Abstract
Three short hydrophobic loops and a conserved undecapeptide at the tip of domain 4 (D4) of the cholesterol-dependent cytolysins (CDCs) mediate the binding of the CDC monomers to cholesterol-rich cell membranes. But intermedilysin (ILY), from Streptococcus intermedius, does not bind to cholesterol-rich membranes unless they contain the human protein CD59. This observation suggested that the D4 loops, which include loops L1–L3 and the undecapeptide, of ILY were no longer required for its cell binding. However, we show here that membrane insertion of the D4 loops is required for the cytolysis by ILY. Receptor binding triggers changes in the structure of ILY that are necessary for oligomerization, but membrane insertion of the D4 loops is critical for oligomer assembly and pore formation. Defects that prevent membrane insertion of the undecapeptide also block assembly of the prepore oligomer, while defects in the membrane insertion of the L1–L3 loops prevent the conversion of the prepore oligomer to the pore complex. These studies reveal that pore formation by ILY, and probably other CDCs, is affected by an intricate and coupled sequence of interactions between domain 4 and the membrane.
The cholesterol-dependent cytolysins (CDCs)2 are poreforming toxins that are widely distributed among Gram-positive pathogens and have significant similarity in their primary sequences (reviewed in Ref. (1)). The crystal structure of two family members, perfringolysin O (PFO) (secreted by Clostridium perfringens) and intermedilysin (ILY) (secreted by Streptococcus intermedius) revealed that the three-dimensional structures are also well conserved (2,3). Their structural similarity is also reflected in a significant degree of functional similarity. The CDCs share a common mechanism in which soluble monomers bind to the membrane and oligomerize into a prepore complex (4–6) of 34–50 monomers (7–9), which is then converted to the β-barrel pore complex by the cooperative membrane insertion (10) of two amphipathic β-hairpins per monomer (11, 12).
The tip of the CDC domain that first interacts with the membrane is domain 4 (D4) (6). D4 contains four loops, the conserved undecapeptide and three short hydrophobic loops (L1, L2, L3) (Fig. 1). Both the undecapeptide (13,14) and the L1–L3 loops (15) have been shown to insert into the membrane surface, thereby anchoring the toxin in a perpendicular orientation (16) and initiating the structural changes that lead to oligomerization and pore-formation (15). Since mutations within the undecapeptide usually interfere with membrane binding (14, 17–22), it is generally accepted that the undecapeptide plays an important role in mediating the interaction of the CDCs with the cholesterol-rich membrane surface. Sekino-Suzuki et al. (20) also showed that mutation of the conserved tryptophan residues in the undecapeptide significantly decreased its affinity for cholesterol-rich erythrocyte membranes. Interestingly, even though the monomers bound at a decreased rate, they still formed the typical doughnut-shaped oligomeric complexes. However, these complexes did not form pores. Instead, these mutants appeared to be trapped in the prepore structure that forms prior to the insertion of the transmembrane β-barrel pore (4, 5,13). These results suggested that the functional role of the undecapeptide is complex. Yet it was not possible to isolate and study specific contributions of the undecapeptide to various stages of the CDC pore-forming mechanism because modifications of the undecapeptide sequence typically also affected the binding of most CDCs to the membrane.
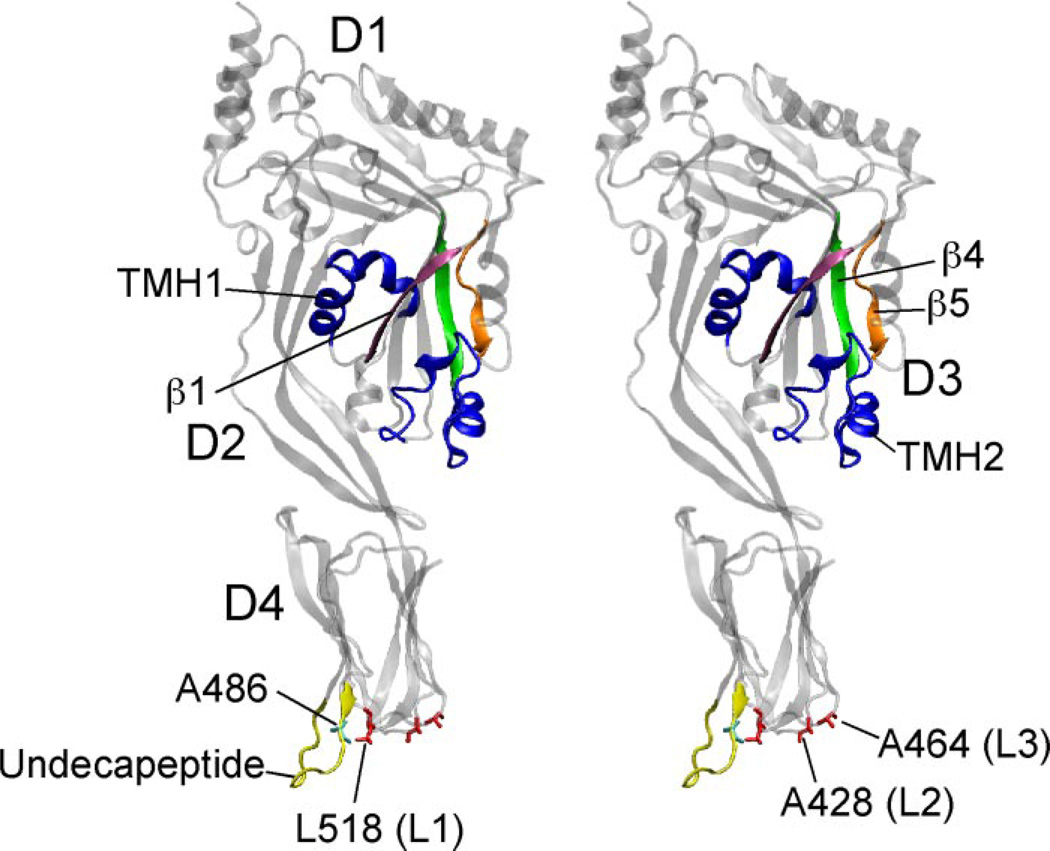
FIGURE 1. Crystal Structure of ILY.
Shown is side-by-side stereographic ribbon representation of the crystal structure of ILY (2). Shown are locations of the D3 transmembrane β-hairpins (blue), β-strands β1, β4, and β5 (magenta, green, and orange, respectively). Also shown are the D4 L1, L2, and L3 loop residues Leu-518, Ala-428, and Ala-464 (respectively) (red), the undecapeptide loop (yellow), and residue Ala-486 in the undecapeptide (cyan). D1–D4, domains 1–4. The ribbon structures were generated using Virtual Molecular Display (31).
ILY shares most of the above properties with other members of the CDC family but differs from those CDCs in two respects: first, ILY is specific for human cells because it binds only to human CD59 (huCD59) (23); second, ILY contains a variant undecapeptide (GATGLAWEPWR) (compare to the consensus sequence (ECTGLAWEWWR)). The evolution of ILY specificity for huCD59 required at least two structural changes: first, structural alterations in the undecapeptide were necessary to prevent the binding of ILY directly to cholesterol-rich membranes; second, mutations in D4, but not the undecapeptide, were necessary to provide the binding site for huCD59 (2, 23–25). These observations suggested the ability of ILY to bind huCD59 functionally replaced the role of the undecapeptide and D4 loops in membrane binding and anchoring the toxin to the cell surface.
