodes.
3.1.1 Splicing dependent localization of oskar
For the majority of localized mRNAs examined to date, all of the information necessary to recapitulate localization is present in the RNA sequence. This is demonstrated by the ability of fluorescently-labeled, in vitro synthesized mRNAs to localize to the correct destination when injected into the appropriate system. However, there are several notable exceptions, and one of the best-understood examples is the oskar RNA in Drosophila oogenesis. Initial work on localization of oskar mapped the localization activity of the transcript to the 3′UTR and showed that these sequences were sufficient to localize a transcript when fused to a reporter RNA (Gunkel et al., 1998; Kim-Ha et al., 1993). However, when the oskar 3′UTR was assayed for localization activity in a mRNA null mutant, it was not sufficient to confer full localization activity (Hachet and Ephrussi, 2004), which led the authors to propose that the 3′ UTR localized by “hitchhiking” with endogenous transcripts (presumably through 3′UTR mediated dimerization of oskar mRNA molecules). Mutagenesis experiments revealed a requirement for splicing of the first oskar intron in concert with the 3′UTR to confer full localization activity, demonstrating that the nuclear history of a transcript can be important for the localization of the RNA in the cytoplasm. A recent study from the Ephrussi lab has now provided a very detailed mechanistic explanation for the requirement for mRNA splicing (Ghosh et al., 2012). Splicing of the first intron creates a stem-loop structure that is absolutely required for oskar localization, but is not sufficient to localize intronless versions of the RNA, indicating that other factors are required for localization. Previous genetic evidence had indicated that components of the Exon Junction Complex (EJC) were required for oskar localization, and using an in vitro system, it was shown that all the components of the EJC are deposited just upstream of the first exon:exon junction created by splicing. These results suggested that formation of the stem-loop must work in concert with deposition of the EJC to effect localization. Taken together these studies suggest that some transcripts will require a very complex series of processing events, and that sequential assembly of trans-acting factors may be required to achieve proper mRNA localization.
3.1.2 Dimerization dependent mRNA localization
While most mRNA localization signals function entirely in cis by recruiting specific proteins to promote localization, another class of zipcodes promote intermolecular mRNA dimerization to achieve RNA localization. The Drosophila bicoid mRNA is one of the best-characterized localized mRNAs and is known to localize to the anterior of the Drosophila oocyte in a microtubule-dependent manner. The localization of bicoid is dependent on a large section of the 3′UTR that is predicted to form a series of stem-loop structures and long regions of double-stranded RNA (dsRNA; (MacDonald, 1990; Macdonald and Kerr, 1998; Macdonald et al., 1993; Macdonald and Struhl, 1988). Localization of bicoid is dependent on a number of proteins including the dsRNA-binding protein Staufen (Ferrandon et al., 1994; St Johnston et al., 1991). While studying the interactions of bicoid with Staufen, it was observed that unpaired loops on the bicoid 3′UTR were important for localization, and that these loops had the potential to pair in trans with other bicoid molecules (Ferrandon et al., 1997). Mutation of bases in the loops abolished localization, but could be restored by complementary mutations in other bicoid molecules. Bicoid also demonstrated the ability to dimerize in vitro, and dimerization ability correlated with localization activity in vivo. Although the dimerization ability of bicoid RNA is important for recruitment of Staufen in embryos and for localization of biocid mRNA to the apical side of early embryos and to embryonic mitotic spindles, it is not required for localization to the anterior of the Drosophila oocyte (Snee et al., 2005). Another recent study has also highlighted the use of RNA dimerization motifs to promote RNA localization. Oskar RNA (described above) is known to be packaged into large RNA granules containing many molecules of oskar RNA (Glotzer et al., 1997);, and as described above, the 3′UTR of oskar RNA can hitchhike to the posterior of the oocyte using the endogenous transcript. To determine if oskar hitchhiking and oligimerization requires RNA:RNA base pairing, the authors investigated the base pairing of oskar in vitro (Jambor et al., 2011). They found that oskar was able to form oligomers in vitro that depended on unpaired bases present in a loop in the 3′UTR, similar to the mechanism used by bicoid. The authors then demonstrated that pairing between different oskar molecules is necessary for the hitchhiking behavior of the oskar 3′UTR. Interestingly, mutation of bases involved in trans pairing did not affect the localization of a full length oskar transcript, demonstrating that dimerization is not absolutely necessary for localization in vivo. While the localization of neither oskar nor bicoid is dependent on dimerization activity, it is possible that dimerization facilitates packaging into higher order particles for more efficient transport or translational control. Another interesting possibility is that pairing of other (as yet unknown) RNAs with the dimerization elements in bicoid or oskar could facilitate their localization within the oocyte..
3.2 Assembly of active RNA localization complexes
In order for an RNA to localize to the correct destination, it must contain the correct cis-acting sequence elements that are processed in a defined temporal and spatial manner as discussed above. In addition to processing of the zipcodes, proteins must be loaded onto the localizing RNA to facilitate assembly of a localization-competent RNP. While studies in a number of genetically-tractable model systems, most notably S. cerevisiae and D. melanogaster, have provided lists of proteins required for the localization of specific mRNAs, a clearly defined understanding of how a localization competent RNP is formed has been lacking. In addition, when the interaction of genetically-identified localization factors with mRNAs is examined in vitro, it has been difficult to reconstitute the exquisite specificity of the interaction (Dienstbier et al., 2009; Du et al., 2008). A biochemical and structural study of the assembly of the ASH1 localization complex has begun to provide the first clear picture of how a series of modest affinity and specificity interactions can act synergistically to promote the high affinity and high specificity interactions observed in vivo (Muller et al., 2011).
In S. cerevisiae, the ASH1 mRNA is transported to the bud tip of the daughter cell through myosin-driven transport along actin cables. Once in the daughter cell, ASH1 represses mating-type switching in the daughter cell. Studies from a number of labs have found that ASH1 contains a dispersed localization sequence consisting of four elements, E1, E2A, E2B and E3 that are implicated in transport of the mRNA. In vivo, the E3 element can mediate efficient transport, suggesting that this some of the localization sequences serve redundant functions (Heym and Niessing, 2012). A series of proteins have also been implicated in mRNA transport: She2 an, a RNA-binding protein that contacts ASH1 in the nucleus; She3, an adaptor for myosin driven transport; and Myo4, the motor for transport(Heym and Niessing, 2012). Additionally, the RNA-binding proteins Puf6, Khd1 and Loc1 are part of the localization complex and required for efficient ASH1 transport, but their roles are less well-defined than She2, She3 and Myo4. Although a nearly complete set of players was known for ASH1 mRNA localization it was not clear until recently how these proteins functioned together for generate a high specificity RNP.
