Abstract
There has been a marked increase in the incidence of autoimmune diseases in the last half-century. While the underlying genetic basis of this class of diseases has recently been elucidated implicating predominantly immune response genes1, changes in environmental factors must ultimately be driving this increase. The newly identified population of interleukin (IL)-17 producing CD4+ helper T cells (Th17 cells) plays a pivotal role in autoimmune diseases2. Pathogenic IL-23 dependent Th17 cells have been shown to be critical for the development of experimental autoimmune encephalomyelitis (EAE), an animal model for multiple sclerosis (MS), and genetic risk factors associated with MS are related to the IL23/Th17 pathway1, 2. However, little is known regarding the environmental factors that directly influence Th17 cells. Here we show that increased salt (sodium chloride; NaCl) concentrations found locally under physiological conditions in vivo dramatically boost the induction of murine and human Th17 cells. High-salt conditions activate the p38/MAPK pathway involving the tonicity-responsive enhancer binding protein (TonEBP/NFAT5) and the serum/glucocorticoid-regulated kinase 1 (SGK1) during cytokine-induced Th17 polarization. Gene silencing or chemical inhibition of p38/MAPK, NFAT5 or SGK1 abrogates the high-salt induced Th17 cell development. The Th17 cells generated under high-salt display a highly pathogenic and stable phenotype characterized by the up-regulation of the pro-inflammatory cytokines GM-CSF, TNFα and IL-2. Moreover, mice fed with a high-salt diet develop a more severe form of EAE, in line with augmented central nervous system infiltrating and peripherally induced antigen specific Th17 cells. Thus, increased dietary salt intake might represent an environmental risk factor for the development of autoimmune diseases through the induction of pathogenic Th17 cells.
While we have recently elucidated many of the genetic variants underlying the risk of developing autoimmune diseases1, the significant increase in disease incidence, particularly of MS and type 1 diabetes, indicate that there have been fundamental changes in the environment that cannot be related to genetic factors. Diet has long been postulated as a potential environmental risk factor for this increasing incidence of autoimmune diseases in developed countries over recent decades3. One such dietary factor, which rapidly changed along with the “western diet” and increased consumption of so called “fast foods” or processed foods, is salt (sodium chloride, NaCl)4, 5. The salt content in processed foods can be more than a 100 times higher in comparison to similar homemade meals5, 6.
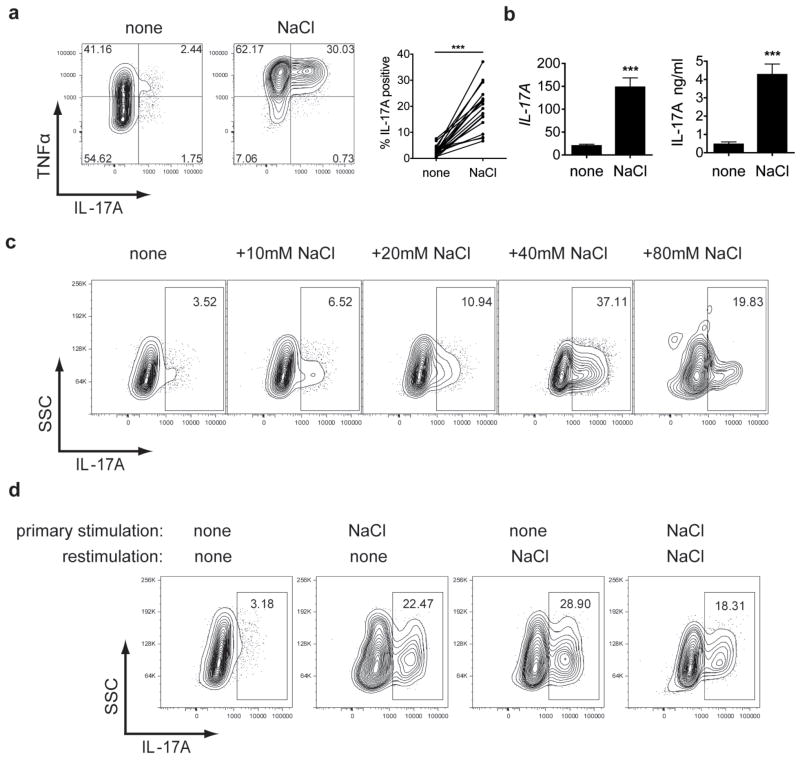
We have shown that excess NaCl uptake can affect the innate immune system7. Macrophages residing in the skin interstitium modulate local electrolyte composition in response to NaCl-mediated extracellular hypertonicity and their regulatory activity provides a buffering mechanism for salt-sensitive hypertension7. Moreover, blockade of the renin-angiotensin system can modulate immune responses and affect EAE8, 9. Thus to investigate whether increased NaCl intake might have a direct effect on CD4+ T cell populations and therefore represents a risk factor for autoimmune diseases, we investigated the effect of NaCl on the in vitro differentiation of human Th17 cells. We induced hypertonicity by increasing NaCl by 10–40mM (high-salt) in the culture medium and thus mimicked concentrations that could be found in the interstitium of animals fed a high-salt diet7. As we previously reported, Th17 promoting conditions for naïve CD4 cells only induced a mild Th17 phenotype10. Surprisingly, stimulation under increased NaCl concentrations dramatically induces naïve CD4 cell expression of IL-17A as determined by flow cytometry (Fig. 1a) or by quantitative PCR with reverse transcription (qRT-PCR) and enzyme-linked immunosorbent assay (ELISA) (Fig. 1b). The effect was dose dependent and an optimum of IL-17A