Are the D4 loops and undecapeptide of ILY still important to the cytolytic mechanism of ILY? Apparently yes, since Pole-khina et al. (2) have shown that mutations within the ILY undecapeptide have a dramatic affect on the cytolytic mechanism of ILY. Furthermore, mutation of ILY Trp-491 to alanine resulted in a mutant that was defective in prepore to pore conversion. This observation suggested the ILY undecapeptide is important for pore formation, even though it is not required for the binding of ILY to the cell surface. In this study we have performed a detailed examination of the contributions of the D4 loops to the cytolytic mechanism of ILY. Our studies revealed that the D4 loops of ILY are critical for pore formation and that each polypeptide segment is required for the proper progression of ILY through specific stages of oligomer assembly and pore formation.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Chemicals
The gene for ILY was cloned into pTrcHisA (Invitrogen) as described previously (25). ILYH242C (cysteine-containing derivative) is similar in activity to native ILY but contains a single engineered cysteine at residue His-242, which is used for site-specific labeling with various sulfhydryl-specific probes. All other mutations that were introduced into ILY were made in the cysteine-less native ILY background unless otherwise stated. All chemicals and enzymes were obtained from Sigma, VWR, and Research Organics except where noted. All fluorescent probes were obtained from Molecular Probes (Invitrogen).
Generation and Purification of ILY and Its Derivatives
Various amino acid substitutions in the gene for ILY were generated by PCR QuikChange mutagenesis (Stratagene) and each derivative sequenced (Oklahoma Medical Research Foundation Core DNA sequencing facility). The expression and purification of recombinant His-tagged ILY and its derivatives from Escherichia coli were carried out as described (12) with the following differences. The crude protein lysate was loaded onto a cobalt loaded metal chelate column (GE Healthcare) and washed with 30 ml of buffer A (10 mM MES, 150 mM NaCl, pH 6.5). The column was then washed with a 20–120 mM gradient of imidazole (in buffer A) in a total volume of 40 ml. The histi-dine-tagged protein was eluted from the column with 36 ml of buffer A containing 0.5 m imidazole. The cationic exchange step was not performed. The eluted protein was dialyzed into buffer (300 mM NaCl, 10 mM MES, 1 mM EDTA, pH 6.5) overnight at 4 °C. The toxin was then stored in 5 mM dithiothreitol and 10% (v/v) sterile glycerol at −80 °C until used.
To determine whether the mutants displayed any structural changes due to the mutations, each was subjected to trypsin digestion and the proteolytic cleavage pattern compared with that of wild type ILY. Wild type and mutant ILY (90 pmol) were incubated with serial 10-fold dilutions from 800 pM to 80 fM trypsin for 30 min at 37 °C. At the end of the incubation the digest was denatured by the addition of SDS-PAGE sample buffer and heating to 100 °C for 5 min. The samples were then separated on a 10% SDS-PAGE gel and stained with Coomassie R-250. Peptide cleavage patterns were then compared with wild type. All mutants used in the present study exhibited identical cleavage patterns to native ILY.
Hemolytic Activity
The hemolytic activity of each labeled or unlabeled ILY derivative on human red blood cells (hRBCs) was determined as described previously (12). The HD50 is defined as the concentration of toxin required to lyse 50% of the hRBCs.
Modification of ILY with Fluorescent Probes
The labeling of ILY cysteine-containing mutants with the environmentally sensitive fluorescent probe iodoacetamido-N,N'-dimethyl-N-(7-nitrobenz-2-oxa-1,3-diazolyl)ethylene diamine (NBD; Molecular Probes) via the sulfhydryl was carried out as described previously (25). The dye-labeled protein samples were made 10% (v/v) in sterile glycerol, quick-frozen in liquid nitrogen, and stored at −80 °C. Proteins were typically labeled at an efficiency of 60–100%.
Human Erythrocyte Ghost Membrane Preparation and Doxyl Stearate Incorporation
Human erythrocyte ghost membranes were prepared and quantified for protein content as described (12, 25). For the collisional quenching experiments, 5- and 16-doxyl-stearic acid was prepared by dissolving the doxyl compounds in MeOH to a final concentration of 10 mg/ml. A stock of 5- and 16-doxyl-stearic acid at a 1:1 ratio (v/v) was made. The hRBC ghost membranes were equilibrated with 200 µg of probe/1 mg of ghost membrane protein. The stearic acid quencher was allowed to partition into the membrane at room temperature for 1 h with periodic mixing (25). Membranes were washed of unincorporated probe.
Fluorescence Measurements
All fluorescence intensity measurements were performed using an SLM-8100 photon-counting spectrofluorimeter as described previously (12). An excitation wavelength of 480 nm and an emission wavelength of 540 nm were used for NBD with a bandpass of 4 nm. Emission scans from 500 to 600 nm for each sample were carried out at a resolution of 1 nm with an integration time of 1 s. Samples containing 180 pmol of total toxin were incubated with hRBC ghost membranes (equivalent to 303.25 µg of membrane protein) or hRBC ghost membranes incorporated with doxyl-stearic acid in PBS (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 3 mM KCl (pH 7.5)) at 37 °C for 5–10 min before making spectral measurements.
Flow Cytometry
Flow cytometry was used to detect the intermolecular interaction of ILY monomers on the membrane surface of hRBCs by fluorescence resonance energy transfer (FRET) as described previously (25). Briefly, ILY or derivatives thereof were labeled via cysteine-substituted Asp-66 in domain 1 of ILY. ILY was then modified via the sulfhydryl group of the cysteine with 5-iodoacetamidofluorescein (donor dye (D)) or tetramethylrhodamine (acceptor dye (A)). Approximately 1 × 106 hRBCs were incubated with 18 pmol of total toxin for 30 min on ice. Total toxin is defined as a 1:1 molar ratio of donor-labeled (D) and unlabeled (U) toxin or donor-labeled and acceptor (A)-labeled toxin. The mixtures were analyzed by FACSCalibur flow cytometer (University of Oklahoma Health Sciences Center) and FLOWJO software (Treestar). Changes in the donor fluorescence due to FRET with the acceptor were determined by comparing donor emission intensity in the presence of unlabeled toxin (D + U) to that of donor emission intensity in the presence of acceptor-labeled toxin (D + A).
SDS-Agarose Gel Electrophoresis (AGE) and Immunoblot Analyses
Denaturing agarose gel electrophoresis was carried out as described previously (12). Briefly, toxin (250 ng) was incubated alone or with hRBCs (1.5 × 106 cells) for 30 min at 37 °C. The samples were solubilized with SDS sample buffer (without heating) and then the monomeric and oligomeric complexes were resolved on a 1.5% SDS-AGE gel and transferred to nitrocellulose paper. Protein bands were recognized by rabbit anti-ILY antibody followed by goat anti-rabbit antibody conjugated to horseradish peroxidase. The bands were visualized using a chemiluminescent substrate (ECL Western blotting detection reagents, Amersham Biosciences/GE Healthcare) and autoradiography.