To understand how She2, She3, Puf6 interact with each other and the ASH1 mRNA localization element to generate a localization complex, the interactions of purified proteins with the ASH1 E3 RNA sequence were examined in vitro (Muller et al., 2011). Consistent with previous results, She2 interacted directly with the ASH1 E3 element (Niessing et al., 2004), but only showed a modest preference for ASH1 E3 compared to nonlocalizing RNAs (~2–10-fold higher affinity than for localizing RNAs). In contrast, the Puf6 protein bound RNAs with a modest affinity and specificity, showing little preference for RNAs containing a Puf6 consensus binding sequence. When assayed in tandem, She2 and Puf6 exhibited no synergism for RNA binding, but were able to form a trimeric complex mediated by RNA as a scaffold. In contrast, She2 and She3 interact directly with one another and also with RNA to form a stoichiometric complex. Although there was no previous indication that She3 was an RNA-binding protein, it bound RNA with a modest affinity and no specificity in vitro. When the affinity and specificity of She2 and She3 were assayed with ASH1 localization elements, it revealed that the complex of She2/She3 has a dramatically higher affinity for RNA containing zipcodes compared to control RNAs This demonstrates conclusively that high affinity and specific recognition of a zipcode requires the concerted action of two low affinity RNA-binding proteins, which is likely to be a paradigm relevant to many other zipcode-binding proteins. Consistent with the previous results, when the She3 surfaces that interact with both She2 and ASH1 RNA were identified and mutated in vivo, they disrupted RNA transport of ASH1 mRNA. This study leads to the first nearly complete mechanistic model for how a transport-competent RNP is formed. First, ASH1 is transcribed in the nucleus where it is bound by She2, Puf6, and Loc1. Following export to the cytoplasm, the interaction of She2 and She3 with each other and with the zipcode drives mRNP remodeling to form a very stable, highly specific mRNP that is competent to interact with Myo4 and be transported to the bud neck. Two important ideas emerge from this study. First, it will be important to obtain nearly complete lists of the factors that interact with localizing mRNAs to understand mRNP complex assembly, and it will be important to use in vitro studies to understand how each factor interacts with the others to promote high affinity and specific zipcode binding. Second, the identification of She3 as a direct RNA binding protein suggests that the proteins that interact directly with localizing mRNAs will not be limited to proteins that have well-known and annotated RNA-binding domains, but will likely encompass a much wider range of proteins. This idea has been confirmed by two recent studies using poly-A purification to identify proteins that interact directly with mRNAs. Both studies found that hundreds of proteins that have no known RNA-binding domains directly contact mRNAs (Baltz et al., 2012; Castello et al., 2012). It will be interesting to extend this type of analysis to localized mRNAs.
4. Motor-driven directed transport of localization competent mRNPs
In order to achieve localization of an mRNA to a distinct cytoplasmic location, an assembled RNP is generally transported along the cytoskeleton by molecular motors. While there are some examples of mRNA localization through diffusion and entrapment or selective protection against degradation, these are the exceptions and most examples of mRNA localization are driven by active transport along the cytoskeleton (Martin and Ephrussi, 2009). Several questions about motor-driven mRNA transport have been addressed by recent studies. First, is the direction of transport determined by the polarity of the cytoskeleton? Second, is the directionality of transport determined by the number and type of motor proteins associated with the RNP? What is the fate of a transport particle and associated motor protein once it has reached its destination?
4.1 Role of cytoskeletal polarity in mRNA localization
In many cell types that utilize mRNA localization, there is a well-defined polarity to the cytoskeleton that facilitates transport of mRNPs to one end of the cytoskeletal filaments. For example, in neuronal cells, the centrosome is located in the cell body near the nucleus, and the microtubule plus ends extend into the axons and dendrites. Thus, kinesin motor proteins are used for transport of cargo to the distal portions of the cell (anteriograde transport) and dynein motors are used to transport cargo back to the cell body (retrograde transport). Another system with a well-defined cytoskeletal polarity is the early Drosophila embryo, where the centrosome is located at the surface of the embryo and the plus ends of microtubules extend into the interior of the embryo. In this system, dynein motors are used to transport mRNP cargo to the surface of the embryo (Gaspar, 2011). However, many systems that utilize mRNA localization do not have a defined polarity of the cytoskeleton, which raises the question as to how directional transport is achieved on a nonpolarized cytoskeleton.
While the Drosophila oocyte is a classic system for the study of mRNA localization it is also a system that has a very poorly defined cytoskeletal polarity (Cooley and Theurkauf, 1994). During stages 8–10 of oogenesis, when many mRNAs are known to localize, the oocyte has a gradient of microtubules with the majority of microtubules nucleated from the anterior of the oocyte. Consistent with this idea, transport of mRNAs from the nurse cells to the oocyte is driven by the dynein motor and associated factors (Gaspar, 2011). An open question in the field of RNA transport has been to determine how transcripts that localize to the posterior of the oocyte achieve this localization. Mutation of the kinesin heavy chain results in a loss of oskar mRNA from the posterior (Brendza et al., 2002; Cha et al., 2002), supporting the idea that oskar localization is driven by kinesin-mediated transport on a polarized cytoskeleton from the anterior to the posterior of the oocyte. However, direct measurement of oskar transport had not been observed until recently.
Movement of oskar localization particles tracked by live imaging revealed a small portion (~15 %) of oskar transcripts moving in a manner consistent with motor driven transport. Interestingly, oskar particles appeared to be moving in all different directions rather than showing a concerted, directed transport to the posterior of the oocyte. However, analysis of hundreds of particles revealed that there was a weak bias in the directionality of transport, with ~57% of all transport particles moving towards the posterior of the oocyte. Furthermore, these movements were almost entirely towards the plus ends of microtubules and were dependent on the action of kinesin. Various mutants known to affect the localization of oskar were found to affect different steps of the localization process, with some proteins involved in establishing the net directionality of transport and others involved in affecting the rate. The conclusion from this study is that over time, an accumulation of mRNA can occur through the use of very small differences in the net direction of transport on a weakly polarized cytoskeleton. The results from this study will likely be applicable to transport of other mRNAs in the Drosophila oocyte as well as RNA transport in other oocyte systems. A notable example of another oocyte system that has a very poorly defined cytoskeletal organization is the Xenopus oocyte. Although many mRNAs are transported to each hemisphere of these oocytes two studies using different methodologies have argued for very different polarity of the oocyte cytoskeleton(Messitt et al., 2008; Pfeiffer and Gard, 1999), which will impact models of how directional transport of mRNAs is achieved. Live imaging of specific mRNAs will likely be required to help define how transport operates in this system.
