Abstract
Background:
With the emerging role of digital imaging in pathology and the application of automated image-based algorithms to a number of quantitative tasks, there is a need to examine factors that may affect the reproducibility of results. These factors include the imaging properties of whole slide imaging (WSI) systems and their effect on the performance of quantitative tools. This manuscript examines inter-scanner and inter-algorithm variability in the assessment of the commonly used HER2/neu tissue-based biomarker for breast cancer with emphasis on the effect of algorithm training.
Materials and Methods:
A total of 241 regions of interest from 64 breast cancer tissue glass slides were scanned using three different whole-slide images and were analyzed using two different automated image analysis algorithms, one with preset parameters and another incorporating a procedure for objective parameter optimization. Ground truth from a panel of seven pathologists was available from a previous study. Agreement analysis was used to compare the resulting HER2/neu scores.
Results:
The results of our study showed that inter-scanner agreement in the assessment of HER2/neu for breast cancer in selected fields of view when analyzed with any of the two algorithms examined in this study was equal or better than the inter-observer agreement previously reported on the same set of data. Results also showed that discrepancies observed between algorithm results on data from different scanners were significantly reduced when the alternative algorithm that incorporated an objective re-training procedure was used, compared to the commercial algorithm with preset parameters.
Conclusion:
Our study supports the use of objective procedures for algorithm training to account for differences in image properties between WSI systems.
Keywords: Quantitative immunohistochemistry, reproducibility, whole slide imaging
BACKGROUND
Digital pathology is an emerging field enabled by recent technological advances in whole slide imaging (WSI) systems, which can digitize whole slides at high resolution in a short period of time. Advantages in the use of digital pathology include telepathology, digital consultation and slide sharing, pathology education, indexing and retrieval of cases, and the use of automated image analysis.[1–3]
The latter might be an important contributor to reducing inter- and intra-observer variability for certain pathology tasks such as the evaluation of HER2/neu (Human Epidermal growth factor Receptor 2) immunohistochemical staining.[4–6] The College of American Pathologists/American Society of Clinical Oncology guidelines recommend image analysis as an effective tool for achieving consistent interpretation of immunohistochemistry (IHC) HER2/neu staining, provided that a pathologist confirms the result.[7] Reducing inter- and intra-observer variability is critical toward improving reproducibility in IHC, along with efforts for improving and standardizing procedures for pre-analytic specimen handling,[8] antibody selection,[9] and staining and scoring methods.[10,11] Image algorithms and computer aids to assist the pathologist have been applied to a number of pathology tasks, though the focus has been on automated quantitative IHC of tissue-based biomarkers.[12–22] In addition to research studies, several commercial image analysis systems are currently available for the evaluation of IHC,[5,23–26] as reviewed by Cregger et al.[27] A number of commercially available imaging systems have received Food and Drug Administration (FDA) premarket approval to quantify biomarker expression as an aid in diagnosis; however, each of these algorithms was verified across a single imaging platform.[28]
An issue that has been under-examined in the general topic of computer-assisted IHC is the variability in image properties between different WSI scanners and the effect of such differences on the performance of computer algorithms. The imaging chain of a WSI system consists of multiple components including the light source, optics and sensor for image acquisition, as well as embedded algorithm systems for auto-focusing, selecting and combining different fields of view in a composite image, image compression and color correction. Details regarding the components of WSI systems can be found in Gu and Ogilvie.[29] Different manufacturers of WSI systems often utilize different components and algorithms in their imaging chain, as reported in the review of 31 commercial systems by Rojo et al.,[30] often resulting in images with different properties as can be seen in the example of Figure 1. Considering the likely application of image analysis tools on datasets extracted from different WSI scanners, those tools would need to be retrained to account for differences in image properties. Similarly, retraining would be necessary for analyzing images acquired with the same scanner but from slides stained at different times and stained with different antibodies or images processed differently using manipulation software. Retraining procedures adjust the required parameters of the algorithms in order to maintain a certain achievable level of performance. Different algorithms can be re-trained in different ways. Some commercial software for image analysis usually have a preset algorithm version and often allow for the operator to manually “tune” them, by adjusting a set of parameters. Other algorithms incorporate operator independent training procedures, such as the algorithm by Keller et al.,[22] which will be utilized in this study.
Figure 1.

Example of a field of view stained with a HER2/neu antibody, extracted from a whole slide image, digitized using: (a) The Aperio-CS (top), (b) The Aperio-T2 (middle), and (c) The Hamamatsu Nanozoomer (bottom) whole slide imaging systems. Images were extracted at ×20
The scope of this work was to quantify the variability between the performances of two different algorithms for the assessment of HER2/neu when applied to image datasets acquired with three different WSI systems. To the best of our knowledge this is the first study focusing on this task. Emphasis was placed on the importance of the retraining aspect of algorithms to allow them to adjust to the different image properties of different datasets. Here, we present the design, implementation and results of our study.
MATERIALS
Description of HER2/neu Whole Slide Set
A dataset of 77 paraffin-embedded breast cancer tissue slides stained with an antibody against the HER2/neu biomarker (HerceptTest™, Dako, CA, USA) were used in our study. Details regarding the preparation of these slides have been described by Masmoudi et al.[18] The slides were acquired from the Department of Pathology and Laboratory Medicine, University of California at Irvine, along with their clinically archived HER2/neu scores (categories of expression), verified by an expert breast pathologist in accordance with the HercepTest™ scoring system. The slides were chosen to have a balanced distribution of 1+, 2+, and 3+ HER2/neu categories. Appropriate ethical approval for the use of this material in research has been previously obtained. No 0+ cases were included in this dataset mainly because there were not enough cases in that category at the time of acquisition and also because focus originally was on developing an algorithm for membrane staining estimation.