RESULTS
The ILYL1–L3 Loops Insert into the Membrane
We previously showed the loops L1, L2, and L3 of PFO insert into the membrane surface and proposed that these loops, along with the undecapeptide of PFO, formed a binding site that recognized and inserted into cholesterol-rich membranes, thereby anchoring PFO to the membrane surface (15). However, since ILY binding to the membrane surface is mediated by an interaction with huCD59 (23), the nature and extent of ILY D4 loop insertion into the bilayer was unknown.
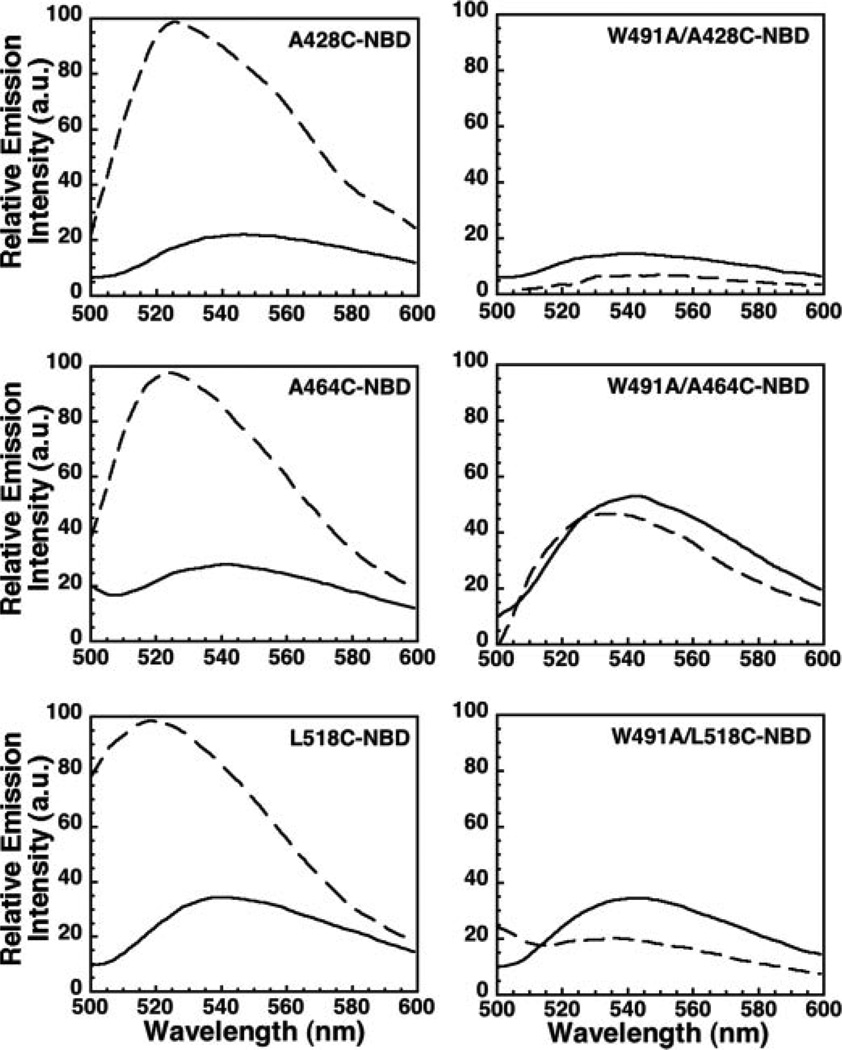
To assess D4 loop insertion into the membrane, a unique cysteine residue was substituted for residues Leu-518, Ala-428, and Ala-464 located, respectively, in loops L1, L2, and L3 of native ILY (Fig. 1). These residues are conserved in both ILY and PFO and have been shown in PFO to insert into the membrane surface (15). Each cysteine-substituted mutant was derivatized with the environmentally sensitive fluorescent probe, NBD. Hemolytic activity of either the cysteine-substituted mutants or their NBD modified forms was similar to native ILY (data not shown). Since NBD emission intensity and lifetime are very different in aqueous and non-aqueous environments, we have frequently used this dye to determine whether the residue to which the dye is attached is exposed to an aqueous milieu or to the nonpolar environment of the membrane bilayer (4–6, 11, 12, 15, 26, 27). When a single probe was positioned in each of the three D4 loops of native ILY, a significant increase in emission intensity was observed upon addition of hRBC ghost membranes (Fig. 2, left panels). The presence of the probe in the hRBC membrane was confirmed by the introduction of a membrane-restricted collisional quencher (a mixture of 5- and 16-doxyl-stearic acid (25)) that significantly quenched the NBD fluorescence (data not shown). This result is similar to that previously observed for PFO (15). These data show that L1-L3 of ILY insert into the membrane, suggesting that ILY binding to the cell surface is not solely stabilized by ILY binding to huCD59.
FIGURE 2. Membrane insertion of the ILY D4 loops is blocked by the Trp-491 to alanine undecapeptide mutant.
Residues Ala-428, Ala-464, or Leu-518 located in loops L2, L3, and L1, respectively, were individually mutated to a cysteine in native ILY and modified with the fluorescent probe NBD (left panels). The same loop mutations were also made in the ILYW491A mutant (ILYA428C/W491A, ILYA464C/W491A, ILYL518C/W491A) and also labeled with NBD (right panels). Each fluorescently modified toxin was incubated alone as a soluble monomer (solid line) or bound to hRBC ghost membranes (dashed line) at 37 °C. Membrane insertion of the NBD-labeled cysteine residues results in an increase in fluorescence intensity of the probe as the monomer makes its transition from theaqueous environment (soluble monomer)tothe membrane-bound state. a.u., arbitrary units.
L1–L3 Insertion Does Not Occur in the ILYW491A Undecapeptide Mutant
Previous studies have shown that the ILY undecapeptide is involved in prepore to pore conversion (25). Therefore, we determined whether this mutation affected the insertion of the L1–L3 loops. The same studies as shown in Fig. 2 (left panels) were repeated, but in the background of the ILYW491A mutation (Fig. 2, right panels). In stark contrast to native ILY, the presence of the W491A mutation abolished the membrane insertion of L1–L3, even though ILY still binds to the membrane surface via huCD59 (data not shown and Ref. 25). Since this mutation of Trp-491 prevents the insertion of the neighboring D4 loops, it appears the structural state of the undecapeptide dictates whether L1–L3 insert into the membrane.
Mutation of Loops L1–L3 Disrupts Pore Forming Activity
We next determined whether mutations in loops L1, L2, and/or L3 would prevent pore formation. Individual mutants were generated in which an aspartate was substituted for residues Leu-518, Ala-428, or Ala-464, which reside in loops L1, L2, and L3, respectively, in native ILY. Charged aspartate is rarely found in the bilayer, and most aspartate carboxylates are deprotonated at pH 7.4 (pKa 3.7). Hence, one would predict that an Asp in a loop would inhibit its insertion into the membrane. The substitution of a single aspartate into each of the L1, L2, and L3 loops reduced ILY cytolytic activity by >98% (Table 1). The introduction of a charged group for each loop did not prevent binding of the toxin to the cell surface via its receptor, as determined by flow cytometry (shown below in the section entitled “FRET-detected monomer-monomer association”). Also, the insertion of the undecapeptide of ILYA464D could not occur if the toxin was not bound to the cell surface (shown below in the section entitled “Membrane insertion of the undecapeptide is required for prepore oligomer formation”).