4.2 Influence of transport complex composition on transport direction
In the simplest case, a localized mRNA will become incorporated into a transport complex that is linked to one molecular motor and will localize to its destination on a cytoskeleton with a well-defined polarity. While this is the case for some localized mRNAs (such as ASH1), localization of an mRNA is usually much more complicated, as illustrated above. A recent study examining the composition and function of the ASH1 localization complex has discovered some unexpected complexity in the assembly of the complex in vivo (Chung and Takizawa, 2010). As discussed above, localization of ASH1 depends on She2, She3 and Myo4. However, Myo4 is a nonprocessive motor protein and would not be expected to mediate transport of a cargo as a monomer (Reck-Peterson et al., 2001). Purification of a transport competent ASH1 mRNP revealed that the transport complex contained four copies of Myo4 and She2, suggesting a role for protein (or RNA) oligomerization in assembly of a transport competent mRNP and providing a molecular explanation for the processive movement of the ASH1 mRNP. Interestingly, using an artificial reporter system that could recruit a defined number of Myo4 motors to a RNA, demonstrated that recruitment of more motor proteins leads to a greater efficiency of mRNA transport, a theme that is consistent with the results discussed below.
As mentioned in the preceeding section, one of the great challenges to understanding the specificity of mRNA transport is that the exact composition and biochemical activity of most transport complexes is not known. Reconstitution of mRNA binding complexes has provided a major insight into recognition of mRNA zipcodes. However, assembly of full transport complexes is considerably more complicated as it requires the inclusion of molecular motors, which are technically challenging to work with in vitro. In Drosophila embryos, certain transcripts (e.g. hairy, fushi tarazu, K10) localize to the apical centrosome of the embryo, and this behavior can be recapitulated by fluorescently labeled transcripts injected into the embryo (Bullock and Ish-Horowicz, 2001). Analysis of injected wild-type and localization-deficient transcripts in vivo demonstrated that both classes of transcripts exhibit fast bidirectional movement along microtubules, consistent with motor-driven transport(Bullock et al., 2006; Bullock et al., 2003). However, localization-competent RNAs exhibited more frequent and longer movements to the minus ends of microtubules, which may result in the net accumulation of the localized mRNA at the minus ends of microtubules (Bullock et al., 2006). In addition, mutation or overexpression of the localization factors Egalitarian and BicaudalD led to slower or faster RNA transport respectively, suggesting that recruitment of a different number of motor proteins could be responsible for the different relative transport activities (Bullock et al., 2006). However, in these experiments it was not possible to directly visualize the motor proteins involved, and as a result it was not possible to prove that differences in motor number account for differences in motility.
A recent study using partially purified mRNPs from Drosophila embryos has begun to provide insight into how directional transport of mRNPs is achieved (Amrute-Nayak and Bullock, 2012). From the above results, it was possible that recruitment of more molecules of dynein or an increased activity of dynein could result in the increased minus end directed processivity observed in vivo. In a clever experiment to study the behavior of localization-competent mRNPs at a single molecule level, mRNAs were incubated with Drosophila embryo extract, selected for microtubule binding, and then analyzed at a single molecule level using TIRF microscopy. Remarkably, behavior of single molecules of the localization-competent and incompetent versions of the K10 RNA in vitro was consistent with movement of the RNAs in vivo. Both RNAs exhibited fast movement to both the plus and minus ends of microtubules, with the localization competent RNA exhibiting an increased frequency and longer runs toward the minus ends. Furthermore, GFP-tagged dynein or a subunit of the dynein accessory dynactin complex was co-visualized with fluorescent mRNA in vitro. Sequential photobleaching was used to measure the number of motors associated with each transport particle and was found to differ by ~50% between a localization-competent RNA and a localization-incompetent RNA. These data confirm the idea that recruitment of more motor proteins is responsible for increased processivity towards the minus end.
Another open question is to determine how many mRNA molecules are present in each mRNP transport particle. Work on oskar localization complexes (Glotzer et al., 1997) and purification of kinesin associated RNPs from neuronal cells (Kanai et al., 2004) suggests that many molecules of RNA may be present in localization complexes, but this has not been tested rigorously. Using sequential photobleaching, the authors showed that each transport complex contains only one molecule of mRNA (Amrute-Nayak and Bullock, 2012). Furthermore, this was confirmed by FISH in Drosophila embryos for two mRNAs that both localize to the apical side of the embryo: h and eve mRNAs do not colocalize, suggesting that transport particles only contain single mRNA molecules. This result is consistent with a recent FISH study in neurons demonstrating little colocalization between different mRNAs in transport complexes (Mikl et al., 2011). Taken together, the single molecule studies of K10 mRNA localization complexes demonstrate that localization complexes can consist of a single mRNA that recruits multiple motor proteins. Interestingly, localization-incompetent mRNAs can also recruit motor proteins, and the role of the localization sequence is to increase the recruitment of one particular type of motor protein to achieve a bias in the directionality of transport. This demonstrates that very subtle changes in protein complex composition can result in dramatic localization changes in vivo. It will be important to extend these studies to determine what other motors are recruited to the RNP (kinesin?) and how many copies of each of the accessory proteins are recruited. A long-term goal will be to understand the molecular mechanism by which a localization element results in the recruitment of additional copies of motor proteins to an mRNA.
4.3 RNA anchoring
Once a transported RNA reaches its destination, the RNA must be anchored in order to retain the localized distribution. In many organisms that localize mRNAs via transport along the microtubule cytoskeleton, the function of RNA anchoring is served by the actin cytoskeleton. However, we understand very little about the molecular mechanisms that transfer an RNA from the microtubule cytoskeleton to actin cytoskeleton. Another mechanism of RNA anchoring is to change the activity of the motor proteins involved in the transport RNP from active motors to static anchors. Two studies of RNA localization in Drosophila have provided clear evidence for this mechanism and begun to shed light on the molecular details of the process (Delanoue and Davis, 2005; Delanoue et al., 2007).
As described above in several previous sections, one of the major types of mRNA transport in Drosophila embryos is the movement of mRNAs involved in developmental pattern formation to the apical centrosome. Movement of these mRNAs occurs in a dynein-dependent manner and requires Egl and BicD for transport. However, little was known about the mechanism of RNA anchoring at the centrosome. It was shown that disruption of the actin cytoskeleton had no effect on RNA transport or anchoring, while disruption of microtubules dramatically delocalized the transported RNA (Delanoue and Davis, 2005). Interestingly, dynein was shown to colocalize with localized, anchored RNAs. In addition, interference with dynein function through a blocking antibody completely disrupted RNA that was already anchored at the cortex. Surprisingly, inhibition of dynein motor activity, but not the dynein complex, blocked active transport but retained RNA anchoring. These results show that incorporation into an RNP can change dynein from an active motor protein to a static anchor once it has reached its destination at the microtubule minus ends. A follow-up study examining the role of dynein in localization of the gurken RNA in the Drosophila oocyte reached a similar conclusion (Delanoue et al., 2007). Interestingly, mutation of the Squid protein, an hnRNP, results in an inability of dynein to change to a static anchor at the destination. However, the molecular details of how mRNPs can change the motor activity of dynein remain unknown. One interesting implication from these studies is that mRNA transport complexes do not necessarily function solely as passive cargo on the cytoskeleton, but have the potential to feedback and change the activity of factors that influence the cytoskeleton, a theme that will be discussed in the last section of this review.