Whole Slide Image Acquisition and Development of Image Datasets
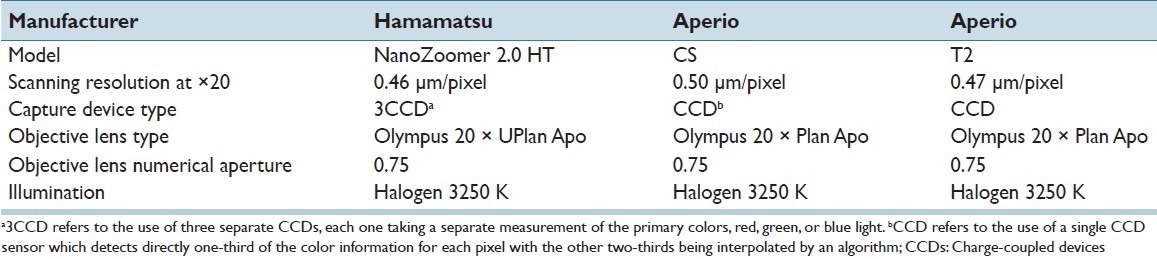
All 77 slides were digitized using three different WSI systems: Aperio ScanScope T2, Aperio Scanscope CS (Aperio Technologies, Vista, California) and Hamamatsu NanoZoomer 2.0 HT (Hamamatsu Photonics, Bridgewater NJ). Table 1 summarizes some basic scanner characteristics for the three systems (Information source for Aperio CS: http://www.aperio.com/lifescience/capture/cs, Information source for Aperio T2: http://web.archive.org/web/20040412140041/ http://www.aperio.com/products-ScanScope_T2.asp, Information source for Hamamatsu NanoZoomer 2.0HT: http://www.hamamatsu.com/jp/en/C9600-13.html. All slides were scanned at ×20 magnification. In a previous study,[4] 241 regions of interest (ROI) were extracted from 64 of the 77 whole slide images digitized with the Aperio ScanScope T2 system for conducting an observer study to compare inter-observer variability with unaided and computer-aided immunohistochemical evaluation. These ROIs were selected by the principal investigator after being trained by the expert pathologist on a different set of images to identify regions of invasive tumor. The size of the ROIs extracted from slides scanned with the Aperio ScanScope T2 was 816 × 646 pixels. For the purpose of this study, corresponding ROIs at the same locations were also extracted from the whole slide images acquired with the other two systems, thus creating three sets of ROI, one for each WSI system. Each ROI image was saved in a color tagged image file format with 8 bits per color channel. Due to differences in pixel size, ROIs from the three scanners had slightly different sizes: ROIs extracted from the Aperio ScanScope CS had a size of 760 × 600 pixels, and ROIs from the Hamamatsu NanoZoomer 2.0 HT had a size of 826 × 656 pixels. The whole slide viewer Image Scope (Aperio Technologies, Vista, California) was used to extract the ROIs. The three sets of 241 ROI each served as the main image datasets. The remaining 13 out of 77 slides were also digitized by each scanner and were used as algorithm development sets as described in the methods section.
Table 1.
Technical characteristics for the three whole slide imaging systems utilized in this study*
For the purpose of algorithm training, ground truth (reference standard) for each ROI in the main dataset in terms of HER2/neu expression was established using a panel of seven pathologists as part of the previously conducted observer study.[4] The pathologist group ranged in training and experience consisting of 5 board certified pathologists (post-board experience ranging from 2 years to 12 years) and 2 residents (1 had completed 2nd year, 1 had completed 3rd year). Each pathologist was asked to select a value for the HER2/neu expression of the image in the range between 1 and 100 using a moving slider and instructions that a value of less or equal to 33 corresponded to a 1+, a value between 33 and 66 corresponded to 2+, and a value larger than 66 corresponded to a 3+. No cases of 0 score were present in our dataset. The mean continuous score from the pathologist panel was processed using the thresholds above to produce a consensus categorical score for each ROI. Using this method, 21 ROIs in the testing image dataset had a categorical expression level of 1+, 137 ROIs had an expression of 2+, and 83 ROIs had an expression of 3+.
METHODS
The two algorithms used in our study for the automated quantitative assessment of HER2/neu were: (a) A commercial algorithm (membrane v9 algorithm, Aperio Technologies, Vista, California), hereinafter referred to as Algorithm 1 and (b) a color histogram-based algorithm previously developed by Keller et al.,[22] hereinafter referred to as Algorithm 2. Each of the two algorithms was used to classify the ROIs in each of the datasets acquired with the three WSI systems in terms of HER2/neu expression. The two algorithms are presented in this section along with a description of our statistical analysis methodology.