TABLE 1. Hemolytic activity of ILY D4 aspartate loop mutants.
The HD50 (hemolytic dose for 50% lysis) was determined for native ILY and the D4 loop mutants. Shown is the relative hemolytic activity of each mutant compared with native toxin.
| Hemolytic activity | |
|---|---|
| CDC | % native ILY |
| ILY | 100 |
| ILYW491A | 0.3 |
| ILYA428D | 0.1 |
| ILYA464D | 1.3 |
| ILYL518D | 0.3 |
Membrane Insertion of the D4 Loops Is Coordinated
The above analyses demonstrated that a dramatic decrease in hemolytic activity occurred if residues Leu-518, Ala-428, and Ala-464 were substituted with aspartate. Since a single aspartate mutation in any single loop blocked pore formation, it is possible that all three loops insert coordinately. Furthermore, having two of three loops intact cannot overcome the disruption of the other loop by mutation. This interdependency suggests that the insertion of the loops is coupled and possibly cooperative. We therefore determined whether an aspartate substitution in one loop interfered with the membrane insertion of the other two loops.
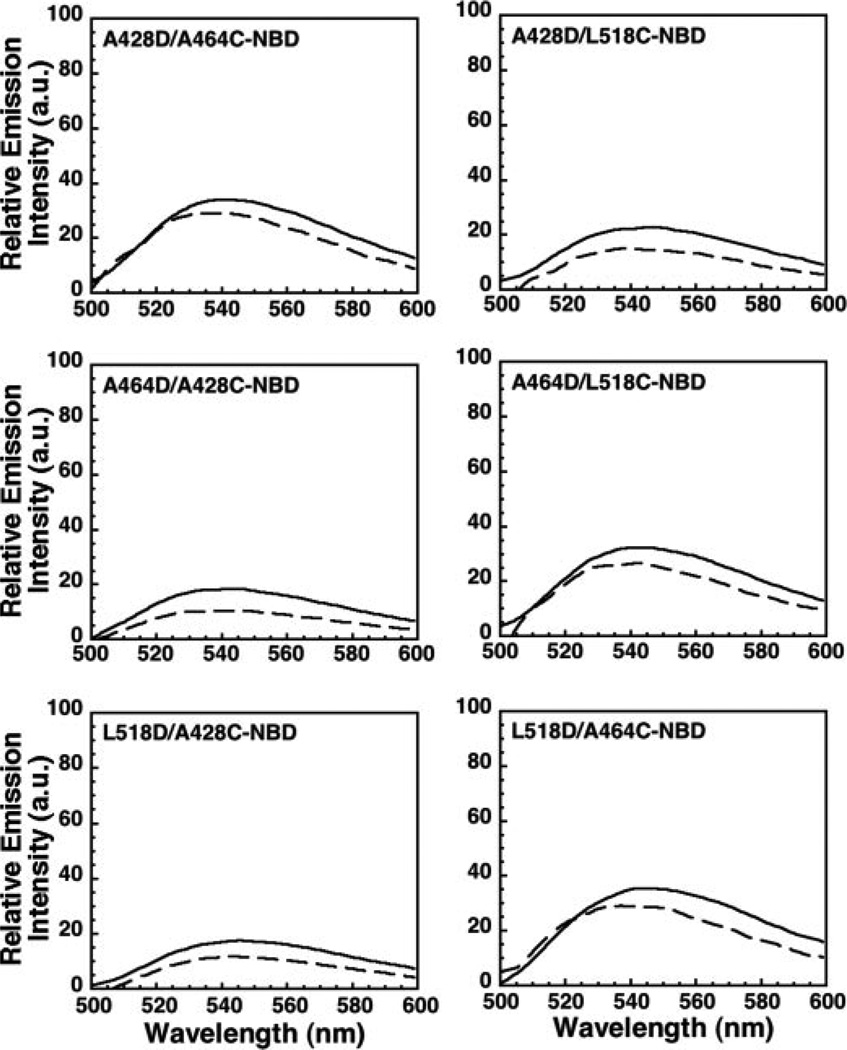
Experimentally, one loop residue was substituted with an aspartate to prevent its insertion. A cysteine residue was then introduced into one of the other two D4 loops and labeled with NBD, and its insertion into the membrane was assessed spectroscopically. For example, two double mutants, ILYL518D/A428C-NBD and ILYL518D/A464C-NBD, were generated so that the insertion (or not) of loops L2 and L3 (containing cysteine substituted Ala-428 or Ala-464) could be detected in the presence of the Leu-518 to aspartate mutation. The membrane-dependent change in the fluorescence intensity of each double mutant was then compared with that of a mutant lacking an aspartate substitution (e.g. ILYA428C-NBD or ILYA464C-NBD; Fig. 2). The results in Fig. 3 show that if the L518D mutation is present, the insertion of L2 and L3 does not occur. The same results were obtained for the ILYA428D and ILYA464D double mutants (Fig. 3). Hence, by disrupting the insertion of one loop the insertion of all loops is prevented.
FIGURE 3. Membrane insertion of the D4 aspartate loop mutants into hRBC ghost membranesiscoupled.
Experiments were performed as in Fig. 2, except that each NBD-labeled cysteine loop mutant also contained an aspartate mutation in a neighboring loop. Compare the membrane-dependent changes in NBD emission intensities in the absence (solid line) and presence (dashed line) of membranes for the double ILY mutants with those observed with the ILY single mutants (Fig. 2).
Since the effect of the aspartate substitutions was the same when these experiments were repeated in high salt (0.5 m NaCl) (data not shown), an electrostatic repulsion between an Asp and negative charges at the surface does not appear to be a major factor in preventing insertion of the loop residues. Instead, it seems most likely that an Asp-containing loop is not exposed to the hydrophobic interior of the bilayer because of the energetic difficulty of inserting a charged carboxyl group into a non-aqueous nonpolar milieu. Since blocking the insertion of one loop prevents the insertion of the other loop residues, loop insertion is apparently coupled and coordinated conformationally. This also extends to the undecapeptide loop because the insertion of L1, L2, or L3 is blocked by the undecapeptide ILYW491A mutation. The insertion of all four loops at the tip of D4 therefore appears to be coupled.
The D4 Loop Mutants Are Conformationally Prepared for Oligomerization
A key structural change that prepares the CDCs for oligomerization is the movement of one β-strand (β5) away from another (β4) in the core β-sheet of domain 3 (D3). This conformational change occurs more than 60 Å above the membrane surface (16) but is triggered by interaction of the tip of D4 with the membrane (28). When β5 disengages from β4, interstrand hydrogen bond formation can occur between β1 and β4 of adjacent monomers, thus initiating oligomer formation. We have previously shown that an NBD probe located at the cysteine-substituted Val-322 in β4 of PFO is buried under β5 in the soluble monomer (28). Upon binding to a membrane, this residue is exposed to the aqueous environment and the fluorescence emission of the NBD is quenched by water. NBD at this site therefore detects the exposure of β4 and its availability for oligomerization.
