Abstract
Halorhodospira halophila is among the most halophilic organisms known. It is an obligately photosynthetic and anaerobic purple sulfur bacterium that exhibits autotrophic growth up to saturated NaCl concentrations. The type strain H. halophila SL1 was isolated from a hypersaline lake in Oregon. Here we report the determination of its entire genome in a single contig. This is the first genome of a phototrophic extreme halophile. The genome consists of 2,678,452 bp, encoding 2,493 predicted genes as determined by automated genome annotation. Of the 2,407 predicted proteins, 1,905 were assigned to a putative function. Future detailed analysis of this genome promises to yield insights into the halophilic adaptations of this organism, its ability for photoautotrophic growth under extreme conditions, and its characteristic sulfur metabolism.
Keywords: halophile, saturated salt, sulfur metabolism, purple sulfur bacterium, phototrophic
Introduction
Halorhodospira halophila is an anoxygenic photosynthetic halophile that was isolated from salt-encrusted mud along the shore of Summer Lake in Oregon [1], and from the hypersaline Wadi Natrun lakes in Egypt [2]. The original name of this organism, Ectothiorhodospira halophila, was modified to Halorhodospira halophila when the genus Ectothiorhodospira was divided into two genera (Ectothiorhodospira and Halorhodospira), and E. halophila was reclassified as a member of the genus Halorhodospira, serving as the type species of the new genus [3]. Over the last decade, the genomes of a number of extremely halophilic Archaea have been sequenced and analyzed, including Halobacterium salinarum [4,5], Haloarcula marismortui [6], Natronomonas pharaonis [7], and Haloquadratum walsbyi [8]. In addition, the genomes of three halophilic Bacteria have become available: Salinibacter ruber [9], Halothermothrix orenii [10], and ‘Halanaerobium hydrogenoformans’ [11]. All of these organisms are obligate chemotrophs. Thus, H. halophila is the first phototrophic extreme halophile to have its genome sequence determined and analyzed. In contrast to other extreme halophiles that grow well in saturated salt concentrations, H. halophila has a high flexibility with respect to the salt concentrations that it tolerates, and grows optimally at all NaCl concentrations from 15% to 35%, with growth down to 3.5% NaCl [12]. In contrast, the above extremely halophilic archaea and S. ruber require 15% NaCl for growth.
H. halophila is of significant interest because it is an obligately anaerobic purple sulfur bacterium, and among the most halophilic organisms known [13]. To date, genome sequences are available for two phototrophic purple sulfur bacteria, Allochromatium vinosum DSM 180 and the H. halophila SL1 genome reported here. H. halophila has very few growth requirements. However, it does need reduced sulfur compounds for growth, as does A. vinosum [14]. Its pathways for both photosynthetic electron transfer [15-17] and nitrogen fixation [18] have attracted attention. In addition, H. halophila contains photoactive yellow protein [19,20]. This is the first member of a novel class of blue light receptors, and triggers a negative phototaxis response in H. halophila [21]. The photoactive yellow protein (PYP) from H. halophila has been studied extensively for its biophysical characteristics [22-24].
The sulfur metabolism of H. halophila is unusual, resulting in the transient accumulation of extracellular sulfur globules via metabolic pathways that are not yet fully resolved [14]. While purple non-sulfur phototrophs such as Rhodobacter sphaeroides and Rhodospirillum rubrum use organic compounds like malate as electron donors, H. halophila obtains electrons from reduced sulfur compounds. The genome sequence of H. halophila promises to reveal insights into its adaptations to hypersaline environments, and to allow a better understanding of its unique combination of metabolic capabilities, combining properties from extreme halophiles, anoxygenic phototrophs, and purple sulfur bacteria.
Classification and features
H. halophila belongs to the Gammaproteobacteria [3] (Table 1). The 16S rRNA gene sequence of H. halophila SL1 reveals closer relationships with H. halochloris and Alkalilimnicola ehrlichii, the other representatives of the Ectothiorhodospiraceae (Figure 1), than with A. vinosum, a purple sulfur bacterium in the Chromatiaceae family, and the haloalkaliphilic chemolithoautotrophic Thioalkalivibrio strains.