induction was achieved by adding 40mM NaCl in the presence of Th17 inducing cytokines (TGF-β1/IL-1β/IL-6/IL-21/IL-23) (Fig. 1c and Supplementary Fig. 1). As expected, TNFα was also induced11 and increasing salt concentrations further led to cell death (data not shown). Nevertheless, adding 40mM NaCl was tolerated by CD4 cells with little impact on growth or apoptosis (Supplementary Fig. 2). We then examined whether the nature of cation, anion, or osmolarity drives the increases in IL-17A secretion. We found that adding 40mM sodium gluconate delivered an almost similar degree of Th17 induction, while mannitol or MgCl2 had only a slight effect. Moreover, 80mM urea, an osmolyte able to pass through cell membranes, had no effect (Supplementary Fig. 3). Thus, the sodium cation was critical for IL-17A induction. We next examined the stability of the salt-induced effect. Naïve CD4 cells that were initially stimulated under high-salt conditions continued to express increased amounts of IL-17A if restimulated under normal salt conditions but could not be further induced with additional salt restimulation (Fig. 1d). This is consistent with the observation that only naïve but not memory CD4 cells respond efficiently to increased salt concentrations (Supplementary Fig. 4). The high-salt effect was also observed when Th17 cells were induced by antigen specific stimulation (Supplementary Fig. 5)12. Furthermore, the effect was largely specific for Th17 cells, since we did not observe comparable outcomes on differentiation of Th1 or Th2 cells (Supplementary Fig. 6).
Figure 1. Sodium Chloride promotes the stable induction of Th17 cells.
a, Naïve CD4 cells were differentiated into Th17 cells in the presence (NaCl) or absence (none) of additional 40mM NaCl and analysed by flow cytometry (FACS) for IL-17A (n=20). b, IL-17A expression was measured by qRT-PCR (left panel, n=10) and ELISA (right panel, n=5). c, Cells were stimulated as in a) under the indicated increased NaCl concentrations and analysed by FACS (one representative experiment of five is shown). d, Cells were stimulated as in a) and were rested in the presence of IL-2. After 1 week, cells were re-stimulated as in a) in the presence or absence of NaCl for another week and analysed by FACS (one representative experiment of five is shown).
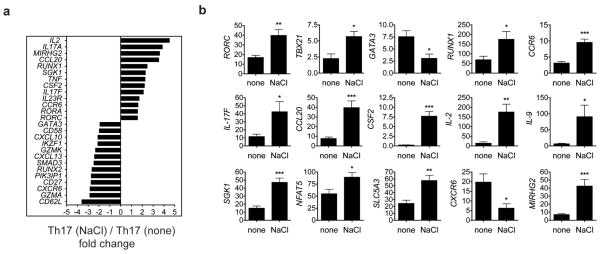
To examine the mechanisms of enhanced IL-17A induction we performed a microarray analysis of naïve CD4 T cells differentiated in the presence or absence of high-salt (Fig. 2a, Supplementary Fig. 8). These data confirmed that cells displayed a stronger Th17 phenotype under high-salt conditions, as most key signatures of Th17 cells2, 13 including CCL20, IL-17F, RORC and IL23R expression were highly upregulated. The analysis of the microarray data and its verification on mRNA or protein expression suggested that high-salt induces a pathogenic type of Th17 cells14. In addition to IL-17A, high NaCl concentration induced the pro-inflammatory cytokines IL-2, TNFα, IL-9 and several chemokines. These cells also upregulated CSF2/GM-CSF which is essential for the pathogenicity of Th17 cells15, 16 and CCR6, which is crucial for Th17 function in autoimmune disease17. Furthermore, MIR155HG, the host gene for the micro RNA miR-155 which is necessary for Th17 induced EAE, was highly induced18. The high-salt-induced Th17 cells also expressed more TBX21/Tbet and less GATA3 and CXCR6 (Fig. 2a,b, Supplementary Fig. 7,8 and data not shown). In total, these observations indicate that increased NaCl concentrations specifically promote the generation of a highly pathogenic Th17 cell type14.
Figure 2. High-salt induced Th17 cells display a pathogenic phenotype.
a, Microarray analysis of naïve CD4 cells differentiated into Th17 cells in the presence (NaCl) or absence (none) of additional 40mM NaCl. Depicted is a selection of 26 up- and down-regulated genes (mean fold change of two independent experiments). b, qRT-PCR analysis of differentially expressed genes in the two groups (n=5–8).