5. Formation and structure of RNA granules
One of the most distinctive features of localized RNAs is their ability to form cytologically visible cytoplasmic structures termed RNA granules (Anderson and Kedersha, 2006). RNA granules are most obviously visible in the germ line of most model organisms (P-granules in C. elegans, polar granules in Drosophila, and germinal granules in Xenopus). These granules are composed of RNAs and RNA-binding proteins that are known to control the formation of germ cells in the developing embryos and were one of the first recognized sites of mRNA localization. Germ Cell Granules (GCG) share many similarities with other types of RNA granules found in somatic cells, such as stress granules and P-granules (Anderson and Kedersha, 2006). Stress granules are large cytoplasmic aggregates of mRNAs and RNA-binding proteins that accumulate upon cellular stress (e.g. heat shock or arsenite treatment) and are thought to function as sites of mRNA storage during cellular response to stress. P-bodies are also visible sites of mRNAs and RNA-binding proteins, but because of the protein composition of P-bodies they are linked to RNA decapping, deadenylation and RNA destruction (Balagopal and Parker, 2009). One common feature of all RNA granules is that they are higher order assemblies of many RNAs and RNA-binding proteins that appear to function in temporary or long term storage of mRNAs. Although RNA granules are visible cytoplasmic structures very little was known about the mechanisms of formation or functions of these structures until recently.
5.1 Mechanisms of RNA granule formation
Insight into the physical properties and formation of RNA granules has come from careful analysis of the composition and behavior of C. elegans P-granules. In an important study, live cell imaging was used to examine the behavior of the C. elegans P-granule component PGL-1 in the gonad and early embryo (Brangwynne et al., 2009). Interestingly, PGL-1 exhibited behavior consistent with a liquid droplet, such as dissolution, condensation and various flow behaviors. Measurement of the viscosity of P-granules suggested that they have a viscosity comparable to glycerol (~1000 times that of water) and colloidal liquids. One of the most interesting properties is regulated dissolution and condensation. At high component concentrations, PGL-1 condensed to form P-granules, but when the effective protein concentration was lowered the granules dissolved. These results suggested that weak interactions between P-granule components might be important for their formation.
The molecular interactions that lead to the liquid-like behavior of P-granules was addressed by two recent studies that explored the molecular requirements for the formation of P-granules (Updike et al., 2011; Updike and Strome, 2009; Voronina and Seydoux, 2010). Both studies made the important observation that several proteins present in P-granules (the Vasa related proteins GHL-1, GHL-2 and GLH-3) contain repeats of the amino acids phenylalanine and glycine, and these were required in GLH-1 for granule formation (Updike et al., 2011). This was informative because FG-repeats are one of the prominent features of the proteins that line the central channel of the nuclear pore (Weis, 2003). It has been hypothesized that weak interactions between the FG-containing nucleoporins are responsible for creating a selective hydrogel-like barrier between the nucleus and the cytoplasm. In fact, it was recently demonstrated that very concentrated solutions of FG-repeats from the yeast nucleoporin Nsp1p form a solid hydrogel through hydrophobic interactions (Frey et al., 2006), which allows only proteins with appropriate characteristics to pass through (Frey and Gorlich, 2007). Although nucleoporins exhibit hydrogel-like properties at high concentrations, while P-granules exhibit liquid-like properties in vivo, the properties of homotypic interaction and selective permeability appeared to suggest a common mechanism. Consistent with a link between the nuclear pore and P-granules, P-granules exhibited a size exclusion of fluorescent dextrans, similar to the nuclear pore (Updike et al., 2011). In addition, RNAi against a subset of nuclear pore proteins disrupted P-granule formation (Voronina and Seydoux, 2010). Furthermore, GFP-fusions of several nuclear pore proteins colocalized with P-granules, and mutation of the C. elegans Nup98 homolog disrupted P-granule formation. Interestingly, the mouse Nup98 also interacted with the mouse Vasa homolog in mouse testes extract, suggesting that nucleoporins and nucleoporin-like interactions may be important for GCG formation in other systems(Voronina and Seydoux, 2010). Taken together, these results suggest that GCG exhibit many properties of nuclear pores and that regulated, homotypic interactions between FG-repeats are important for granule coalessence.
Biochemical insight into the mechanism of RNA granule formation recently came from two studies analyzing a compound causing cellular differentiation (Han et al., 2012; Kato et al., 2012). While analyzing proteins that bound to a differentiation-inducing drug, the authors noticed that the addition of this compound to cellular extracts caused the aggregation and precipitation of many cellular proteins. Identification of the precipitated proteins revealed that a substantial fraction were known RNA-binding proteins. Furthermore, analysis of the coprecipitated RNAs from cellular extracts identified a large number of mRNAs known to exhibit distinct subcellular localizations, suggesting that regulated protein aggregation may have a physiological role. Mapping the aggregation-promoting activity of precipitated RNA-binding proteins identified low complexity (LC) sequence domains that were both necessary and sufficient for protein aggregation. The low complexity regions contained repeats of tyrosine, serine and glycine, similar to the FG repeats found in nuclear pore proteins. Also similar to the FG-repeats found in nucleoporins, the LC domains of RNA-binding proteins were able to form hydrogels in very concentrated solutions in vitro. Interestingly, hydrogels could recruit RNAs as fusion proteins and from cellular extracts, demonstrating that LC-based sequence aggregation can be used to concentrate RNA and RNA-binding proteins into granules. Importantly, mutation of serine residues within the LC sequences blocked hydrogel formation and also blocked localization of the proteins to stress granules formed in tissue culture cells (Kato et al., 2012). Analysis of the interaction of LC regions from various RNA binding proteins revealed that homotypic interactions were favored, but that there were also differing degrees of heterotypic affinities between LC regions from different proteins. This gradient of affinities could suggest a possible mechanism for generating RNA granules of mixed compositions, containing different RNAs and RNA-binding proteins.
While these studies were preformed primarily in vitro, the parallels with P-granule formation suggest a compelling model for RNA granule formation. RNA binding proteins bound to target RNAs interact with one another via homotypic interactions between repetitive low complexity sequences. These interactions are promoted by high local protein concentrations that might be facilitated by transport of many copies of RNPs to a specific destination. Once the proteins are aggregated, they are relatively static and likely prevent translation of the RNAs present within the granules. Importantly, RNPs can be released from the granules either by lowering effective protein concentration, a mechanism known to occur in C. elegans (Brangwynne et al., 2009), or by phosphorylation of the self-association domains (Han et al., 2012). While this is a compelling model that fits with in vivo evidence from many different model systems, these ideas will all need to be tested in vivo.