Table 1. Classification and general features of H. halophila SL1 according to the MIGS recommendations [25].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [26] | |
| Phylum Proteobacteria | TAS [27] | ||
| Class Gammaproteobacteria | TAS [28,29] | ||
| Order Chromatiales | TAS [28,30] | ||
| Family Ectothiorhodospiraceae | TAS [31] | ||
| Genus Halorhodospira | TAS [32-34] | ||
| Species Halorhodospira halophila | TAS [32,33] | ||
| MIGS-7 | Subspecific genetic lineage | DSM 244T | |
| Gram stain | negative | NAS | |
| Cell shape | spiral | TAS [1] | |
| Motility | motile | TAS [1] | |
| Sporulation | non-sporulating | NAS | |
| Temperature range | mesophilic | NAS | |
| Optimum temperature | 47°C | TAS [1] | |
| Carbon source | CO2, succinate, acetate | TAS [35] | |
| Energy source | photosynthesis | TAS [1] | |
| MIGS-6 | Habitat | salt lake mud | TAS [1] |
| MIGS-6.3 | Salinity | Extreme halophile | TAS [1] |
| MIGS-22 | Oxygen | anaerobe | TAS [1] |
| MIGS-15 | Biotic relationship | free living | NAS |
| MIGS-14 | Pathogenicity | none | NAS |
| MIGS-4 | Geographic location | Summer Lake, Lake County, OR | TAS [1] |
| MIGS-5 | Sample collection time | about 1967 | TAS [1] |
| MIGS-4.1 | Latitude | not reported | |
| MIGS-4.2 | Longitude | not reported | |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
aEvidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [36]. If the evidence code is IDA, then the property should have been directly observed, for the purpose of this specific publication, for a live isolate by one of the authors, or an expert or reputable institution mentioned in the acknowledgements.
Figure 1.

Phylogram representation of a phylogenetic tree highlighting the position of Halorhodospira halophila strain SL1 relative to other organisms of interest, including members of the Ectothiorhodospiraceae, as well as additional strains that were included for comparison purposes, based on environmental and functional considerations. The strains (type=T) and their corresponding GenBank accession numbers (and coordinates) for 16S rRNA genes are: H. halophila strain SL1T, CP00544:380025-381562; Alkalilimnicola ehrlichii strain MLHE-1, CP00453:369818-369894; Thioalkalivibrio sp. HL-EbGR7, CP001339:2548250-2549775; Thioalkalivibrio sp. K90mix, CP001905:423231-424758; Allochromatium vinosum DSM 180T, CP001896:112452-113967; Ectothiorhodospira halochloris M59152; Burkholderia phytofirmans PsJN, CP001052:1541578-1543101; Desulfovibrio vulgaris subsp. vulgaris strain Hildenborough, AE017285:105921-107426; Rhodobacter sphaeroides 2.4.1, CP000143:1-1464; Rhodospirillum rubrum ATCC 11170, CP000230: 192528-194004; Escherichia coli B strain REL606, CP000819: 226609-228150. The 16S rRNA sequences were aligned by MUSCLE [37]. The tree was determined by the maximum likelihood model of PhyML [38] and rendered with TreeDyn [39], using the “one click” pipeline of the Phylogeny.fr web resource [40].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing to better understand its halophilic adaptations, its unusual sulfur metabolism, its photosynthetic pathways, and to provide a framework for better understanding signaling pathways for photoactive yellow protein. The complete genome sequence has been deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). Table 2 presents the project information and its association with MIGS version 2.0 compliance [25].
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | 40kb, 8kb, 3kb |
| MIGS-29 | Sequencing platforms | Sanger |
| MIGS-31.2 | Fold coverage | 12× |
| MIGS-30 | Assemblers | phrap |
| MIGS-32 | Gene calling method | Critica |
| Genbank ID | CP000544 | |
| Genbank Date of Release | January 12, 2012 | |
| GOLD ID | Gc00492 | |
| Project relevance | extremophile |
Growth conditions and DNA isolation
H. halophila SL1 strain DSM 44T was obtained from Deutsche Sammlung vor Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany, and were grown in DSMZ 253 medium. The cells were grown anaerobically and photosynthetically by placing them in 20 ml glass culture tubes completely filled with growth medium and sealed with screw caps. The tubes were kept at 42ºC in a water bath and illuminated with 70 W tungsten light bulbs. Chromosomal DNA was purified from the resulting cell cultures using the CTAB procedure.
Genome sequencing and assembly
The random shotgun method was used in sequencing the genome of H. halophila SL1. Large (40 kb), median (8 kb) and small (3 kb) insert random sequencing libraries were sequenced for this genome project with an average success rate of 88% and average high-quality read lengths of 750 nucleotides. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher (unpublished, C. Han) or by transposon bombing of bridging clones (EZ-Tn5 <P6Kyori/KAN-2> Tnp Transposome kit, Epicentre Biotechnologies). Gaps between contigs were closed by editing, custom primer walks or PCR amplification. The completed genome sequence of H. halophila SL1 contains 36,035 reads, achieving an average of 12-fold sequence coverage per base with error rate less than 1 in 100,000.