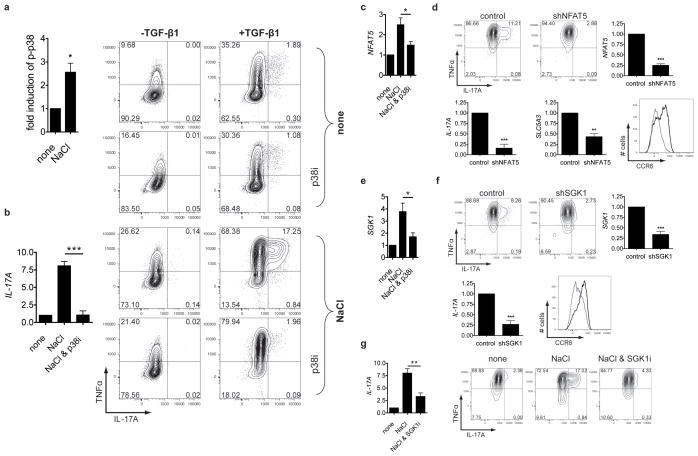
We then examined the pathways whereby high-salt induced this inflammatory phenotype. It has been shown that increased NaCl concentrations associated with augmented hypertonicity could induce immune system activation11, 19. Moreover, it is known that hypertonic stress in mammals is sensed through p38/MAPK, a homolog to HOG1, the ancient yeast hypertonic stress response element19. The key translator of this cascade is the osmosensitive transcription factor NFAT520, 21. Analysis of the microarray dataset indicated the stimulation of both inflammatory and classic hypertonicity induced pathways. The CD4 cells expressed high levels of the NFAT5 targets SGK122 and the Sodium/myo-inositol cotransporter SLC5A3 (Fig. 2a,b and Supplementary Fig. 7,8)21, 23. Therefore, we hypothesized that increased NaCl leads to phosphorylation of p38/MAPK that activates other downstream targets, including NFAT5. The phosphorylation of p38/MAPK was indeed increased in the presence of high-salt (Fig. 3a and Supplementary Fig. 9a) and was accompanied by induction of NFAT5 (Fig. 3c). We then determined whether inhibition of the p38/MAPK pathway influenced the effect. SB202190, an inhibitor of p38/MAPK21 (p38i) only partially decreased NFAT5 mRNA induction (Fig. 3c). However, SB202190 sharply reduced Th17 polarization (Fig. 3b). In line with these findings, siRNA mediated knockdown of MAPK14 in CD4 cells led to less IL-17A production (Supplementary Fig. 9b). High-salt could also promote p38/MAPK activation via the release of ATP24. However, by interfering with this pathway we could not observe significant changes on Th17 differentiation (data not shown).
Figure 3. The induction of Th17 cells by sodium chloride depends on p38/MAPK, NFAT5 and SGK1.
a, Naive CD4+ cells were stimulated in the presence (NaCl) or absence (none) of additional 40 mM NaCl and were analysed by FACS for phosphorylated p38 (p-p38; n = 5). b, Naïve CD4 cells were differentiated into Th17 cells as indicated in the presence or absence of NaCl and SB202190 (p38i) and analysed by qRT-PCR as depicted in the bar graph (n=7) or by FACS (the left row shows cells differentiated in the absence of TGF-β1). c, Naïve CD4 cells were stimulated for 3h in the presence or absence of NaCl and SB202190 and analysed by qRT-PCR for NFAT5 (n=4). d, Cells were transduced with NFAT5 specific (shNFAT5) or control shRNA (control), stimulated like in b) and analysed by FACS. The bar graphs depict qRT-PCR analyses of NFAT5, IL-17A and SLC5A3 (n=5). CCR6 was analysed by FACS (black histogram: control, grey histogram: shNFAT5, one representative experiment of four is shown). e, Cells were stimulated like in c), but analysed by qRT-PCR for SGK1 (n=4). f, Cells were transduced with a shRNA specific for SGK1 (shSGK1) or a control shRNA (control) and activated like in b), and analysed by FACS. Expression of SGK1 and IL-17A was determined by qRT-PCR (n=5). CCR6 was analysed by FACS (black histogram: control, grey histogram: shSGK1, one representative experiment of four is shown). g, Cells were cultured like in b), but in the presence or absence of the SGK1 inhibitor GSK650394 (SGK1i) and analysed by FACS. The bar graph shows qRT-PCR for IL-17A under similar conditions (n=5). FACS and qRT-PCR (relative expression) data depicted in bar graphs were normalised to controls.
Our data indicate that NFAT5 is involved in this NaCl induced inflammatory pathway. Since it was previously shown that NFAT5 influences responses of immune cells under similar conditions7, 20, 21, we silenced NFAT5 by a short hairpin RNA (shRNA) in naïve CD4 cells. As expected, NFAT5 silencing reduced SLC5A3 expression, but also decreased IL-17A and CCR6 expression (Fig. 3d). A direct downstream target of NFAT5 is SGK122. Besides being activated by tonicity dependent signals25, 26, SGK1 expression is also regulated by TGF-β27 and glucocorticoids28. As SGK1 activation can be regulated by p38/MAPK26, 29 and NFAT522, and was strongly upregulated in the microarray, it was of interest to examine whether this kinase plays a role in high-salt mediated Th17 polarization. SGK1 was indeed upregulated in naïve CD4 cells after stimulation with high-salt conditions. To confirm that SGK1 is regulated by p38/MAPK dependent signals, expression of SGK1 was measured in the presence of SB202190 (Fig. 3e). The addition of p38i reduced NaCl induced SGK1 mRNA expression, consistent with previous reports in other systems22, 26, 29. Moreover, shRNA-mediated silencing of SGK1 significantly decreased IL-17A production in high-salt exposed cells and led to diminished CCR6 expression (Fig. 3f). In line with these observations, pharmacological blockade of SGK1 produced similar, albeit less pronounced results compared to SB202190 (Fig. 3g).