Algorithm 1: Aperio Membrane v9 Algorithm
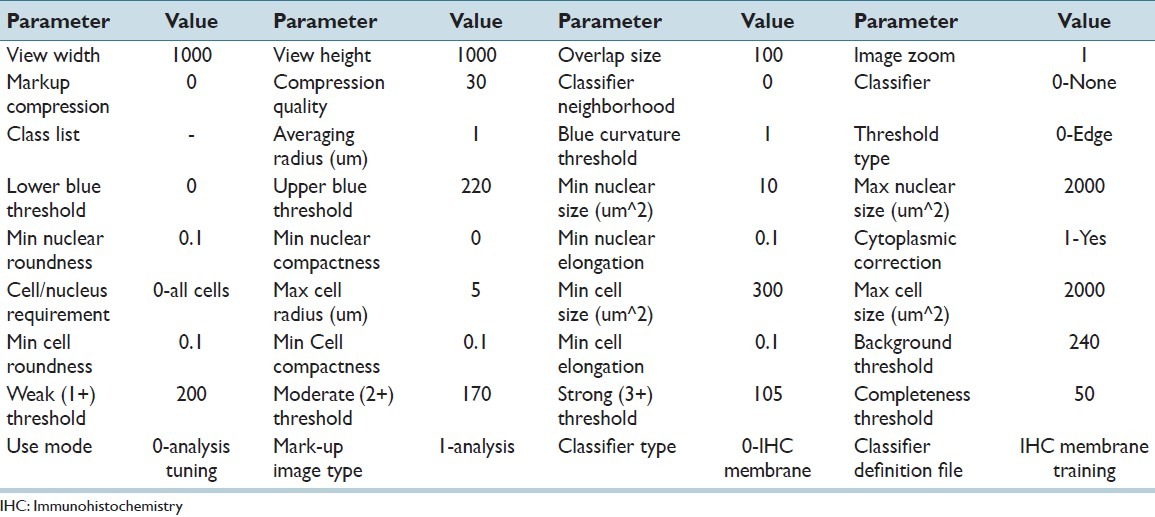
The first algorithm employed was the commercial Aperio membrane v9 algorithm. Based on the manufacturers′ description, the algorithm detects the membrane staining for individual tumor cells in selected regions and quantifies the intensity and completeness of the membrane staining. Tumor cells are individually classified as 0, 1+, 2+, and 3+ based on their membrane staining intensity and completeness. The overall HER2/neu score for a region is then calculated based on the percentages of 0, 1+, 2+, and 3+ cells according to the HER2/neu scoring scheme. The algorithm uses a large number of parameters that include: Average radius, blue curvature threshold, threshold type, lower blue threshold, upper blue threshold, min nuclear size, max nuclear size, min nuclear roundness, min nuclear compactness, and min nuclear elongation. The user can tailor the algorithm to adapt to various cell morphologies and scanning conditions. However, considering the large number of parameters and the lack of an objective method for parameter selection, manual tuning would be an ad-hoc procedure that could introduce subjective criteria. Instead, in this study we utilized the pre-tuned version of the algorithm (membrane v9) that was provided by the manufacturer. It should be noted that for clinical practice the manufacturer recommends that a pathologist consult the score of the algorithm as well as a markup image highlighting the detected cell features before finalizing a HER2/neu score. All the parameters in the algorithm version that were used in this study, Algorithm 1, are tabulated in Table 2. Algorithm 1 was run on the three main datasets, producing categorical HER2/neu scores for each ROI.
Table 2.
Parameter values of the membrane v9 algorithm for the quantitative assessment of HER2/neu expression, as used in this study
Algorithm 2: Keller Histogram-based Method
The second classifier used in our study incorporated an automated algorithm to allow re-training based on the color properties of different training sets. Technical details are presented by Keller et al.[22] Briefly, the algorithm first generated a color-palette containing the colors present in the development datasets, based on ROIs from the 13 slides that were not part of the main dataset. Palette generation was based on a fuzzy c-means clustering method, implemented using the MATLAB fuzzy toolbox. The pre-set number of colors in the palette was chosen as 128, which was previously shown to be adequate.[22] A palette was generated for each of the three WSI datasets. Based on the palette, a normalized color histogram was calculated for each image, representing the frequency that pixels in an image had each of the particular colors in the color palette. The color histogram values served as input features to a linear classifier which was trained and tested on the main datasets acquired from each of the three WSI systems using a leave one out cross validation (LOOCV) procedure. During the LOOCV procedure, all cases except one are used for algorithm training (consisting of adjusting the weights of a linear function to maximize performance) and the remaining case is used for algorithm testing. The procedure was repeated as many times as the size of the dataset so that all ROIs were used for algorithm testing. With this procedure, the greatest possible amount of data is used for algorithm training. The mean score from the pathologist panel (1-100) was used as ground truth from the pathologist panel was used during the training process. At the end of LOOCV, the testing score (algorithm output in the range of 1-100) of each ROI was transformed to a categorical value using the thresholds described previously in the manuscript (a value of less or equal to 33 corresponded to a 1+, a value between 33 and 66 corresponded to 2+, and a value larger than 66 corresponded to a 3+).
Statistical Analysis
The extracted scores from the two algorithms on data from the three WSI systems resulted in six datasets which were analyzed with the following procedures. Since absolute truth (reference standard) regarding the actual HER2/neu score was not available, agreement analysis was utilized. Two well-known measures of agreement were utilized, the Kendall’s tau-beta and percent correct agreement. Kendall’s tau-beta is a rank-based metric which calculates the difference in the fraction of concordant and discordant pairs while correcting for ties.[31] The range of Kendall’s tau_beta is (-1-1), where 1 indicates the readers are always concordant (perfect agreement),-1 indicates they are always discordant (perfect disagreement), and 0 indicates no agreement. The same metric has been used in the previously published reader study on the same dataset,[4] thus providing a useful reference of performance. Kendall’s tau-beta and the standard error in the measurement were computed based on the definitions outlined by Woolson and Clarke[32] The second figure of merit used in our analyses was percent correct agreement which was further broken down into (a) overall percent correct agreement, defined as the percentage of cases for which the scores from two distributions coincided, and (b) category-specific correct agreement (for 1+, 2+, and 3+), defined as the percentage of cases for which a specific score was observed in both distributions divided by the number of scores in that category observed in either distribution. Confidence intervals (CIs) for percent correct agreement were calculated using bootstrap analysis.