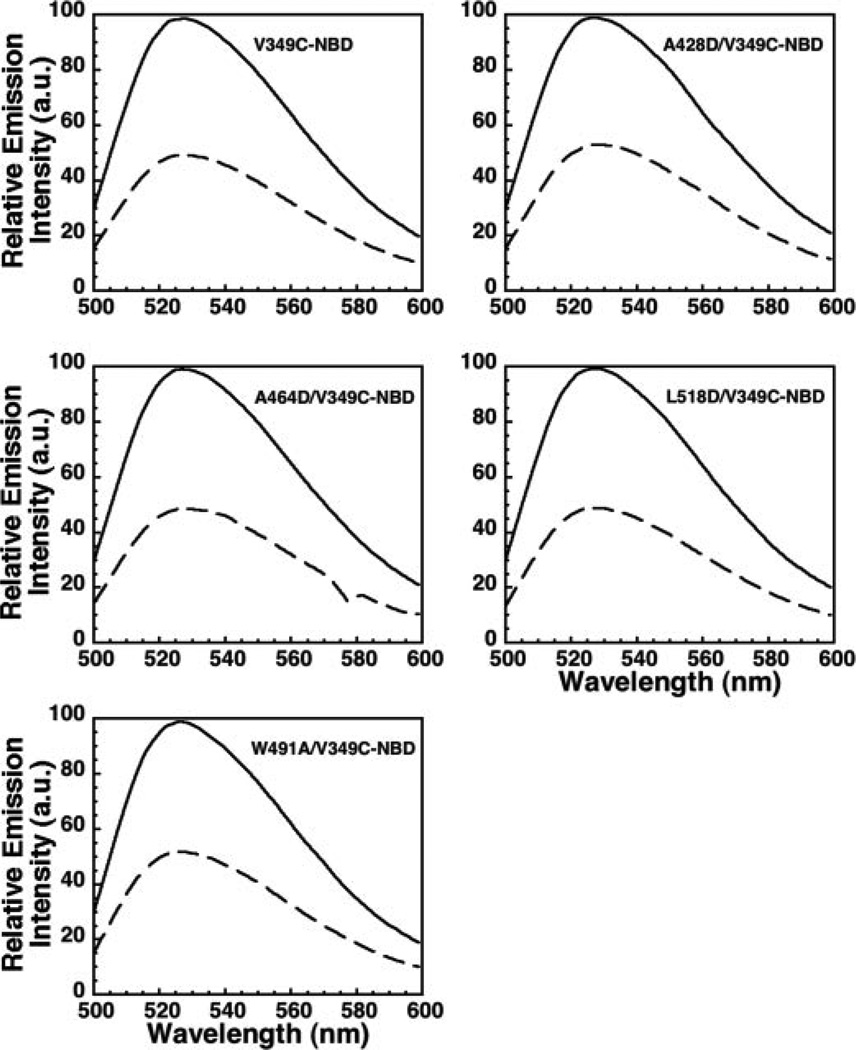
To determine whether β5 was moving away from β4 in the various mutants, we introduced a cysteine for Val-349 (analogous to Val-322 in PFO) in native and mutant ILY and then labeled each with NBD. When each of these proteins bound to the membrane, the extent of NBD quenching was nearly identical (Fig. 4). Thus, the conformational change required for monomer oligomerization was not affected by mutations that block the insertion of the D4 loops. Receptor binding alone is therefore sufficient to trigger this conformational change in ILY, preparing the ILY toxin monomers for oligomerization.
FIGURE 4. Receptor binding triggers disengagement of β5 from β4.
A cysteine was introduced into native ILY and its D4 loop mutants at Val-349 and modified with NBD. NBD emission intensity scans are shown for each ILY species in solution (solid line) and bound to the membrane (dashed line).
Mutation of the D4 Loops of ILY Affect Both Oligomer Assembly and Prepore-to-Pore Conversion
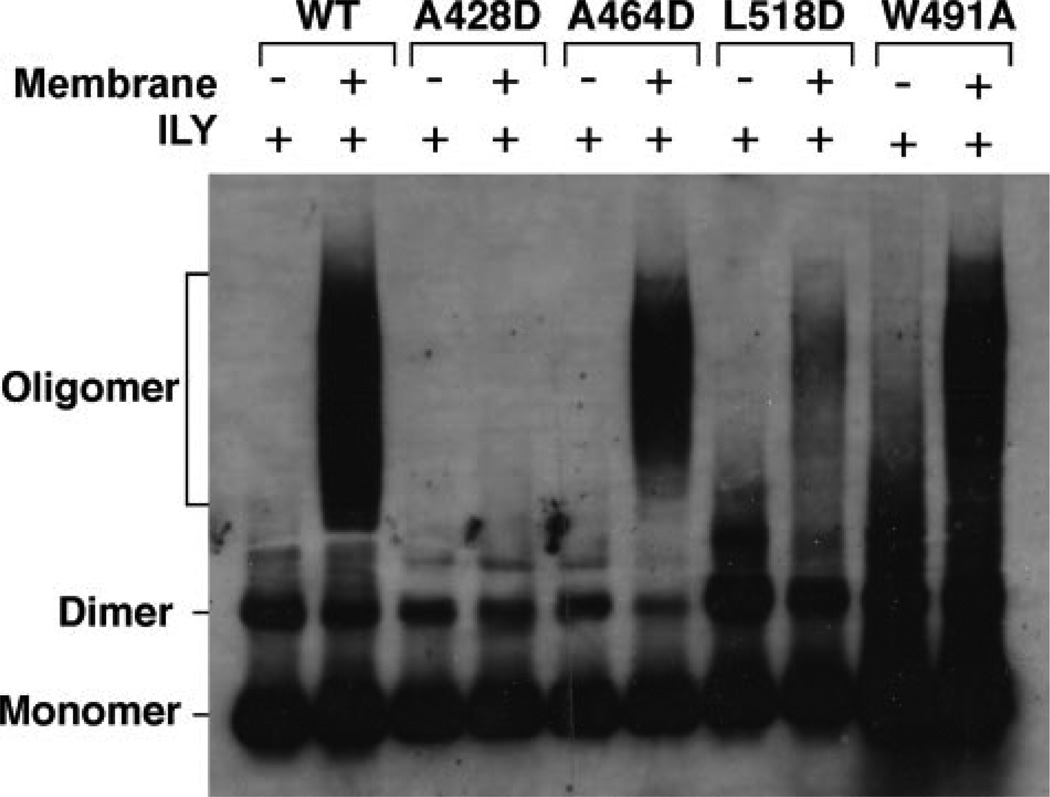
Since monomer binding to the receptor primes ILY for oligomerization via β4 exposure, we next examined whether the insertion of the D4 loops was necessary for oligomerization. SDS-AGE was used to detect the formation of the SDS-resistant prepore oligomer (5) for each D4 loop mutant on hRBCs (Fig. 5). An SDS-resistant prepore complex was formed by ILYA464D and ILYW491A, whereas SDS-resistant prepores were not detected for ILYA428D and were barely detected for ILYL518D. Thus, the ILYA428D and ILYL518D mutations affected assembly of the oligomer, while the ILYA464D and ILYW491A mutation blocked the conversion of the prepore to pore (cf. Table 1).