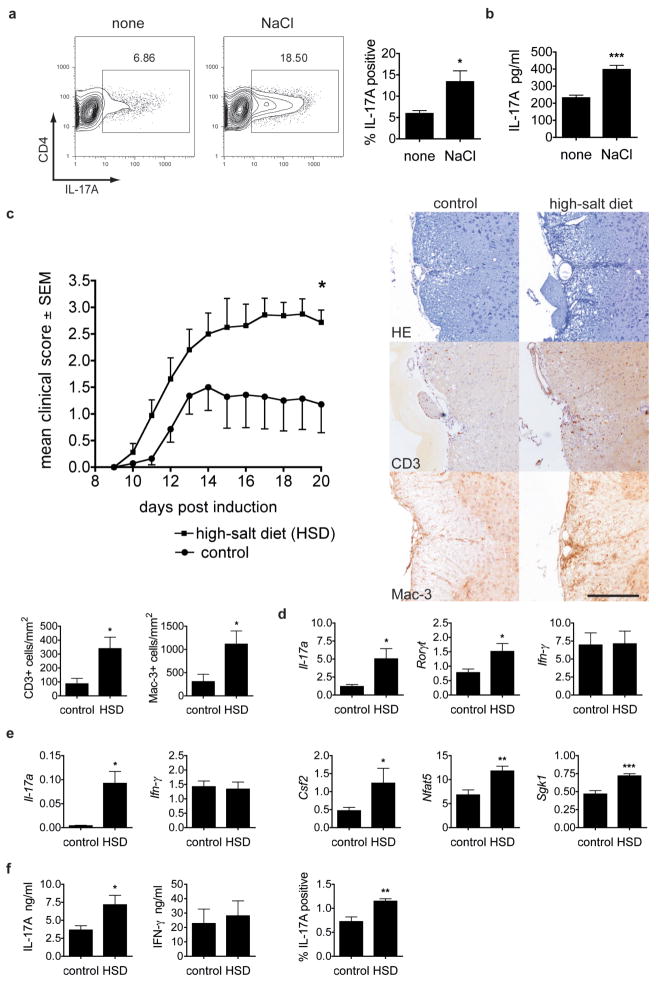
The rather dramatic in vitro effects of high-salt on naïve human CD4 cells prompted us to examine the effects of increased dietary NaCl in an in vivo system. We first adapted the human culture system to various murine Th17 differentiation models and made similar observations of increased Th17 induction and the accompanying phenotype (Fig. 4 and Supplementary Fig. 10). High-salt conditions did not significantly alter proliferation or cell death. Moreover, the effect was specific for Th17 conditions, since there was no enhancement of Th1 or Th2 differentiations (Supplementary Fig. 11,12 and data not shown). High-salt induced expression of Nfat5, Sgk1 and IL-17A was dependent on p38/MAPK. Enhanced Th17 differentiation could be blocked by SB202190 and gene deletion of p38α decreased Il-17a, Nfat5 and Sgk1 induction (Supplementary Fig. 10,13). Since the high-salt effect on Th17 cells appeared similar among species, we examined whether dietary NaCl influenced EAE. High-salt diet (HSD) accelerated onset and increased severity of the disease (Fig. 4c) while blood pressure was not affected (Supplementary Fig. 14). Animals on HSD displayed significantly higher numbers of CNS infiltrating CD3+ and Mac-3+ cells compared to controls (Fig. 4c). IL-17A expressing CD4 cells in CNS infiltrates almost doubled in frequency and accordingly, we detected increased Il-17a and Rorc mRNA expression in spinal cords (Fig. 4d and Supplementary Fig. 15). In contrast to Ifn-γ, we found augmented expression of Il-17a and Csf2 and higher levels of Nfat5 and Sgk1 in the spleens of HSD EAE mice compared to controls (Fig. 4e). Interestingly, splenocytes from EAE mice fed HSD showed enhanced IL-17A but not IFN-γ or Th2 cytokine expression upon antigen restimulation, indicating increased in vivo induction of antigen specific Th17 cells (Fig. 4f and data not shown). Consistent with in vitro data, HSD induced effect was dependent on p38/MAPK, since in vivo administration of SB202190 inhibited salt-induced increases in the frequency of TH17 cells infiltrating the CNS (Supplementary Fig. 15b,c).