5.2 Function of RNA granules
While the vast majority of evidence from a variety of model systems supports the model that RNA granules are sites of mRNA storage, several studies of the cytoplasmic polyadenylation binding protein (CPEB) from Aplysia californicia suggest that sites of RNA granules could also be sites of high enzymatic activity that promote protein translation. CPEB proteins are proteins that recognize the Cytoplasmic Polyadenylation Element (CPE) in the 3′UTR of mRNAs under posttranscriptional control (Richter, 2007). CPEB proteins are implicated in both repression of mRNAs and activation of the mRNA through extension of poly-A tail length in response to a signaling cue (Pique et al., 2008). In Aplysia, the CPEB protein contains an unusual N-terminal extension that has some similarity to prion forming domains from yeast (Si et al., 2003). The N-terminus of CPEB behaves as a prion in yeast and appears to form amyloid-like aggregates. Interestingly, aggregation of Aplysia CPEB is required for full CPEB activity when expressed in yeast, and blocking prion-like aggregates when expressed in neurons interferes with synaptic function (Si et al., 2010). These results suggest that there is a role for regulated aggregation in promoting the CPEB-mediated cytoplasmic polyadenylation (Si et al., 2003). It will be important to determine if regulated aggregation of other RNA-binding proteins plays a role in the activation of other RNA processing enzymes, or if the majority of RNA granules are sites of RNA storage. Interestingly, amyloid-like aggregation of other RNA-binding proteins has been observed, but in these cases it has been associated with loss of function of the aggregated proteins. These data suggest that amyloid-like aggregation of RNA-binding proteins could contribute to neurodegenerative diseases (Sun et al., 2011). In the future, it will be important to distinguish between regulated aggregation of RNA-binding proteins, such as that observed in vitro and in C. elegans P-granules, and the irreversible, prion-like protein aggregation that could result in deleterious consequences to the organism.
6. Translational regulation of localized mRNAs
Once an mRNA has been localized and anchored at a specific destination, it needs to be activated for translation in response to an external, internal or developmental signaling cue. Translational control of localized mRNAs has recently been discussed in an excellent and comprehensive review (Besse and Ephrussi, 2008) and will not be covered here in detail. However, I will summarize one study to illustrate one of the best-understood examples of translational control of a localized mRNA.
One of the first examples of a localized mRNA is the β-actin transcript from chick fibroblasts. The transcript was shown to localize to the leading edge of motile fibroblasts and the localization activity mapped to a small region of the 3′UTR termed the zipcode. The zipcode of β-actin mRNA is bound by the ZBP-1 protein that is required for mRNA localization. Interference with mRNA localization has been shown to affect cell migration, but it was not understood how the interaction of the zipcode with ZBP-1 controlled β-actin translation (Condeelis and Singer, 2005). ZBP-1 acts as a zipcode-dependent translational repressor in vitro in rabbit reticulocyte lysate and in vivo in neuronal cells (Huttelmaier et al., 2005). RNA interference.
Interestingly, ZBP-1 contains a tyrosine residue near one of the RNA-binding domains that resides within a consensus sequence for the Src protein kinase, and ZBP-1 is in fact phosphorylated by Src in vitro and in vivo. Phosphorylation of this residue reduced binding of ZBP-1 to the β-actin transcript, demonstrating that phosphorylation by Src can serve as a signal to trigger translational activation. Consistent with this model, knockdown of ZBP-1 in neuronal cell culture resulted in a decrease in neuronal outgrowth, which could be rescued by wild type ZBP-1, but not by a non-phosphorylatable ZBP-1 variant. Taken together, these results suggest that ZBP-1 associates with β-actin transcripts in the nucleus during transcription, and remains bound to the transcript during transport in the cytoplasm. Once in the cytoplasm, ZBP-1 suppresses the translation of β-actin mRNA until a signal to release the RNA is received in the form of phosphorylation by the Src tyrosine kinase. Importantly, both premature translation induced by ZBP-1 RNAi and failure to relieve translational inhibition result in failures of neuronal outgrowth. This study demonstrates that temporal and spatial coupling of mRNA localization to translational control is critical for the function of the encoded protein (Huttelmaier et al., 2005). The paradigm of signaling-induced mRNA release is likely to be applicable to other RNA-binding proteins, and also fits well with the observation that phosphorylation of serine residues in LC regions can release RNA-binding proteins from RNA granules. It will be important to understand the signaling pathways and proteins that contribute to release of RNA from mRNP transport complexes to fully understand local translational control.
7. Noncoding roles of localized RNAs
This review has been based on the idea that most localized RNAs are mRNAs and that the primary function of restricting the localization of the mRNA is to control the temporal and spatial expression pattern of the protein. However, there is increasing evidence that some localized mRNAs function as RNA rather than or in addition to serving as a template for protein production. This topic has recently been reviewed (Kloc et al., 2011) and will be discussed briefly to highlight the additional possible roles of localized RNAs.
One of the first hints for non-protein coding roles for localized RNAs came from the characterization of the Xlsirst (Xenopus laevis short interspaced repeats) family of RNAs in Xenopus. Xlsirst are a family of ~80 bp repeats that are highly transcribed during oogenesis. Interestingly, sense strand versions of this repeat are localized to the Balbiani Body and vegetal cortex of the oocyte in pattern nearly indistinguishable form other well-characterized localized mRNAs (e.g. Vg1 and Xcat2; (Kloc et al., 1993). When the function of the Xlsirts was explored by removal using antisense deoxyoligonucleotides (ODN) it was found that Xlsirts were required for the proper localization of the Vg1 transcript to the vegetal cortex of the oocyte (Kloc and Etkin, 1994). Since Xlsirts are a tandemly repeated RNAs that lack protein coding capacity this was a direct demonstration that localized, noncoding RNAs could serve a function, in this case to promote the localization of a protein-coding RNA. The idea that localized RNAs can function as RNA rather than as a protein gained further support from study of the Xenopus localized mRNA VegT (Heasman et al., 2001). VegT is another mRNA that is localized to the vegetal cortex of the oocyte and encodes a T-box transcription factor important for development (Zhang et al., 1998). Interestingly, destruction of the VegT RNA by injection of ODN resulted in delocalization of a number of mRNAs from the vegetal cortex (including Vg1), but blocking protein production from the mRNA did not result in the same phenotype. These data demonstrate that in addition to Xlsirts, the VegT mRNA also plays a role in promoting mRNA localization, independently of protein coding capacity.
Insight into how Xlsirts and VegT could function to promote mRNA localization has come from a careful analysis of the cytoskeleton in oocytes. In Xenopus, many mRNAs that are localized to the vegetal cortex are anchored at the cortex by association with either the actin or keratin cytoskeletons. Destruction of Xlsirt and VegT revealed that the cortical cytokeratin cytoskeleton was affected, while actin remained unaffected(Kloc et al., 2005). Consistent with this result, both mRNAs localized to the vicinity of cytokeratin filaments, suggesting that they may act directly to promote proper keratin assembly. Thus, RNAs can act to influence mRNA localization through modulation of cytoskeletal structure.