FIGURE 5. Oligomer formation by the ILY aspartate loop mutants.
The ILY D4 loop mutants and native ILY were assessed for their ability to form an SDS-resistant oligomeric prepore complex using SDS-AGE in the presence or absence of hRBCs. Shown is an autoradiograph of the monomer, dimer and oligomer bands present on the membrane surface of intact hRBCs identified by anti-ILY antibody.
FRET-detected Monomer-Monomer Association
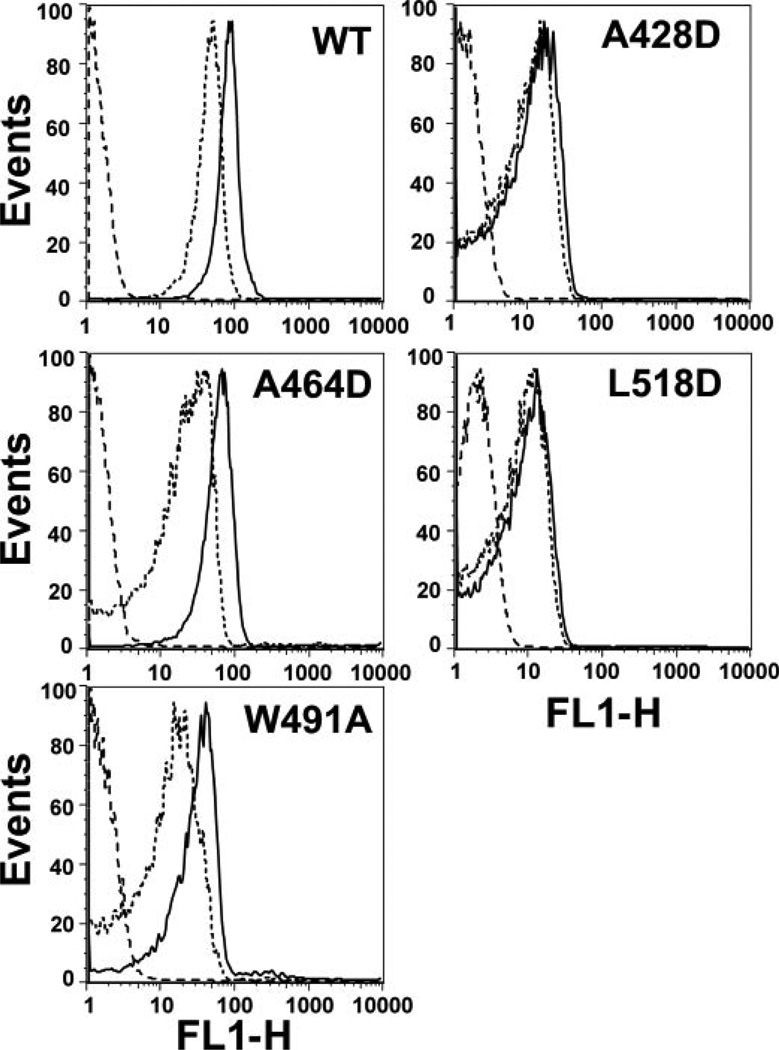
Since ILYA428D and ILYL518D may form an oligomer that is sensitive to SDS, we determined whether ILY monomers associate using FRET on intact hRBCs. This is a non-destructive approach for evaluating the proximity of CDC monomers to each other (29). We previously developed a flow cytometry-based FRET assay to measure monomer-monomer associationon intact hRBCs (25) and here use the same approach to determine whether the monomers of native ILY and the various mutants associated on the cell surface. As shown in Fig. 6, native ILY, ILYW491A, and ILYA464D exhibited donor quenching due to FRET, as expected from the data in Fig. 5, whereas ILYA428D and ILYL518D did not exhibit detectable FRET. Hence, receptor-bound monomers of ILYA428D and ILYL518D do not form appreciable levels of either SDS-resistant or SDS-sensitive oligomer on the membrane surface.
FIGURE 6. FRET detected monomer association on intact hRBCs.
A cysteine was introduced into each mutant or native ILY at Asp-66. Samples of each were labeled with either 5-iodoacetamidofluorescein (donor fluorophore) or tetramethylrhodamine (acceptor fluorophore), and equimolar amounts of donor-labeled ILY and of either acceptor-labeled ILY (D+A, dotted line) or unlabeled ILY (D+U, solid line) were bound to hRBC. Donor emission intensity is shown on the horizontal axes. Since donor emission intensity decreases in D+A samples, FRET occurs, a shift in the D+A intensity peak to the left relative to the D+U intensity peaks shows that the monomers are on average located close enough (<90 Å) to exhibit FRET. No significant FRET occurs in samples that have very similar D+Aand D+U intensity profiles (25). The autofluorescence of the hRBCs alone is also shown (dashed line). FL1-H, fluorescence detector 1 height.
Membrane Insertion of the Undecapeptide Is Required for Prepore Oligomer Formation
The above studies show that two classes of D4 loop mutants exist: mutation of Trp-491 to alanine or Ala-464 to aspartate blocks the prepore to pore transition, whereas mutation of either Ala-428 or Leu-518 to aspartate blocks formation of the prepore oligomer. In all cases, the insertion of L1, L2, and L3 was blocked by the mutations. The structural basis for these two phenotypic effects was therefore not due to the insertion state of L1, L2, L3. However, the insertion state of the undecapeptide in the L1, L2, and L3 mutants had not been resolved. We therefore determined whether the undecapeptide inserted into the membrane for each mutant and native ILY. The membrane insertion of the undecapeptide was monitored by introducing a cysteine residue for Ala-486 of the undecapeptide, modifying it with NBD, and assessing if its emission intensity increased (i.e. entered a nonpolar environment).