Figure 4. High-salt diet induces Th17 cells in vivo and exacerbates experimental autoimmune encephalomyelitis.
a, Naïve murine CD4 cells were stimulated with radiated APC, anti-CD3, IL-6 and TGF-β1 in the presence (NaCl) or absence (none) of additional 40mM NaCl and were analysed by FACS (n=3). b, IL-17A secretion (ELISA) of primary splenocytes, stimulated by anti-CD3 in the presence or absence of NaCl (n=6). c, Mean clinical scores of EAE in HSD animals (squares) or controls (dots, pooled data of two independent experiments with 12 animals). Histological analyses show sections of the spinal cord stained with hematoxylin and eosin (HE), anti-CD3 and anti-Mac-3 for control or HSD animals (scale bar=100 μM) and were quantified for CD3 and Mac-3 (bar graphs, n=5–6). d, Spinal cord from EAE animals was analysed by qRT-PCR (n=5–6). e, Splenocytes from EAE animals were analysed by qRT-PCR (n=4–7). f, Splenocytes from EAE animals were re-stimulated with MOG for 2 days and supernatants were analysed for IL-17A and IFN-γ by ELISA (n=7–8) or cells were analysed for IL-17A by FACS (n=4). qRT–PCR data are depicted as relative expression.
In this investigation, we found that modest increases in NaCl could stimulate an almost logarithmic in vitro induction of IL-17A in naïve CD4 cells mediated through p38/MAPK, NFAT5 and SGK1. Importantly, the addition of 40mM of NaCl to Th17 differentiation cultures not only increased IL-17A expression but also led to a pathogenic phenotype of Th17 cells. In line with these findings, common salt added to the diet of mice led to severe worsening of EAE accompanied by increased numbers of Th17 cells. What might be the physiologic role for the effect of high-salt on the induction of inflammatory Th17 cells? The concentration of Na+ in plasma is approximately 140mM, similar to standard cell culture media. Less well appreciated is that in the interstitium and lymphoid tissue, considerably higher Na+ concentrations between 160mM and even as high as 250mM can be encountered7, 20, the “high-salt” conditions we found to induce inflammatory Th17 cells. Thus, this may be a mechanism for decreasing immune activation in the blood while favoring an inflammatory response in lymphoid tissues or with migration of cells into tissue. In this context it could be expected that other immune cells can react on high-salt conditions as well and potentially contribute to the effects observed in vivo.
Do these data indicate that increased salt intake is the long sought-after environmental factor associated with the epidemic of autoimmune disease? While these data present an attractive hypothesis, the direct causality of salt intake and incidence of autoimmune disease are yet to be demonstrated. That is, no in vitro observation can prove causality; instead, our data indicate that clinical trials with severe curtailment of salt intake for individuals at risk for developing autoimmune disease are required. Clinical scenarios, where a dietary salt restriction protocol could be tested are MS or psoriasis, both autoimmune diseases with strong Th17 components2. Additionally, excess salt content in diet should be investigated as a potential environmental risk factor for autoimmune diseases. However, this study would be difficult in Western cultures where the application of a true low-salt diet, representing the conditions by which homo sapiens were environmental selected in Africa, is difficult to achieve. Nevertheless, although there might be additional mechanisms contributing to the observed effects, the pathways identified in this study may offer new targets for the treatment of autoimmune diseases with interference in the p38/MAPK, NFAT5 and SGK1 pathways in blocking the generation of pathogenic Th17 cells.
Methods
Antibodies, recombinant cytokines and reagents
The following mAbs and reagents were used as follows: for surface staining, anti-CD4 (RPA-T4), anti-CD45RO (UCHL1), anti-CD45RA (HI100), anti-CD25 (M-A251), anti-CD127 (hIL-7R-M21), anti-CCR6 (11A9) and AnnexinV all from BD Biosciences (San Jose, CA) and for intracellular staining anti-IL-17A (eBio64DEC17) and anti-TNF-α (MAb11), anti-IFN-γ (4S.B3), anti-IL-2 (MQ1-17H12), anti-RORC (AFKJS-9), anti-GATA3 (TWAJ) and anti-Tbet (eBio4B10) from eBioscience (San Diego, CA) and anti-pp38 (36/p38) (BD Biosciences) and anti-GM-CSF (BVD2-21C11) from Biolegend (San Diego, CA); for T cell stimulation, anti-CD3 (UCHT1) and anti-CD28 (28.2) from BD Biosciences. Recombinant human TGF-β1 was purchased from eBioscience, recombinant human IL-1β, IL-6, IL-4, IL-12 and IL-23 and neutralizing anti-IFN-γ (25718) and anti-IL-4 (3007) were purchased from R&D Systems (Minneapolis, MN), and recombinant human IL-21 was purchased from Cell Sciences (Canton, MA). CFSE was obtained from Invitrogen (Carlsbad, CA).