The agreement measures described above were used to quantify variability in the quantitative assessment of HER2/neu as a function of WSI system and automated algorithm, and the interaction of these variables. Moreover, the extracted HER2/neu scores were compared to those derived from a panel of seven pathologists.
RESULTS
Table 3 shows the overall raw distribution of scores for each HER2/neu category as determined by the pathologist panel, Algorithm 1, and Algorithm 2, across images generated by the three scanners. It should be noted that, based on the pathologist panel scores, this dataset is skewed toward 2+ and 3+ slides. Results show large differences between the score distributions across the WSI systems as well as between the algorithms. For example, the number of cases scored as 1+ by Algorithm 1 varied from 24 to 65 between the datasets from the three WSI systems, whereas the number of 3+ scores varied from 75 to 98. In comparison, the range in scores when Algorithm 2 was applied to the datasets was narrower, varying from 13 to 14 for 1+ and 78-83 for the 3+ category.
Table 3.
Classification score distribution in HER2/neu categories (1+, 2+, 3+) from pathologist panel, Algorithm 1 applied on image data from the three scanners, and Algorithm 2 applied on image data from the three scanners
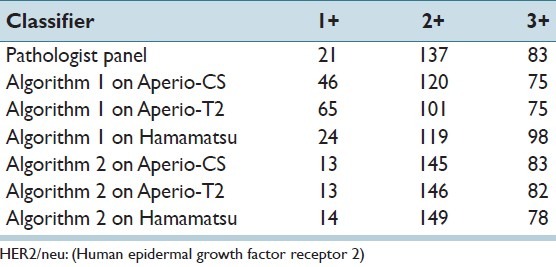
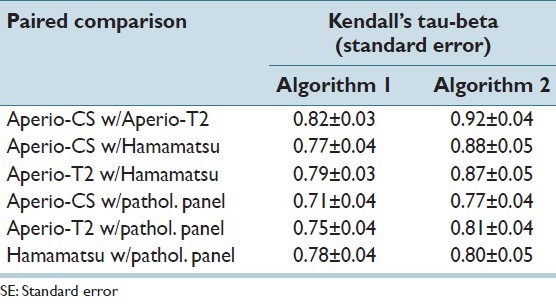
Table 4 presents pair-wise agreement results between each pair of WSI system using the Kendall’s tau-beta metric. Results were tabulated for each of the two automated algorithms. For the sake of comparison, inter-observer agreement on the same dataset was 0.70 (95% CI 0.64-0.76).[4] It can be seen from the table that: (a) Agreement between the algorithm scores for the three systems derived from any of the two algorithms was equal or better to the inter-observer agreement on the same cases, and (b) agreement between the scores from each pair of WSI systems was improved when Algorithm 2 was used.
Table 4.
Pair-wise agreement values using Kendall’s tau-beta (±SE) between algorithm classification results as well as between algorithms and scores from pathologist panel
In addition to overall agreement, it is useful to see a HER2/neu category-specific breakdown of agreement. Table 5 shows percent agreement between paired scores from the three WSI systems. Overall percent agreement is shown, as well as percent correct agreement for each of the 1+, 2+, and 3+ categories. It is shown in this table that, similarly to the Kendall’s tau-beta analysis, overall agreement was improved when Algorithm 2 was used, with the improvement being statistically significant for all three paired comparisons. The biggest, and statistically significant, improvement in agreement when using Algorithm 2 was seen for the 2+ cases. For both algorithms, the lowest agreement across all systems was observed for the 1+ category. It can also be noted that the CIs are much larger for the 1+ category, which was expected due to the small number of cases in that category.
Table 5.
Percent correct agreement between the scores derived from each algorithm applied to image data from the three scanners. Overall percent agreement along with percent agreement on each of the 1+, 2+, and 3+ categories are shown for each of Algorithm 1 and Algorithm 2
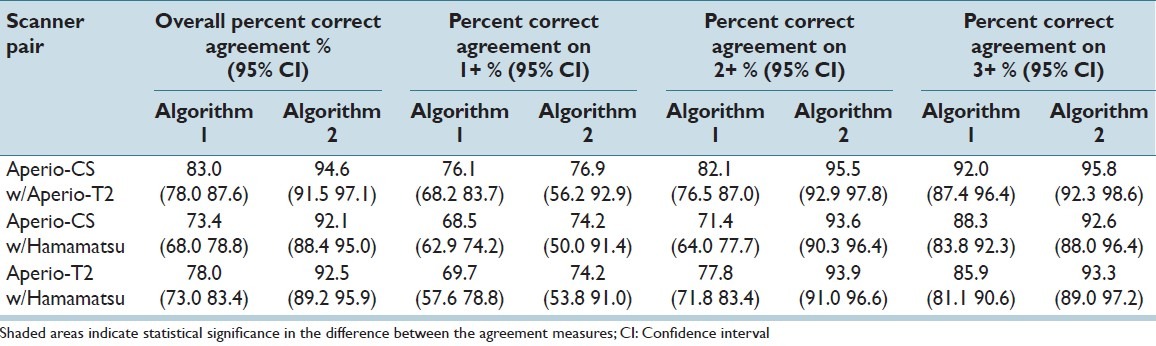
Table 6 tabulates percent correct agreement between algorithm scores and the pathologist panel scores, across the three WSI systems. It can be seen that similar to the results from Table 5, overall agreement is improved with Algorithm 2 for all scanner-panel pairs, even though results are statistically significant for only one pair. Using Algorithm 2, agreement with the pathologist panel becomes more consistent across the three WSI systems, varying in the narrow range of 83.4-86.3, whereas for Algorithm 1 agreement with the pathologist panel varied between 73.9 and 83.8. It can also be seen that agreement with the pathologists for the 1+ category was low compared to the other categories, especially for Algorithm 2, possibly due to small number of 1+ cases that were available for algorithm training. Figure 2 shows two example ROIs, one where the majority of pathologists (four out of seven) scored it as 1+ whereas Algorithm 2 scored that case as 2+ Figure 2a, and another where the majority of pathologists (six out of seven) scored it as 2+ whereas Algorithm 2 scored it as a 1+. Algorithm 2 would benefit from a larger number of 1+ examples as well as examples of 0+ which were absent in this dataset. Additionally, the performance of Algorithm 2 should improve with the addition of a feature describing membrane completeness as the one described in.[18] The algorithm currently utilizes only color information.