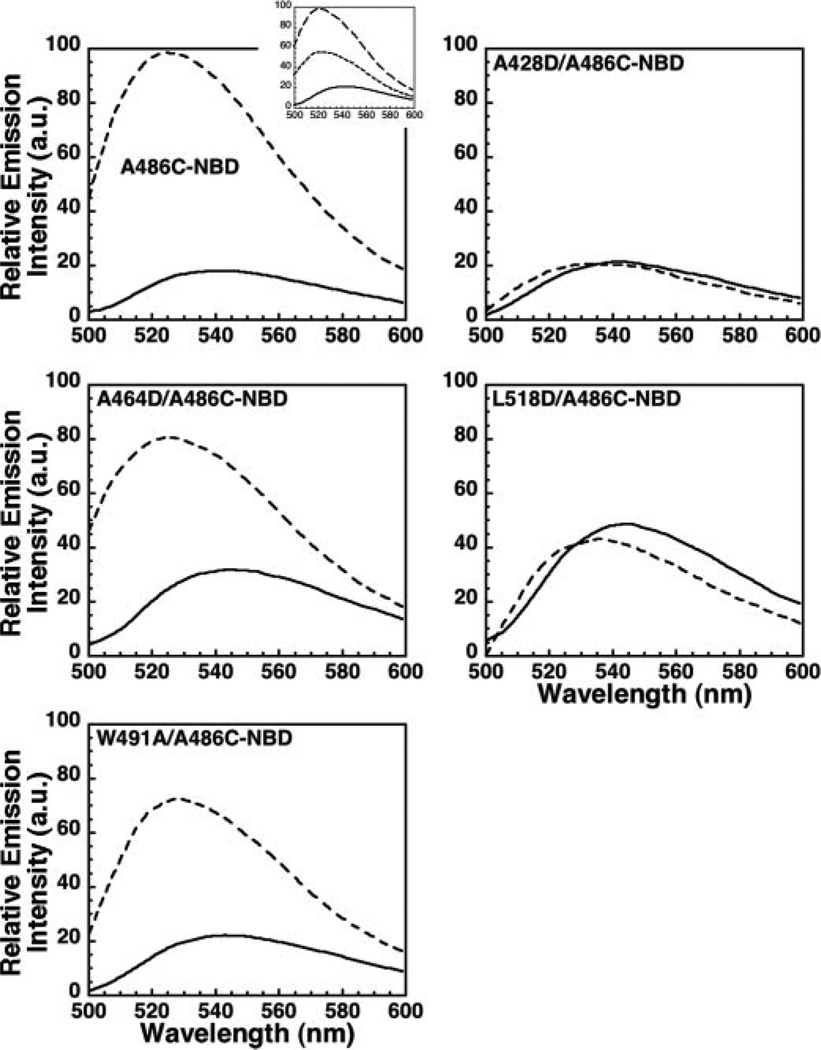
As shown in Fig. 7, the NBD emission intensity increased significantly when ILYA486C-NBD was incubated with hRBC ghost membranes. The introduction of the membrane-restricted collisional quencher, doxyl-stearic acid, into the membrane (25) significantly quenched NBD fluorescence, confirming its membrane location (Fig. 7, inset). Similar results were observed for the ILYW491A and ILYA464D mutants. In contrast, no change in the NBD emission was observed for ILYA428D and ILYL518D. Hence, membrane insertion of the undecapeptide correlates with prepore oligomer formation, while insertion of the L1–L3 loops correlates with prepore to pore conversion.
FIGURE 7. Membrane insertion of the undecapeptide.
Cysteine was substituted for Ala-486 in the ILY undecapeptide and labeled with NBD in background of native ILY, ILYA428D, ILYA464D, ILYL518D, and ILYW491A. Each NBD-labeled protein was incubated in the absence (solid line) or presence (dashed line) of hRBC ghost membranes, and the change in emission intensity was measured as each toxin bound to the membrane. Inset, to determine whether the probe conjugated to residue Ala-486 inserted into the bilayer, 5- and 16-doxyl-stearic acid (200 µg/1 mg of membrane protein) was incorporated into the membrane. Doxyl-stearic acid is a collisional quencher of NBD fluorescence. ILYA486CNBD was incubated in the absence (solid line) or in the presence of membranes with (dotted line) or without (dashed) quencher.
DISCUSSION
Most CDCs are thought to bind to the cell surface via cholesterol-rich domains (reviewed in (30)), and this interaction is mediated by the short hydrophobic loops and the undecapeptide at the tip of domain 4 (6, 15, 17, 20). ILY is unlike most CDCs in that it specifically binds to huCD59 and does not bind to cholesterol-rich cell membranes in the absence of huCD59 (23). Therefore, it was not clear whether the D4 loops, which bind and anchor PFO to the cholesterol-rich membrane surface, were still required for ILY pore formation. Here we have shown that receptor binding by ILY localizes it on the cell surface and triggers the structural change in D3 required for oligomerization. These steps, however, are not sufficient to generate a functional pore. Membrane insertion of the D4 loops also must occur to assemble and form a functional membrane oligomer and pore. Therefore the D4 loops appear to be involved in the pore forming mechanism of all CDCs. Since binding to a protein receptor localizes ILY at the membrane surface, the D4 loops of ILY are not required for membrane recognition and binding, as is the case with other CDCs.
The exposure of β4 in ILY D3, which is necessary for the oligomerization of membrane bound monomers (28), did not require the membrane insertion of the D4 loops. ILY binding to huCD59 was sufficient to trigger the disengagement of β5 from β4, thereby making the receptor-bound monomers available for oligomerization via the interaction of β1 and β4 of adjacent monomers. However, subsequent steps in pore formation, the assembly of the prepore oligomer and conversion of the prepore complex to a functional pore, were accompanied by the insertion of ILY D4 loops. ILY molecules mutated in the undecapeptide or in L3 could oligomerize into an SDS-resistant prepore complex but could not make the structural transition to a functional pore. Mutations in either L1 or L2 blocked association of the receptor-bound ILY molecules and the formation of the prepore complex. These two classes of mutants therefore exhibited two distinct phenotypes affecting two major steps in pore formation.
Interestingly, these two phenotypes differ in the extent of undecapeptide insertion into the membrane. In the mutants that formed little or no oligomer (ILYA428D and ILYL518D), perhaps because β5 did not move sufficiently far from β4 in these two mutants, neither the undecapeptide nor the D4 loops inserted into the membrane. In contrast, in mutants that oligomerized into a SDS-resistant prepore complex (ILyW491A and ILYA464D), the undecapeptide, but not L1-L3, inserted into the membrane. Thus the formation of an SDS-stable oligomer was accompanied by the insertion of the undecapeptide into the membrane, but this structural change was not sufficient to effect pore formation. Pore formation only occurs if L1, L2, and L3 are also exposed to the nonpolar core of the bilayer.
A possible mechanism for L1–L3 and undecapeptide involvement in oligomer assembly and prepore to pore conversion is revealed by studies with PFO. Following oligomerization of PFO monomers into a prepore complex, the cooperative (or concerted) membrane insertion of two D3 amphipathic β-hairpins per monomer occurs to form a transmembrane β-barrel (4, 11, 12). The formation of in-register hydrogen bonds between β1 of one PFO monomer and β4 (Fig. 1) of an adjacent monomer is critical for the membrane insertion of these two β-hairpins (10, 28). Mutations in PFO β1 or β4 that alter the correct pairing of the hydrogen bonds between these strands blocks the prepore to pore conversion of PFO (10, 28). The prepore oligomer can apparently tolerate a fraction of incorrectly paired hydrogen bonds between β1 and β4 (10). However, if the fraction of monomers with incorrectly paired hydrogen bonds exceeds a certain threshold, the prepore cannot make the transition to the pore (10). These combined data suggest that membrane insertion of the D4 loops (L1–L3 and the undecapeptide) may play a role in properly orienting the monomers at the membrane surface, thereby increasing the probability of correctly pairing hydrogen bonds between β1 and β4 of adjacent monomers.
If this model is correct, the ILY mutants that lack all D4 loop contacts with the membrane surface presumably fail to oligomerize because the probability is low that two monomers bound only to the receptor will contact each other in such a way that allows β1–β4 hydrogen bonding. The membrane insertion of the undecapeptide presumably stabilizes the monomer on the membrane sufficiently so that ILY monomers can form inter-molecular hydrogen bonds between β1 and β4. Although intermolecular hydrogen bonds can form, the extent of out-of-register hydrogen bonding may be sufficiently high to prevent the prepore to pore transition. In this scenario, ILY is then correctly oriented at the membrane only by simultaneous interactions with huCD59, L1, L2, L3, and undecapeptide that allow adjacent monomers to form intermolecular hydrogen bonds in the proper register to spontaneously generate a transmembrane β-barrel.