Human cell isolation and stimulation
Peripheral blood was obtained from healthy control volunteers in compliance with Institutional Review Board protocols. PBMCs were separated by Ficoll-Paque PLUS (GE Healthcare, Piscataway, NJ) gradient centrifugation. Untouched total CD4+ T cells were isolated from PBMC by negative selection via the CD4+ T cell isolation kit II (Miltenyi Biotec, Auburn, CA). Naive (CD45RA+CD45RO−CD25−CD127+) and memory (CD45RA−CD45RO+CD25−CD127+) CD4+ T cells were sorted by high-speed flow cytometry with a FACS Aria (BD Biosciences) to a purity >98% as verified by postsort analysis. Dead cells were excluded by propidium iodide (BD Biosciences). CD14+ monocytes were isolated by positive selection with CD14 microbeads (Miltenyi Biotec). Cells were cultured in 96-well round-bottom plates (Costar, Cambridge, MA) at 5×104 cells/well in serum-free X-VIVO15 medium (BioWhittaker, Walkersville, MD), and stimulated with plate-bound anti-CD3 (10 μg ml−1) and soluble anti-CD28 (1 μg ml−1) antibodies. Where indicated, recombinant TGF-β1 (5ng ml−1), IL-1β (12.5ng ml−1), IL-6 (25ng ml−1), IL-21 (25ng ml−1), or IL-23 (25ng ml−1) or additional 10–80mM NaCl was added of the cultures. For Th1 and Th2 differentiation naïve cells were stimulated as described above but in the presence of IL-12 (10ng ml−1) and anti-IL-4 (10 μg ml−1) (for Th1 cells) or with IL-4 (10ng ml−1) and anti-IFN-γ (10 μg ml−1) (for Th2 cells). In some experiments the specific inhibitors SB202190 (Sigma Aldrich, St Louis, MO) or GSK65039430 (Tocris Bioscience/R&D Systems) at concentrations of 5 μM or 1 μM respectively, were added to the cultures. Co-cultures of CD14+ monocytes and T cells were performed as described before12. In brief, monocytes were pulsed for 3h with C. albicans (GREER, Lenoir, NC) and irradiated (45 Gy) before T cell co-culture (ratio of 1:2). Total CD4+ T cells were co-cultured for 12 days and naïve CD4+ T cells were co-cultured for 7 days in the presence of additional cytokine cocktail (TGF-β1, IL-1β, IL-6, IL-21 and IL-23). Recombinant human IL-2 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) and was used for restimulation experiments at 20U ml−1. Cells were cultured for the indicated periods of time.
Flow cytometry and cytokine detection
Cells were analysed by flow cytometry if not specified elsewhere after a culture period between 7 and 8 days. For surface staining, cells were stained with the respective antibodies for 20 minutes in PBS containing 0.5% FCS and 2mM EDTA before analysis. For intracellular staining, cells were stimulated for 4–5 h with PMA (50ng ml−1) and ionomycin (250ng ml−1; both from Sigma-Aldrich) in the presence of GolgiPlug (BD Biosciences), fixed and made permeable (Fix/Perm; eBioscience) according to the manufacturer’s instructions, and stained with the respective antibodies for intracellular cytokine detection for 30–45min. Prior fixation, cells were stained with the LIVE/DEAD cell kit (Invitrogen, Carlsbad, CA) to exclude dead cells. For measurement of phosphorylated p38 (p-p38), cells were stimulated for 20 minutes before cells were fixed (Cytofix buffer, BD Biosciences) and made permeable (Phosflow Perm Buffer III, BD Biosciences) according to the manufacturer’s instructions and stained for 30–45 minutes with anti-pp38. Data were acquired on a LSR II (BD Biosciences) and analysed with FlowJo software (TreeStar, Ashland, OR). Culture supernatants were taken on day 6 and measured by ELISA for secretion of IL-17A (eBioscience) according to the manufacturer.
Real-time PCR
Cells for RNA isolation were harvested if not specified elsewhere between 6 and 7 days of culture and RNA was isolated using the Absolutely RNA 96 Microprep Kit (Agilent Technologies, Palo Alto, CA) or RNeasy micro kit (Qiagen, Valencia, CA) and converted to cDNA via reverse transcriptase by random hexamers and Multiscribe RT (TaqMan Gold RT-PCR kit, Applied Biosystems, Foster City, CA). All primers were purchased from Applied Biosystems. All reactions were performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems). The values are represented as the difference in Ct values normalized to β2-microglobulin for each sample as per the following formula: relative RNA expression = (2−dCt) × 103.
shRNA and siRNA mediated gene silencing
Lentiviral particles expressing shRNAs were obtained from the library of The RNAi Consortium (TRC)31. Lentiviral transduction of human T cells was carried out as described before32. In brief, 5×104 human naïve CD4+ T cells/well were stimulated for 24h prior infection. Cells were then transduced with viral particles containing a vector expressing the indicated specific shRNA or as controls a vector expressing an unspecific shRNA or expressing GFP. Transduction was mediated at a multiplicity of infection (MOI) of 5 by centrifugation at 2250 rpm for 30 min at room temperature in the presence of 3 μg ml−1 polybrene (Millipore, Billerica, MA). After 48h puromycin (Invitrogen) was added to the cultures at a concentration of 0.5 μg ml−1 to select for successfully transduced cells and was controlled by flow cytometry for GFP and propidium iodide. The specific RNAi Consortium clones were TRCN0000020019 for NFAT5 and TRCN0000040175 for SGK1. For siRNA transfections control siRNA (ON-TARGETplus non targeting#1) and a pool of 4 specific siRNA for MAPK14 (ON-TARGETplus SMARTpool#1432) were obtained from Thermo Scientific Dharmacon (Lafayette, CO). Cells were transfected by using Human T Cell Nucleofector Kit and a Nuclofector II device as recommended by the manufacturer (Lonza/Amaxa, Allendale, NJ).