Table 6.
Percent correct agreement between the scores derived from each algorithm applied to image data from each of the three scanners and the scores from the pathologist panel. Overall percent agreement along with percent agreement on each of the 1+, 2+, and 3+ categories are shown for each of Algorithm 1 and Algorithm 2

Figure 2.

Example of two regions of interests, one where the majority of pathologists (four out of seven) scored it as 1+ whereas Algorithm 2 scored that case as 2+ (a), and another where the majority of pathologists (six out of seven) scored it as 2+ whereas Algorithm 2 scored it as a 1+ (b). Images were extracted at ×20
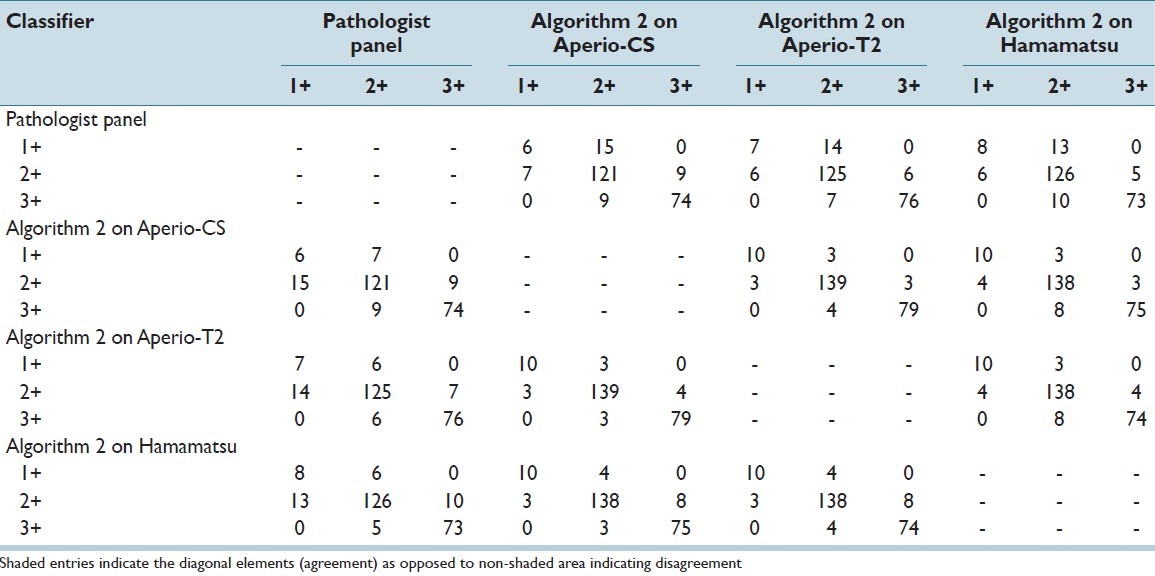
Finally, Table 7 and Table 8 show contingency tables of category specific agreement and disagreement between the scores derived using Algorithms 1 and 2 respectively, across WSI systems and the pathologist panel. When analyzed with Algorithm 1, paired comparisons showed that of all cases classified as 2+ using images from one system (Hamamatsu), 23.3% and 33.3% of them were classified as 1+ when using images from the Aperio-CS and Aperio-T2 systems respectively. Similarly, of all cases classified as 3+ using images from one system (Hamamatsu), 30.2% and 22.9% of them were classified as 2+ when using images from Aperio-CS and Aperio-T2 systems respectively. Using Algorithm 2, the percentage of cases classified as 2+ using images from one system and 1+ using images from the other two systems was reduced to 2.0% and 2.0% respectively, whereas the percentage of cases classified as 3 + using images from one system and 2+ by the other two systems was reduced to 3.8% and 5.1% respectively.
Table 7.