Genome annotation
Identification of putative protein-encoding genes and initial automated annotation of the genome was performed by the Oak Ridge National Laboratory genome annotation pipeline. Additional gene prediction analysis and functional annotation was performed within the IMG platform [41].
Genome properties
The genome is 2,678,452 bp long and comprises one circular chromosome with 67% GC content (Figure 2). For the main chromosome, 2,493 genes were predicted, 2,407 of which are protein-coding genes. A total of 1,905 of protein coding genes were assigned to a putative function, with the remaining annotated as hypothetical proteins. In addition, 31 pseudo genes were identified. The properties and the statistics of the genome are summarized in Tables 3-4.
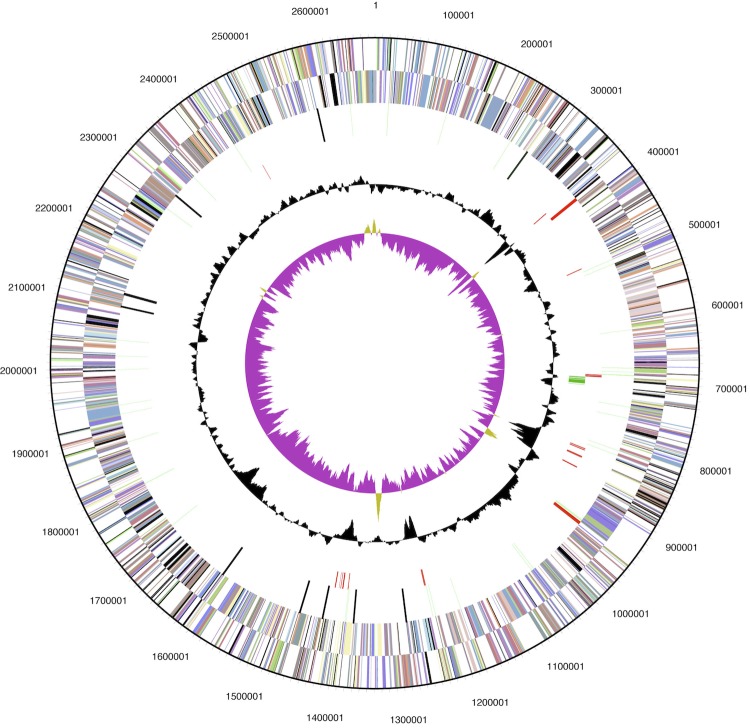
Figure 2.
Graphical circular map of the genome. From outside to the center: Circle 1, genes on forward strand (colored by COG categories); Circle 2, genes on reverse strand (colored by COG categories); Circle 3, RNA genes (tRNAs green, rRNAs red, other RNAs black); Circle 4, mobile element genes; Circle 5, CRISPR-associated protein genes; Circle 6, GC content; Circle 7, GC skew.
Table 3. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 2,678,452 | 100.00% |
| DNA coding region (bp) | 2,437,391 | 91% |
| DNA G+C content (bp) | 1,794,562 | 67% |
| Total genes | 2493 | |
| RNA genes | 63 | 2.65% |
| rRNA operons | 2 | |
| Protein-coding genes | 2,407 | 96.55% |
| Pseudo genes | 31 | 1.24% |
| Genes in paralog clusters | 204 | 8.19% |
| Genes assigned to COGs | 1,457 | 58.44% |
| Genes with signal peptides | 499 | 20.02% |
| Genes with transmembrane helices | 554 | 22.22% |
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 147 | 5.9 | Translation |
| A | 1 | 0.0 | RNA processing and modification |
| K | 86 | 3.5 | Transcription |
| L | 125 | 5.0 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 36 | 1.4 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 29 | 1.2 | Defense mechanisms |
| T | 156 | 6.3 | Signal transduction mechanisms |
| M | 144 | 5.8 | Cell wall/membrane biogenesis |
| N | 93 | 3.7 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 80 | 3.2 | Intracellular trafficking and secretion |
| O | 103 | 4.1 | Posttranslational modification, protein turnover, chaperones |
| C | 168 | 6.7 | Energy production and conversion |
| G | 76 | 3.1 | Carbohydrate transport and metabolism |
| E | 158 | 6.3 | Amino acid transport and metabolism |
| F | 46 | 1.9 | Nucleotide transport and metabolism |
| H | 152 | 6.1 | Coenzyme transport and metabolism |
| I | 72 | 2.9 | Lipid transport and metabolism |
| P | 122 | 4.9 | Inorganic ion transport and metabolism |
| Q | 37 | 1.5 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 222 | 8.9 | General function prediction only |
| S | 167 | 6.7 | Function unknown |
| - | 493 | 19.8 | Not in COGs |
a The total is based on the total number of protein coding genes in the annotated genome.