Additional support for the idea of a structural role for RNA in cytoskeletal assembly has come from study of the mitotic and meiotic spindle. Using Xenopus laevis egg extracts to study meiosis II spindles, it was demonstrated that proper assembly of the spindle was dependent on the presence of RNA (Blower et al., 2005). RNaseA treatment of extracts resulted in dramatic changes to spindle microtubule organization, while inhibition of translation using a variety of translation inhibitors did not change spindle morphology, demonstrating that RNA plays a structural role in microtubule assembly. Interestingly, immunodepletion of Rae1, a protein component of an RNA complex, also resulted in dramatic defects in spindle assembly in vitro, perhaps providing a starting point to understand how RNA can promote spindle assembly. The structural role of RNA in spindles assembly has also been confirmed in a permeabilized cell assay using human cells (Hussain et al., 2009), suggesting that a structural role for RNA in mediating microtubule assembly is likely to be conserved in many organisms.
Although these studies suggest that localized RNAs will play direct, protein coding-independent roles, the molecular details of these processes remain unclear. In the case of Xlsirts and VegT, we know which RNAs act to promote cytokeratin assembly, but we do not know how these RNAs exert their effects, which proteins they bind, or how they affect cytokeratin assembly and maintenance. In the case of the role of RNA in microtubule assembly: we know only that RNA is required for spindle assembly, but we do not know which RNAs promote microtubule assembly or whether proteins other than the Rae1 complex are involved. In both cases it will be important to understand the RNAs that are required for each of these events, and the RNA-binding proteins that are affected. This knowledge will need to be integrated with an understanding of how the RNA-binding proteins affect cytoskeletal dynamics.
One final example of a protein coding-independent role of a localized RNA comes from study of the oskar RNA in Drosophila. During Drosphila embryogeneis, the oskar protein is required for proper segmentation patterning. However, the classic genetic alleles of oskar were loss of protein alleles, but did not lead to loss of the mRNA. Analysis of new alleles of oskar that completely lacked mRNA production showed stronger phenotypes, with -null animals not producing eggs and exhibiting an early oogenesis arrest (Jenny et al., 2006). Interestingly, this oogenesis arrest could be rescued by a protein null allele, or by the oskar 3′ UTR, suggesting that the oskar mRNA plays a translation-independent role early in oogenesis. There is currently no understanding of how the oskar 3′UTR functions to promote early oogenesis in the absence of protein.
Taken together, these studies support the notion that localization of RNAs (both coding and noncoding) can serve functions in addition to the well-documented spatial and temporal control of protein synthesis. Given the recent discovery of thousands of noncoding RNAs (Wang and Chang, 2011) and the fact that both mRNAs and ncRNAs can play structural roles, it seems likely that many new functions of localized RNAs that are independent of protein coding capacity are yet to be discovered.
8. Perspective
As detailed above, the localization of an mRNA to a specific destination within a cell or embryo is a widely utilized mechanism for the spatial and temporal control of protein expression. Mechanistically, it is an extremely complicated process involving cotranscriptional processing, linkage to transport complexes, anchoring at a destination and ultimately relief of translational inhibition. While genetics and cytology in several prominent model organisms have provided a basic picture of how mRNA localization is regulated many open questions remain. Below I have highlighted several interesting questions that will be addressed in the future.
8.1 Cis-acting localization sequences
As described in the preceding section, many individual cis-acting sequences have been described that direct mRNAs to specific destinations. However, many of the localization sequences that have been discovered are large and poorly defined. In addition, there are very few cases where there is obvious homology between defined localization sequences, which has dramatically limited understanding the logic of zipcodes that sort mRNAs within a cell. A technical aspect that has hampered the field is that mapping of localization elements is a labor intensive process that is generally accomplished one mRNA at a time. In order to achieve better predictive ability for defining zipcodes computationally, we will need much larger datasets of localization elements. Given the recent advances in identification of protein binding sites within RNAs by high throughput sequencing (Darnell, 2010) it is possible that these technologies can be applied to localized mRNAs to identify a much larger set of zipcodes. This, coupled with improved methods of RNA structure determination and structural homology prediction (Parisien and Major, 2008) may lead to dramatic increases in our ability to detect and predict mRNA localization elements.
8.2 Biochemical reconstitution of mRNA localization complexes
Genetic and cell biological studies in a wide range of model organisms have provided a list of localized RNAs and proteins that are likely to interact with these RNAs, however we lack a detailed understanding of the molecular interactions that select certain RNAs from transport and the mechanisms that achieve selective transport. The past year has seen several studies that have begun to provide biochemical insight into both RNA recognition and biased directional transport. Future studies will likely continue to try to understand how combinations of low specificity interactions lead to high specificity RNA binding and how subtle biases in transport complex composition lead to dramatic biases in RNA distribution in a cell or embryo. Finally, the ability to reconstitute certain aspects of RNA granule formation in vitro should provide the ability to understand the regulated self-association of RNA-binding proteins that will provide a platform for exploring the functional implications of these interactions for RNA transport, translational control and RNA anchoring in vivo.
8.3 Noncoding functions of localized RNAs
Recent studies of the transcriptomes of a number of organisms have discovered hundreds to thousands of RNAs with little to no protein coding capacity (Ulitsky et al., 2011). Current research has focused on the hypothesis that many/most of these noncoding RNAs will regulate nuclear processes (Wang and Chang, 2011), although there has been no systematic study of the localization of these RNAs. Given the studies of RNAs such as Xlsirts it seems likely that many ncRNAs will have cytoplasmic functions and many may function through localization to specific compartments of the cell. Future studies will likely focus on which ncRNAs are localized to specific compartments within the cell and how they function. It will be important to understand which proteins bind to ncRNAs and how RNA-binding changes the activity of the bound protein or complex. Given that some mRNAs also appear to function in a translation-independent manner it will also be important to differentiate between RNA and protein-coding functions of localized mRNAs.