Contingency table of classification results comparing the scores of the pathologist panel, and Algorithm 1 applied to the three scanners
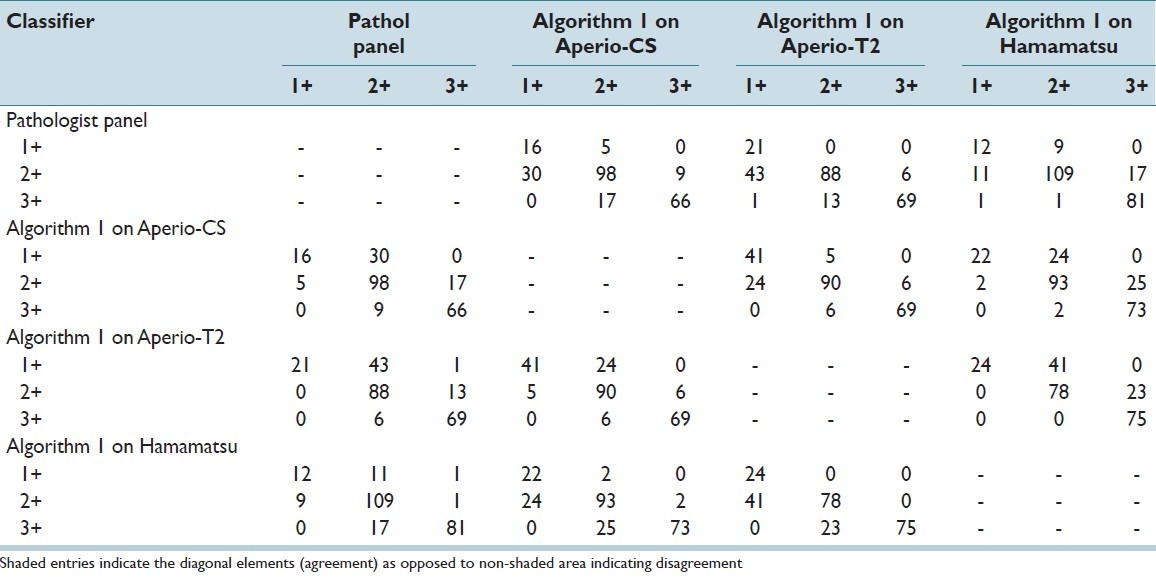
Table 8.
Contingency table of classification results comparing the scores of the pathologist panel, and Algorithm 2 applied to the three scanners
DISCUSSION
Automated image analysis for the quantitative assessment of tissue-based biomarkers is becoming an increasingly significant part of pathology practice. In a survey conducted in 2008 in the United States, image analysis of HER2/neu was utilized by 33% of 720 participating pathology laboratories.[33] This percentage has likely increased since then, considering the clearance of more image analysis software by the U.S. FDA and the number of digital pathology systems installed worldwide. The use of image analysis for quantitative IHC is already predominant in some European countries; an example is Kalmar County Hospital in Sweden, which has reported scanning an estimated 120,000 histopathology slides over the last 2 years and using digital pathology in 75% of their diagnostic cases.[1,34] For image analysis to be effective in clinical practice, results need to be reproducible across different digital pathology systems and at least as good as the pathologist. The results of our study showed that inter-scanner agreement in the assessment of HER2/neu for breast cancer in selected fields of view when analyzed with any of the two algorithms examined in this study was equal or better than the inter-observer agreement previously reported on the same set of data. Still, several discrepancies were observed between the algorithm results on data from different scanners. Such discrepancies could lead to unnecessary follow-up exams, or mistrust in a biomarker test. More importantly they could lead to wrong decisions regarding cancer treatment for individual patients. Our study showed that the discrepancies mentioned above were significantly reduced when, compared to a particular commercial algorithm with pre-set parameters, an alternative algorithm was used that incorporated an objective re-training procedure. By re-training, the algorithm adjusted its parameters to the different imaging properties of the three WSI systems, specifically color differences in this case, resulting in more consistent results.
It is understood that the commercial algorithm examined in this study could be trained or tuned, possibly resulting in an improved performance. However, the large number of tunable parameters listed in Table 2, and the lack of a procedure for objective algorithm training would make it very difficult to train the algorithm in a reproducible manner. Our study supports the inclusion of such procedures so that image analysis algorithms can adjust to differences in image properties between different systems. It is also understood that other algorithms or algorithm versions from Aperio might incorporate objective training procedures. The algorithms used in our study were used as examples of the importance of objective algorithm training and parameter optimization.
In addition to reduced inter-scanner variability, the use of the Keller algorithm incorporating parameter re-training resulted in improved overall agreement of the algorithm results with the consensus scores from the pathologist panel. It should be noted that using either image analysis algorithm inter-scanner variability was less or equal to inter-reader variability reported in a previous study on the same data,[4] further re-enforcing the potential for efficient use of automated image analysis.
The discrepancies observed in this study between the images produced from the three different WSI systems could be attributed to a number of different factors, including different calibration methods, light bulb age, optics, and camera firmware with embedded compression or color management algorithms. It was beyond the scope of this work to quantify the effect of such factors with a thorough technical assessment. In related work, we are currently developing color phantoms to enable such comparisons toward objective technical assessment of digital pathology systems.
Other limitations of our study included a relatively small number of 1+ cases and the analysis of only selected fields of view as opposed to whole slide images. In addition, the pathologist panel scores used in algorithm training and in some of our analyses were derived from images extracted from only one of the three WSI systems (Aperio-T2), which may have led to a certain bias in creating a reference standard. Finally, the performance of Algorithm1 on the Hamamatsu image data might have been affected by the fact that the algorithm was originally developed and optimized using images digitized with Aperio scanners that may share unique characteristics compared to image acquired using Hamamatsu scanners. Despite the aforementioned limitations and technical differences of the three scanners, our study shows that for this particular task of HER2/neu assessment, an algorithm that incorporated an objective procedure for re-training was able to maintain a similar performance across data from the three scanners. This finding might not apply for different pathology tasks such as primary diagnosis with digital pathology.
CONCLUSION
Our study supports the use of objective algorithm training to account for differences in image properties between WSI systems. With appropriate algorithm parameter optimization, image analysis can provide valuable assistance in the quantitative assessment of tissue-based biomarkers.