Conclusion
H. halophila is among the most halophilic eubacteria known. Further analysis and characterization of its genome will provide insights into the mechanisms it uses to adapt to hypersaline environments.
Acknowledgements
This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle, and Oak Ridge National Laboratory under contract DE-AC05-00OR22725.
References
- 1.Raymond JC, Sistrom WR. The isolation and preliminary characterization of a halophilic photosynthetic bacterium. Arch Mikrobiol 1967; 59:255-268 10.1007/BF00406339 [DOI] [PubMed] [Google Scholar]
- 2.Imhoff JF, Hashwa F, Trüper HG. Isolation of extremely halophilic phototrophic bacteria from the alkaline Wadi Natrun, Egypt. Arch Hydrobiol 1978; 84:381-388 [Google Scholar]
- 3.Imhoff JF, Suling J. The phylogenetic relationship among Ectothiorhodospiraceae: A reevaluation of their taxonomy on the basis of 16S rDNA analyses. Arch Microbiol 1996; 165:106-113 10.1007/s002030050304 [DOI] [PubMed] [Google Scholar]
- 4.Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, et al. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci USA 2000; 97:12176-12181 10.1073/pnas.190337797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeiffer F, Schuster SC, Broicher A, Falb M, Palm P, Rodewald K, Ruepp A, Soppa J, Tittor J, Oesterhelt D. Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 2008; 91:335-346 10.1016/j.ygeno.2008.01.001 [DOI] [PubMed] [Google Scholar]
- 6.Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, et al. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res 2004; 14:2221-2234 10.1101/gr.2700304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falb M, Pfeiffer F, Palm P, Rodewald K, Hickmann V, Tittor J, Oesterhelt D. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res 2005; 15:1336-1343 10.1101/gr.3952905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, Pfeiffer F, Oesterhelt D. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 2006; 7:169. 10.1186/1471-2164-7-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mongodin EF, Nelson KE, Daugherty S, Deboy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, et al. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA 2005; 102:18147-18152 10.1073/pnas.0509073102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavromatis K, Ivanova N, Anderson I, Lykidis A, Hooper SD, Sun H, Kunin V, Lapidus A, Hugenholtz P, Patel B, et al. Genome analysis of the anaerobic thermohalophilic bacterium Halothermothrix orenii. PLoS ONE 2009; 4:e4192 10.1371/journal.pone.0004192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown SD, Begemann MB, Mormile MR, Wall JD, Han CS, Goodwin LA, Pitluck S, Land ML, Hauser LJ, Elias DA. Complete genome sequence of the haloalkaliphilic, hydrogen-producing bacterium Halanaerobium hydrogeniformans. J Bacteriol 2011; 193:3682-3683 10.1128/JB.05209-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deole R, Challacombe J, Raiford DW, Hoff WD. An Extremely Halophilic Proteobacterium Combines a Highly Acidic Proteome with a Low Cytoplasmic Potassium Content. J Biol Chem 2012. [DOI] [PMC free article] [PubMed]
- 13.Ollivier B, Caumette P, Garcia JL, Mah RA. Anaerobic bacteria from hypersaline environments. Microbiol Rev 1994; 58:27-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frigaard NU, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol 2009; 54:103-200 10.1016/S0065-2911(08)00002-7 [DOI] [PubMed] [Google Scholar]
- 15.Leguijt T, Engels PW, Crielaard W, Albracht SPJ, Hellingwerf KJ. Abundance, subunit composition, redox properties, and catalytic activity of the cytochrome bc1 complex from alkaliphilic and halophilic, photosynthetic members of the family Ectothiorhodospiraceae. J Bacteriol 1993; 175:1629-1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leguijt T, Hellingwerf KJ. Characterization of reaction center antenna complexes from bacteriochlorophyll a containing Ectothiorhodospiraceae. Biochim Biophys Acta 1991; 1057:353-360 10.1016/S0005-2728(05)80147-1 [DOI] [Google Scholar]