Microarray analysis
Cells for microarray analysis were harvested at day 7 of culture and total RNA was isolated using Trizol reagent according to the manufacturer (Invitrogen). Expression data was generated by using GeneChip Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA) at the Yale Center for Genome Analysis (YCGA). For analysis, the data was normalized using the GenePattern software33 with the Robust Multi Array (RMA) algorithm34. The COMBAT software was used to remove batch effects. Fold change was computed between the average expression levels of each probeset in samples with the different conditions. To avoid spurious fold levels due to low expression values a small constant (c=50) was added to the expression values. Only cases where more than 50% of the four possible pair-wise comparisons were over a cut-off of 1.5 fold change were reported. A Z-score was computed as additional filter by comparing the mean of the expression levels in the NaCl treated samples to the expression levels in the control samples. Only cases with a corresponding p-value lower than 0.05 were reported.
Western blotting
Western blotting was performed as described before35. Phospho-p38 was detected by using anti-phospho-p38 (Cell Signaling Technology, Boston, MA). Anti-β-Actin and anti-SGK1 antibodies were obtained from Cell Signaling Technolgy and anti-NFAT5 antibodies were purchased from Pierce/Thermo Scientific (Rockford, IL). Primary antibodies were detected by peroxidase-conjugated streptavidin (Jackson Immuno Research, West Grove, PA), secondary anti-rabbit-HRP conjugated (Cell signalling Technology or Jackson Immuno Research) and secondary anti-mouse-HRP conjugated (Bio-Rad, Hercules, CA) antibodies.
Mice, EAE induction, high-salt diet and blood pressure analysis
C57BL/6J mice were purchased from Harlan (Borchen, Germany) and housed at the in-house animal care facility of the University of Erlangen under standardized conditions. EAE induction was done as described before9. Briefly, male mice were immunized with 200 μg MOG (35–55) (Charite, Berlin, Germany) in an equal amount of complete Freund’s adjuvant and received 200ng pertussis toxin (List Biochemicals, Campbell, CA) intraperitonally (i.p.) on days 0 and 2 post induction (p.i). The clinical evaluation was performed on a daily bases by a 5 point scale ranging from 0, no clinical sign; 1, limp tail; 2, limp tail, impaired righting reflex, and paresis of one limb; 3, hind limb paralysis; 4, hind limb and forelimb paralysis; 5, moribund. Mice received normal chow and tap water ad libitum (control group) or sodium-rich chow containing 4% NaCl (SSNIFF, Soest, Germany) and tap water containing 1% NaCl ad libitum (high-salt group). Inhibition of p38/MAPK in vivo was done as described before36. In brief, mice were maintained on a control or high-salt diet and either received 1mg/kg/d SB202190 (TOCRIS) i.p. or vehicle from day −3 p.i. of EAE. Brain leukocytes were isolated by percoll gradient centrifugation on day 17 post EAE induction, stimulated by PMA/ionomycin and analysed by flow cytometry for IL-17A and CD4 expression. Mx-Cre+/p38αfl/fl mice37 maintained on C57BL/6 background were a kind gift of Dr. Jean-Pierre David. Mice were injected with 13mg/kg/body-weight Polyinosinic-polycydidylic acid (poly(I:C), Sigma-Aldrich) on days 0, 2, 6 and were sacrificed on day 8 for isolation of splenocytes. Blood pressure analysis was performed by the tail cuff method as described previously9. All animal experimentation was performed in accordance to the German animal protection law.
Histology
On day 20 p.i., mice were perfused with 4% paraformaldehyde and then the lumbar, thoracal and cervical part of their spinal cord was embedded in paraffin. Spinal cord cross sections were stained with hematoxylin/eosin to assess inflammation. T cells were labelled by anti-CD3 (Serotec, Wiesbaden, Germany), macrophages/microglia by anti-Mac-3 (BD Biosciences) and IL-17 positive cells by anti-IL-17 (Abcam, Cambridge, MA).