ACKNOWLEDGMENT
The authors would like to thank Drs. Brandon Gallas, Weijie Chen, and Adam Wunderlich at the Division of Imaging and Applied Mathematics, OSEL/CDRH/FDA for useful conversations and reviews of this manuscript. The authors would also like to acknowledge the support of the U.S. Food and Drug Administration’s Office of Women’s Health.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2013/4/1/19/115879
REFERENCES
- 1.Pantanowitz L, Valenstein PN, Evans AJ, Kaplan KJ, Pfeifer JD, Wilbur DC, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform. 2011;2:36. doi: 10.4103/2153-3539.83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstein RS, Graham AR, Richter LC, Barker GP, Krupinski EA, Lopez AM, et al. Overview of telepathology, virtual microscopy, and whole slide imaging: Prospects for the future. Hum Pathol. 2009;40:1057–69. doi: 10.1016/j.humpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Al-Janabi S, Huisman A, Van Diest PJ. Digital pathology: Current status and future perspectives. Histopathology. 2012;61:1–9. doi: 10.1111/j.1365-2559.2011.03814.x. [DOI] [PubMed] [Google Scholar]
- 4.Gavrielides MA, Gallas BD, Lenz P, Badano A, Hewitt SM. Observer variability in the interpretation of HER2/neu immunohistochemical expression with unaided and computer-aided digital microscopy. Arch Pathol Lab Med. 2011;135:233–42. doi: 10.1043/1543-2165-135.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom K, Harrington D. Enhanced accuracy and reliability of HER-2/neu immunohistochemical scoring using digital microscopy. Am J Clin Pathol. 2004;121:620–30. doi: 10.1309/Y73U-8X72-B68T-MGH5. [DOI] [PubMed] [Google Scholar]
- 6.Nassar A, Cohen C, Agersborg SS, Zhou W, Lynch KA, Albitar M, et al. Trainable immunohistochemical HER2/neu image analysis: A multisite performance study using 260 breast tissue specimens. Arch Pathol Lab Med. 2011;135:896–902. doi: 10.5858/2010-0418-OAR1.1. [DOI] [PubMed] [Google Scholar]
- 7.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American society of clinical oncology/College of American pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 8.Werner M, Chott A, Fabiano A, Battifora H. Effect of formalin tissue fixation and processing on immunohistochemistry. Am J Surg Pathol. 2000;24:1016–9. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 9.Thomson TA, Hayes MM, Spinelli JJ, Hilland E, Sawrenko C, Phillips D, et al. HER-2/neu in breast cancer: Interobserver variability and performance of immunohistochemistry with 4 antibodies compared with fluorescent in situ hybridization. Mod Pathol. 2001;14:1079–86. doi: 10.1038/modpathol.3880440. [DOI] [PubMed] [Google Scholar]
- 10.Gancberg D, Järvinen T, di Leo A, Rouas G, Cardoso F, Paesmans M, et al. Evaluation of HER-2/NEU protein expression in breast cancer by immunohistochemistry: An interlaboratory study assessing the reproducibility of HER-2/NEU testing. Breast Cancer Res Treat. 2002;74:113–20. doi: 10.1023/a:1016146130767. [DOI] [PubMed] [Google Scholar]
- 11.Zu Y, Steinberg SM, Campo E, Hans CP, Weisenburger DD, Braziel RM, et al. Validation of tissue microarray immunohistochemistry staining and interpretation in diffuse large B-cell lymphoma. Leuk Lymphoma. 2005;46:693–701. doi: 10.1080/10428190500051844. [DOI] [PubMed] [Google Scholar]
- 12.Hall BH, Ianosi-Irimie M, Javidian P, Chen W, Ganesan S, Foran DJ. Computer-assisted assessment of the human epidermal growth factor receptor 2 immunohistochemical assay in imaged histologic sections using a membrane isolation algorithm and quantitative analysis of positive controls. BMC Med Imaging. 2008;8:11. doi: 10.1186/1471-2342-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi AS, Sharangpani GM, Porter K, Keyhani S, Morrison C, Basu AS, et al. Semi-automated imaging system to quantitate Her-2/neu membrane receptor immunoreactivity in human breast cancer. Cytometry A. 2007;71:273–85. doi: 10.1002/cyto.a.20374. [DOI] [PubMed] [Google Scholar]
- 14.Skaland I, Øvestad I, Janssen EA, Klos J, Kjellevold KH, Helliesen T, et al. Comparing subjective and digital image analysis HER2/neu expression scores with conventional and modified FISH scores in breast cancer. J Clin Pathol. 2008;61:68–71. doi: 10.1136/jcp.2007.046763. [DOI] [PubMed] [Google Scholar]
- 15.Lehr HA, Jacobs TW, Yaziji H, Schnitt SJ, Gown AM. Quantitative evaluation of HER-2/neu status in breast cancer by fluorescence in situ hybridization and by immunohistochemistry with image analysis. Am J Clin Pathol. 2001;115:814–22. doi: 10.1309/AJ84-50AK-1X1B-1Q4C. [DOI] [PubMed] [Google Scholar]
- 16.Matkowskyj KA, Schonfeld D, Benya RV. Quantitative immunohistochemistry by measuring cumulative signal strength using commercially available software photoshop and matlab. J Histochem Cytochem. 2000;48:303–12. doi: 10.1177/002215540004800216. [DOI] [PubMed] [Google Scholar]