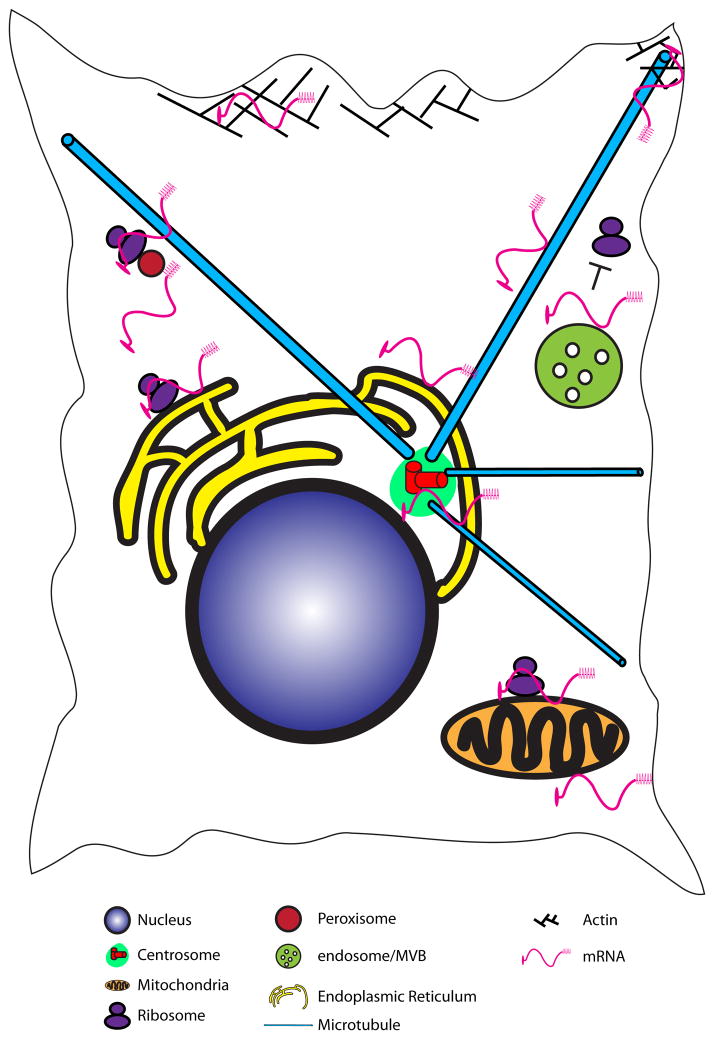
Figure 1. Sites of intracellular mRNA localization.
Cartoon of a typical eukaryotic cell depicting sites with identified localized mRNAs. Translation-dependent localization is depicted as a mRNA associated with a ribosome at a destination, while translation-independent localization is depicted as mRNA localization in the absence of a ribosome.
Acknowledgments
The author would like to thank Judith Sharp and Ashwini Jambhekar for comments on the manuscript. Work on RNA localization in my lab is supported by a grant from the NIH (5 R01 GM086434-05) and from a Career Award in the Biomedical Science from the Burroughs Wellcome Fund.
References
- Adesnik M, Lande M, Martin T, Sabatini DD. Retention of mRNA on the endoplasmic reticulum membranes after in vivo disassembly of polysomes by an inhibitor of initiation. J Cell Biol. 1976;71 (1):307–313. doi: 10.1083/jcb.71.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol. 2012;14 (4):416–423. doi: 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172 (6):803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14 (7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21 (3):403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46 (5):674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Basyuk E, Galli T, Mougel M, Blanchard JM, Sitbon M, Bertrand E. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev Cell. 2003;5 (1):161–174. doi: 10.1016/s1534-5807(03)00188-6. [DOI] [PubMed] [Google Scholar]
- Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrugge M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci. 2012;125 (Pt 11):2740–2752. doi: 10.1242/jcs.101212. [DOI] [PubMed] [Google Scholar]
- Becht P, Konig J, Feldbrugge M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci. 2006;119 (Pt 23):4964–4973. doi: 10.1242/jcs.03287. [DOI] [PubMed] [Google Scholar]
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9 (12):971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179 (7):1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121 (2):223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324 (5935):1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Saxton WM, Duffy JB. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr Biol. 2002;12 (17):1541–1545. doi: 10.1016/s0960-9822(02)01108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414 (6864):611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16 (14):1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22 (10):2484–2494. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell. 2012;149 (6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cha BJ, Serbus LR, Koppetsch BS, Theurkauf WE. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat Cell Biol. 2002;4 (8):592–598. doi: 10.1038/ncb832. [DOI] [PubMed] [Google Scholar]
- Chen Q, Jagannathan S, Reid DW, Zheng T, Nicchitta CV. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol Biol Cell. 2011;22 (14):2646–2658. doi: 10.1091/mbc.E11-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407 (6805):765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- Chung S, Takizawa PA. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae. J Cell Biol. 2010;189 (4):755–767. doi: 10.1083/jcb.200912011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell. 2005;97 (1):97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- Cooley L, Theurkauf WE. Cytoskeletal functions during Drosophila oogenesis. Science. 1994;266 (5185):590–596. doi: 10.1126/science.7939713. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Blugeon C, Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol. 2000;20 (21):7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10 (2):149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XA, Zhang H, Palazzo AF. p180 Promotes the Ribosome-Independent Localization of a Subset of mRNA to the Endoplasmic Reticulum. PLoS Biol. 2012;10 (5):e1001336. doi: 10.1371/journal.pbio.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1 (2):266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M, van Breukelen B, Wittink FR, Menke FL, Weisbeek PJ, Van den Ackerveken G. Membrane-associated transcripts in Arabidopsis; their isolation and characterization by DNA microarray analysis and bioinformatics. Plant J. 2006;46 (4):708–721. doi: 10.1111/j.1365-313X.2006.02724.x. [DOI] [PubMed] [Google Scholar]
- Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122 (1):97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell. 2007;13 (4):523–538. doi: 10.1016/j.devcel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276 (5315):1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet. 2000;25 (1):58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23 (13):1546–1558. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollar G, Struckhoff E, Michaud J, Cohen RS. Rab11 polarization of the Drosophila oocyte: a novel link between membrane trafficking, microtubule organization, and oskar mRNA localization and translation. Development. 2002;129 (2):517–526. doi: 10.1242/dev.129.2.517. [DOI] [PubMed] [Google Scholar]
- Du TG, Jellbauer S, Muller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9 (8):781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du TG, Schmid M, Jansen RP. Why cells move messages: the biological functions of mRNA localization. Semin Cell Dev Biol. 2007;18 (2):171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Eliscovich C, Peset I, Vernos I, Mendez R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat Cell Biol. 2008;10 (7):858–865. doi: 10.1038/ncb1746. [DOI] [PubMed] [Google Scholar]
- Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol. 1981;91 (3 Pt 2):227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79 (7):1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16 (7):1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130 (3):512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314 (5800):815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 2011;17 (8):1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar I. Microtubule-based motor-mediated mRNA localization in Drosophila oocytes and embryos. Biochem Soc Trans. 2011;39 (5):1197–1201. doi: 10.1042/BST0391197. [DOI] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2 (3):E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Marchand V, Gaspar I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol. 2012;19 (4):441–449. doi: 10.1038/nsmb.2257. [DOI] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11 (9):1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95 (2 Pt 1):463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer JB, Saffrich R, Glotzer M, Ephrussi A. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr Biol. 1997;7 (5):326–337. doi: 10.1016/s0960-9822(06)00156-4. [DOI] [PubMed] [Google Scholar]
- Gunkel N, Yano T, Markussen FH, Olsen LC, Ephrussi A. Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes Dev. 1998;12 (11):1652–1664. doi: 10.1101/gad.12.11.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428 (6986):959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Hamill D, Davis J, Drawbridge J, Suprenant KA. Polyribosome targeting to microtubules: enrichment of specific mRNAs in a reconstituted microtubule preparation from sea urchin embryos. J Cell Biol. 1994;127 (4):973–984. doi: 10.1083/jcb.127.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149 (4):768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hann BC, Poritz MA, Walter P. Saccharomyces cerevisiae and Schizosaccharomyces pombe contain a homologue to the 54-kD subunit of the signal recognition particle that in S. cerevisiae is essential for growth. J Cell Biol. 1989;109 (6 Pt 2):3223–3230. doi: 10.1083/jcb.109.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Wessely O, Langland R, Craig EJ, Kessler DS. Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Dev Biol. 2001;240 (2):377–386. doi: 10.1006/dbio.2001.0495. [DOI] [PubMed] [Google Scholar]