At this point, we do not know whether the interactions of the D4 loops with the bilayer actively trigger the transitions of ILY from one state to another in pore formation. It is also possible that D4 loop insertion into the membrane is a passive or secondary result of other structural changes elsewhere in ILY. However, given both the active recognition and binding of most CDCs to membrane surfaces via D4 (6,15,17, 20) and also the apparently instantaneous conformational linkage of D4 to D3 in PFO (28), it seems likely that the insertion of the D4 loops into the bilayer is required for oligomerization and actively triggers the insertion of the β-hairpins into the bilayer.
In summary, ILY has diverged from the other CDCs by taking advantage of a protein-protein interaction to effect membrane targeting. However, our data suggest that many of the interactions required for pore formation have been retained, including the requirement for specific D4 loop interactions with the membrane to promote oligomerization and pore formation.
Footnotes
This work was supported by National Institutes of Health Grant AI037657 (to R. K. T.).
The abbreviations used are: CDC, cholesterol-dependent cytolysin; PFO, perfringolysin O; ILY, intermedilysin; MES, 4-morpholineethanesulfonic acid; hRBC, human red blood cell; NBD, iodoacetamido-N,N’-dimethyl-N-(7-nitrobenz-2-oxa-1,3-diazolyl)ethylene diamine; AGE, agarose gel electrophoresis; FRET, fluorescence resonance energy transfer; D, donor dye; U, unlabeled; A, acceptor.
REFERENCES
- 1.Alouf JE. In: Bacterial Toxins: A Comprehensive Sourcebook. 2nd Ed. Alouf J, Freer J, editors. London: Academic Press; 1999. [Google Scholar]
- 2.Polekhina G, Giddings KS, Tweten RK, Parker MW. Proc. Natl. Acad. Sci. U. S. A. 2005;102:600–605. doi: 10.1073/pnas.0403229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 4.Hotze EM, Wilson-Kubalek EM, Rossjohn J, Parker MW, Johnson AE, Tweten RK. J. Biol. Chem. 2001;276:8261–8268. doi: 10.1074/jbc.M009865200. [DOI] [PubMed] [Google Scholar]
- 5.Shepard LA, Shatursky O, Johnson AE, Tweten RK. Biochemistry. 2000;39:10284–10293. doi: 10.1021/bi000436r. [DOI] [PubMed] [Google Scholar]
- 6.Heuck AP, Hotze E, Tweten RK, Johnson AE. Mol. Cell. 2000;6:1233–1242. doi: 10.1016/s1097-2765(00)00119-2. [DOI] [PubMed] [Google Scholar]
- 7.Tilley SJ, Orlova EV, Gilbert RJ, Andrew PW, Saibil HR. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 8.Olofsson A, Hebert H, Thelestam M. FEBS Lett. 1993;319:125–127. doi: 10.1016/0014-5793(93)80050-5. [DOI] [PubMed] [Google Scholar]
- 9.Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. EMBO J. 2004;23:3206–3215. doi: 10.1038/sj.emboj.7600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hotze EM, Heuck AP, Czajkowsky DM, Shao Z, Johnson AE, Tweten RK. J. Biol. Chem. 2002;277:11597–11605. doi: 10.1074/jbc.M111039200. [DOI] [PubMed] [Google Scholar]
- 11.Shatursky O, Heuck AP, Shepard LA, Rossjohn J, Parker MW, Johnson AE, Tweten RK. Cell. 1999;99:293–299. doi: 10.1016/s0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- 12.Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, Ryan KR, Johnson AE, Tweten RK. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- 13.Heuck AP, Tweten RK, Johnson AE. J. Biol. Chem. 2003;278:31218–31225. doi: 10.1074/jbc.M303151200. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura M, Sekino-Suzuki N, Mitsui K, Ohno-Iwashita Y. J Biochem. (Tokyo) 1998;123:1145–1155. doi: 10.1093/oxfordjournals.jbchem.a022054. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran R, Heuck AP, Tweten RK, Johnson AE. Nat. Struct. Biol. 2002;9:823–827. doi: 10.1038/nsb855. [DOI] [PubMed] [Google Scholar]
- 16.Ramachandran R, Tweten RK, Johnson AE. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7139–7144. doi: 10.1073/pnas.0500556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto M, Ohno-Iwashita Y, Ando S. Eur. J.Biochem. 1987;167:425–430. doi: 10.1111/j.1432-1033.1987.tb13355.x. [DOI] [PubMed] [Google Scholar]
- 18.Ohno-Iwashita Y, Iwamoto M, Mitsui K, Ando S, Iwashita S. J. Biochem. (Tokyo) 1991;110:369–375. doi: 10.1093/oxfordjournals.jbchem.a123588. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura M, Sekino N, Iwamoto M, Ohno-Iwashita Y. Biochemistry. 1995;34:6513–6520. doi: 10.1021/bi00019a032. [DOI] [PubMed] [Google Scholar]
- 20.Sekino-Suzuki N, Nakamura M, Mitsui KI, Ohno-Iwashita Y. Eur. J. Biochem. 1996;241:941–947. doi: 10.1111/j.1432-1033.1996.00941.x. [DOI] [PubMed] [Google Scholar]
- 21.Iwamoto M, Morita I, Fukuda M, Murota S, Ando S, Ohno-Iwashita Y. Biochim. Biophys. Acta. 1997;1327:222–230. doi: 10.1016/s0005-2736(97)00061-8. [DOI] [PubMed] [Google Scholar]
- 22.Shimada Y, Maruya M, Iwashita S, Ohno-Iwashita Y. Eur. J. Biochem. 2002;269:6195–6203. doi: 10.1046/j.1432-1033.2002.03338.x. [DOI] [PubMed] [Google Scholar]
- 23.Giddings KS, Zhao J, Sims PJ, Tweten RK. Nat. Struct. Mol. Biol. 2004;12:1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- 24.Nagamune H, Ohkura K, Sukeno A, Cowan G, Mitchell TJ, Ito W, Ohnishi O, Hattori K, Yamato M, Hirota K, Miyake Y, Maeda T, Kourai H. Microbiol. Immunol. 2004;48:677–692. doi: 10.1111/j.1348-0421.2004.tb03479.x. [DOI] [PubMed] [Google Scholar]
- 25.Giddings KS, Johnson AE, Tweten RK. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11315–11320. doi: 10.1073/pnas.2033520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- 27.Crowley KS, Reinhart GD, Johnson AE. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- 28.Ramachandran R, Tweten RK, Johnson AE. Nat. Struct. Mol. Biol. 2004;11:697–705. doi: 10.1038/nsmb793. [DOI] [PubMed] [Google Scholar]
- 29.Harris RW, Sims PJ, Tweten RK. J. Biol. Chem. 1991;266:6936–6941. [PubMed] [Google Scholar]
- 30.Alouf JE. Int. J. Med. Microbiol. 2000;290:351–356. doi: 10.1016/S1438-4221(00)80039-9. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:27–28. doi: 10.1016/0263-7855(96)00018-5. 33–38. [DOI] [PubMed] [Google Scholar]