- 17.Hatanaka Y, Hashizume K, Kamihara Y, Itoh H, Tsuda H, Osamura RY, et al. Quantitative immunohistochemical evaluation of HER2/neu expression with HercepTestTM in breast carcinoma by image analysis. Pathol Int. 2001;51:33–6. doi: 10.1046/j.1440-1827.2001.01162.x. [DOI] [PubMed] [Google Scholar]
- 18.Masmoudi H, Hewitt SM, Petrick N, Myers KJ, Gavrielides MA. Automated quantitative assessment of HER-2/neu immunohistochemical expression in breast cancer. IEEE Trans Med Imaging. 2009;28:916–25. doi: 10.1109/TMI.2009.2012901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mofidi R, Walsh R, Ridgway PF, Crotty T, McDermott EW, Keaveny TV, et al. Objective measurement of breast cancer oestrogen receptor status through digital image analysis. Eur J Surg Oncol. 2003;29:20–4. doi: 10.1053/ejso.2002.1373. [DOI] [PubMed] [Google Scholar]
- 20.Divito KA, Berger AJ, Camp RL, Dolled-Filhart M, Rimm DL, Kluger HM. Automated quantitative analysis of tissue microarrays reveals an association between high Bcl-2 expression and improved outcome in melanoma. Cancer Res. 2004;64:8773–7. doi: 10.1158/0008-5472.CAN-04-1387. [DOI] [PubMed] [Google Scholar]
- 21.Elhafey AS, Papadimitriou JC, El-Hakim MS, El-Said AI, Ghannam BB, Silverberg SG. Computerized image analysis of p53 and proliferating cell nuclear antigen expression in benign, hyperplastic, and malignant endometrium. Arch Pathol Lab Med. 2001;125:872–9. doi: 10.5858/2001-125-0872-CIAOPA. [DOI] [PubMed] [Google Scholar]
- 22.Keller B, Chen W, Gavrielides MA. Quantitative assessment and classification of tissue-based biomarker expression with color content analysis. Arch Pathol Lab Med. 2012;136:539–50. doi: 10.5858/arpa.2011-0195-OA. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Saboorian MH, Frenkel EP, Haley BB, Siddiqui MT, Gokaslan S, et al. Assessment of HER-2/neu status in breast cancer. Automated Cellular Imaging System (ACIS)-assisted quantitation of immunohistochemical assay achieves high accuracy in comparison with fluorescence in situ hybridization assay as the standard. Am J Clin Pathol. 2001;116:495–503. doi: 10.1309/TMUW-G4WB-LXJ2-FUDN. [DOI] [PubMed] [Google Scholar]
- 24.Ciampa A, Xu B, Ayata G, Baiyee D, Wallace J, Wertheimer M, et al. HER-2 status in breast cancer: Correlation of gene amplification by FISH with immunohistochemistry expression using advanced cellular imaging system. Appl Immunohistochem Mol Morphol. 2006;14:132–7. doi: 10.1097/01.pai.0000150516.75567.13. [DOI] [PubMed] [Google Scholar]
- 25.Tawfik OW, Kimler BF, Davis M, Donahue JK, Persons DL, Fan F, et al. Comparison of immunohistochemistry by automated cellular imaging system (ACIS) versus fluorescence in-situ hybridization in the evaluation of HER-2/neu expression in primary breast carcinoma. Histopathology. 2006;48:258–67. doi: 10.1111/j.1365-2559.2005.02322.x. [DOI] [PubMed] [Google Scholar]
- 26.Luftner D, Henschke P, Kafka A, Anagnostopoulos I, Wiechen K, Geppert R, et al. Discordant results obtained for different methods of HER-2/neu testing in breast cancer - A question of standardization, automation and timing. Int J Biol Markers. 2004;19:1–13. doi: 10.1177/172460080401900101. [DOI] [PubMed] [Google Scholar]
- 27.Cregger M, Berger AJ, Rimm DL. Immunohistochemistry and quantitative analysis of protein expression. Arch Pathol Lab Med. 2006;130:1026–30. doi: 10.5858/2006-130-1026-IAQAOP. [DOI] [PubMed] [Google Scholar]
- 28.Conway C, Dobson L, O′Grady A, Kay E, Costello S, O’shea D. Virtual microscopy as an enabler of automated/quantitative assessment of protein expression in TMAs. Histochem Cell Biol. 2008;130:447–63. doi: 10.1007/s00418-008-0480-1. [DOI] [PubMed] [Google Scholar]
- 29.Gu J, Ogilvie RW. Virtual microscopy and virtual slides in teaching, diagnosis, and research. Boca Raton, Florida: CRC Press; 2005. [Google Scholar]
- 30.Rojo MG, García GB, Mateos CP, García JG, Vicente MC. Critical comparison of 31 commercially available digital slide systems in pathology. Int J Surg Pathol. 2006;14:285–30. doi: 10.1177/1066896906292274. [DOI] [PubMed] [Google Scholar]
- 31.Kendall M. Rank correlation methods. London, UK: Charles Griffin and Co. Ltd; 1948. [Google Scholar]
- 32.Woolson RF, Clarke WR. Statistical Methods for the Analysis of Biomedical Data. New York: Wiley; 1987. pp. 260–5. [Google Scholar]
- 33.Nakhleh RE, Grimm EE, Idowu MO, Souers RJ, Fitzgibbons PL. Laboratory compliance with the American Society of Clinical Oncology/college of American Pathologists guidelines for human epidermal growth factor receptor 2 testing: A College of American Pathologists survey of 757 laboratories. Arch Pathol Lab Med. 2010;134:728–34. doi: 10.5858/134.5.728. [DOI] [PubMed] [Google Scholar]
- 34.Thorstenson S. Digital pathology system. Case study. Adv Lab. 2010;19:69. [Google Scholar]