- Heym RG, Niessing D. Principles of mRNA transport in yeast. Cell Mol Life Sci. 2012;69 (11):1843–1853. doi: 10.1007/s00018-011-0902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326 (5957):1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20 (1):4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186 (1):27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438 (7067):512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324 (5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147 (4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, St Johnston D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445 (7127):554–558. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13 (5):625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Brunel C, Ephrussi A. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA. 2011;17 (12):2049–2057. doi: 10.1261/rna.2686411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Hachet O, Zavorszky P, Cyrklaff A, Weston MD, Johnston DS, Erdelyi M, Ephrussi A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133 (15):2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43 (4):513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149 (4):753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J Biol Chem. 1974;249 (10):3297–3303. [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol. 1975;65 (1):1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119 (1):169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127 (2):441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. beta-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136 (6):1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Etkin LD. The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol. 2004;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Delocalization of Vg1 mRNA from the vegetal cortex in Xenopus oocytes after destruction of Xlsirt RNA. Science. 1994;265 (5175):1101–1103. doi: 10.1126/science.7520603. [DOI] [PubMed] [Google Scholar]
- Kloc M, Foreman V, Reddy SA. Binary function of mRNA. Biochimie. 2011;93 (11):1955–1961. doi: 10.1016/j.biochi.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Kloc M, Spohr G, Etkin LD. Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science. 1993;262 (5140):1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132 (15):3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420 (6916):682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45 (3):407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131 (1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, Nakahara K, Preall JB, Bellare P, Sontheimer EJ, Carthew RW. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11 (9):1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Simone B, Liu G. Mis-localization of Arp2 mRNA impairs persistence of directional cell migration. Exp Cell Res. 2011;317 (6):812–822. doi: 10.1016/j.yexcr.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193 (1):7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald PM. bicoid mRNA localization signal: phylogenetic conservation of function and RNA secondary structure. Development. 1990;110 (1):161–171. doi: 10.1242/dev.110.1.161. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Kerr K. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol Cell Biol. 1998;18 (7):3788–3795. doi: 10.1128/mcb.18.7.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Kerr K, Smith JL, Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118 (4):1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Struhl G. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336 (6199):595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- Margeot A, Blugeon C, Sylvestre J, Vialette S, Jacq C, Corral-Debrinski M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21 (24):6893–6904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136 (4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15 (3):426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikl M, Vendra G, Kiebler MA. Independent localization of MAP2, CaMKIIalpha and beta-actin RNAs in low copy numbers. EMBO Rep. 2011;12 (10):1077–1084. doi: 10.1038/embor.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453 (7191):115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci. 2005;118 (Pt 11):2425–2433. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, Urlaub H, Jansen RP, Niessing D. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 2011;9 (4):e1000611. doi: 10.1371/journal.pbio.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Niessing D, Huttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 2004;119 (4):491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8 (10):381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Springer M, Shibata Y, Lee CS, Dias AP, Rapoport TA. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5 (12):e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452 (7183):51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- Pfeiffer DC, Gard DL. Microtubules in Xenopus oocytes are oriented with their minus-ends towards the cortex. Cell Motil Cytoskeleton. 1999;44 (1):34–43. doi: 10.1002/(SICI)1097-0169(199909)44:1<34::AID-CM3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132 (3):434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JS, Lambert JD. Spiralian quartet developmental potential is regulated by specific localization elements that mediate asymmetric RNA segregation. Development. 2010;137 (23):4039–4049. doi: 10.1242/dev.055269. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J Cell Biol. 2001;153 (5):1121–1126. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287 (8):5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell. 2004;15 (11):5064–5074. doi: 10.1091/mbc.E04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32 (6):279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, MDIC, Suprenant KA, Bonder EM. Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 2005;11 (5):646–656. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3 (6):e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jaedicke A, Du TG, Jansen RP. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr Biol. 2006;16 (15):1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463 (7280):464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- Sharp JA, Plant JJ, Ohsumi TK, Borowsky M, Blower MD. Functional analysis of the microtubule-interacting transcriptome. Mol Biol Cell. 2011;22 (22):4312–4323. doi: 10.1091/mbc.E11-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A. 2003;100 (20):11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140 (3):421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115 (7):879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10 (12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee MJ, Arn EA, Bullock SL, Macdonald PM. Recognition of the bcd mRNA localization signal in Drosophila embryos and ovaries. Mol Cell Biol. 2005;25 (4):1501–1510. doi: 10.1128/MCB.25.4.1501-1510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6 (5):363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66 (1):51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9 (4):e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suprenant KA. Microtubules, ribosomes, and RNA: evidence for cytoplasmic localization and translational regulation. Cell Motil Cytoskeleton. 1993;25 (1):1–9. doi: 10.1002/cm.970250102. [DOI] [PubMed] [Google Scholar]
- Suprenant KA, Dean K, McKee J, Hake S. EMAP, an echinoderm microtubule-associated protein found in microtubule-ribosome complexes. J Cell Sci. 1993;104 (2):445–450. doi: 10.1242/jcs.104.2.445. [DOI] [PubMed] [Google Scholar]
- Suprenant KA, Tempero LB, Hammer LE. Association of ribosomes with in vitro assembled microtubules. Cell Motil Cytoskeleton. 1989;14 (3):401–415. doi: 10.1002/cm.970140310. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147 (7):1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Hachey SJ, Kreher J, Strome S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol. 2011;192 (6):939–948. doi: 10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Strome S. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics. 2009;183 (4):1397–1419. doi: 10.1534/genetics.109.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9 (6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Voronina E, Seydoux G. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development. 2010;137 (9):1441–1450. doi: 10.1242/dev.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981a;91 (2 Pt 1):557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981b;91 (2 Pt 1):551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91 (2 Pt 1):545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43 (6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112 (4):441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148 (4):752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94 (4):515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2009;106 (47):19848–19853. doi: 10.1073/pnas.0910754106. [DOI] [PMC free article] [PubMed] [Google Scholar]