- 17.Lieutaud C, Alric J, Bauzan M, Nitschke W, Schoepp-Cothenet B. Study of the high-potential iron sulfur protein in Halorhodospira halophila confirms that it is distinct from cytochrome c as electron carrier. Proc Natl Acad Sci USA 2005; 102:3260-3265 10.1073/pnas.0407768102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuihiji H, Yamazaki Y, Kamikubo H, Imamoto Y, Kataoka M. Cloning and characterization of nif structural and regulatory genes in the purple sulfur bacterium, Halorhodospira halophila. J Biosci Bioeng 2006; 101:263-270 10.1263/jbb.101.263 [DOI] [PubMed] [Google Scholar]
- 19.Meyer TE. Isolation and characterization of soluble cytochromes, ferredoxins and other chromophoric proteins from the halophilic phototrophic bacterium Ectothiorhodospira halophila. Biochim Biophys Acta 1985; 806:175-183 10.1016/0005-2728(85)90094-5 [DOI] [PubMed] [Google Scholar]
- 20.Meyer TE, Yakali E, Cusanovich MA, Tollin G. Properties of a water-soluble, yellow protein isolated from a halophilic phototrophic bacterium that has photochemical activity analogous to sensory rhodopsin. Biochemistry 1987; 26:418-423 10.1021/bi00376a012 [DOI] [PubMed] [Google Scholar]
- 21.Sprenger WW, Hoff WD, Armitage JP, Hellingwerf KJ. The Eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorption spectrum of the photoactive yellow protein. J Bacteriol 1993; 175:3096-3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cusanovich MA, Meyer TE. Photoactive yellow protein: A prototypic PAS domain sensory protein and development of a common signaling mechanism. Biochemistry 2003; 42:4759-4770 10.1021/bi020690e [DOI] [PubMed] [Google Scholar]
- 23.Hellingwerf KJ, Hendriks J, Gensch T. Photoactive Yellow Protein, a new type of photoreceptor protein: Will this "yellow lab" bring us where we want to go? J Phys Chem A 2003; 107:1082-1094 10.1021/jp027005y [DOI] [Google Scholar]
- 24.Kumauchi M, Hara M, Stalcup P, Xie A, Hoff WD. Identification of six new photoactive yellow proteins: diversity and structure-function relationships in a bacterial blue light photoreceptor. Photochem Photobiol 2008; 84:956-969 10.1111/j.1751-1097.2008.00335.x [DOI] [PubMed] [Google Scholar]
- 25.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 28.Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235-2238 10.1099/ijs.0.64108-0 [DOI] [PubMed] [Google Scholar]
- 29.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 30.Imhoff J. Order I. Chromatiales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1-3. [Google Scholar]
- 31.Imhoff JF. Reassignment of the Genus Ectothiorhodospira Pelsh 1936 to a New Family, Ectothiorhodospiraceae fam. nov., and Emended Description of the Chromatiaceae Bavendamm 1924. Int J Syst Bacteriol 1984; 34:338-339 10.1099/00207713-34-3-338 [DOI] [Google Scholar]
- 32.Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 62. Int J Syst Bacteriol 1997; 47:915-916 10.1099/00207713-47-3-915 [DOI] [PubMed] [Google Scholar]
- 33.Imhoff JF, Süling J. The phylogenetic relationship among Ectothiorhodospiraceae: a reevaluation of their taxonomy on the basis of 16S rDNA analyses. Arch Microbiol 1996; 165:106-113 10.1007/s002030050304 [DOI] [PubMed] [Google Scholar]
- 34.Hirschler-Réa A, Matheron R, Riffaud C, Mouné S, Eatock C, Herbert RA, Willison JC, Caumette P. Isolation and characterization of spirilloid purple phototrophic bacteria forming red layers in microbial mats of Mediterranean salterns: description of Halorhodospira neutriphila sp. nov. and emendation of the genus Halorhodospira. Int J Syst Evol Microbiol 2003; 53:153-163 10.1099/ijs.0.02226-0 [DOI] [PubMed] [Google Scholar]
- 35.Raymond JC, Sistrom WR. Ectothiorhodospira halophila: a new species of the genus Ectothiorhodospira. Arch Mikrobiol 1969; 69:121-126 10.1007/BF00409756 [DOI] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003; 52:696-704 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 39.Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 2006; 7:439. 10.1186/1471-2105-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M and others. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465-9. PMID:18424797 [DOI] [PMC free article] [PubMed]
- 41.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 2012; 40(Database issue):D115-D122 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]