Murine T cell cultures
Splenic T cells from EAE animals were restimulated with 20 μg ml−1 MOG (35–55) peptide for 48h, for intracellular IL-17A detection, monensin (BD Biosciences) was added to the cultures for additional 6h. Splenic T cells from naïve mice were stimulated with 1 μg ml−1 plate-bound anti-CD3 (17A2, BD Biosciences) and 1 μg ml−1 soluble anti-CD28 (37.51, BD Biosciences) for 48h. For Th17 cell differentiation, spleen and lymph nodes cells from 10 week old 2D2 mice38 were pooled and CD4+CD62L+ naïve T cells were isolated by magnetic cell sorting (Miltenyi Biotec). Cells were cultured at 2×106 cells ml−1 and stimulated for 4 days with of 2×107 irradiated (30 Gray) syngenic splenocytes ml−1 and 1 μg ml−1 anti-CD3 (2C11, BD Biosciences) in the presence of TGF-β1 (5ng ml−1) and IL-6 (20ng ml−1) and where indicated of additional 40mM NaCl. For APC free Th17 differentiations naïve T cells were sorted as CD4+CD62L+CD44loCD25− and stimulated by plate bound anti-CD3 (2 μg ml−1) and anti-CD28 (2 μg ml−1) in the presence of IL-6 (40ng ml−1) and TGF-β1 (1ng ml−1) or IL-6 (40ng ml−1) and IL-23 (10ng ml−1). (all from R&D Systems) and were cultured for 4 days. In some experiments 10 μM SB202190 (TOCRIS) were added to the cultures. For Th1 differentiation, naïve CD4+ T cells were cultured for 96h with anti-CD3, anti-CD28, IL-12 (20ng ml−1) (BioLegend) and anti-IL-4 (10 μg ml−1) (1B11, BioLegend). To monitor proliferation, cells were labelled with fixable proliferation dye (eBioscience) according to manufacturers protocol. For intracellular flow cytometry, cells were stimulated for 4h with PMA/ionomycin in the presence of monensin and stained for CD4 (RM4-5, eBioscience) and intracellular IL-17A (eBio17B7, eBioscience), IFN-γ (XMG1.2, eBioscience), Tbet (4B10, eBioscience) or RORC/RORγt (AFKJS-9, eBioscience), excluding dead cells by a fixable viability dye (eBioscience). For murine gene expression analysis, mRNA was prepared using PeqLab Gold HP total RNA kit (PeqLab, Erlangen, Germany) and cDNA was prepared using superscriptTM II reverse transcriptase (Invitrogen). RNA was isolated from EAE animals at day 14 p.i.. Reactions were performed on a 7900 Sequence Detection System (Applied Biosystems). Primers were obtained from Applied Biosystems and target expression was normalized to β-actin expression. For cytokine secretion analysis, cells were stimulated as indicated and supernatants were harvested after three days of culture. Monoclonal antibody pairs and recombinant cytokine standards were purchased from R&D systems (IL-17A, IFN-γ).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software Inc., La Jolla, CA). Data were analysed by unpaired t-test in case of two groups and by one-way ANOVA using Tukey’s post-hoc test in multiple groups. Data tested against a specified value were analysed by one-sample t-test. EAE was analysed using non-parametric Mann-Whitney test. Data are presented if not indicated elsewhere as mean±s.e.m. p<0.05 were considered to be statistically significant (*p<0.05, **p<0.01, ***p<0.001).
Supplementary Material
Acknowledgments
The authors would like to thank Siddheshvar Bhela, Mansoor Zaidi, Petra Quass, Markus Mroz and Silvia Seubert for technical assistance and Friedrich C. Luft for critical reading of the manuscript. We are grateful to Jean-Pierre David and Stefan Teufel for providing Mx-Cre+ p38αfl/fl mice. This work was supported by a National MS Society Collaborative Research Center Award CA1061-A-18, National Institutes of Health Grants P01 AI045757, U19 AI046130, U19 AI070352, and P01 AI039671 and by a Jacob Javits Merit award (NS2427) from the National Institute of Neurological Disorders and Stroke, the Penates foundation and the Nancy Taylor Foundation for Chronic Diseases, Inc. (to D.A.H.). R.A.L. was supported by the ELAN programme, University of Erlangen. D.N.M. was supported by the German Research Foundation (DFG) and the German Center for Cardiovascular Research (DZHK). J.T. was supported by the Interdisciplinary Center for Clinical Research at University of Erlangen and the German Research Foundation.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature. The microarray data sets are deposited in Gene Expression Omnibus database under accession number GSE42569.
Author contributions. M.K. designed the study, planned and performed experiments, analysed data and wrote the manuscript. A.M. planned and performed experiments, analysed data and wrote the manuscript. J.T. and H.K. interpreted data and supported the work with key suggestions and editing the manuscript. N.Y. analysed data. R.A.L. planned experiments, analysed data and wrote the manuscript. D.N.M. designed the study, planned experiments, analysed data and wrote the manuscript. D.A.H. designed the study, planned experiments, analysed data, and wrote the manuscript. M.K., D.N.M and D.A.H. co-directed the project.
References
- 1.IMSGC and WTCCC. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 3.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis. Part II: Noninfectious factors. Ann Neurol. 2007;61:504–13. doi: 10.1002/ana.21141. [DOI] [PubMed] [Google Scholar]
- 4.McGuire S Institute of Medicine. 2010. Strategies to Reduce Sodium Intake in the United States. Washington, DC: The National Academies Press. Adv Nutr. 2010;1:49–50. doi: 10.3945/an.110.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appel LJ, et al. The importance of population-wide sodium reduction as a means to prevent cardiovascular disease and stroke: a call to action from the American Heart Association. Circulation. 2011;123:1138–43. doi: 10.1161/CIR.0b013e31820d0793. [DOI] [PubMed] [Google Scholar]
- 6.Brown IJ, Tzoulaki I, Candeias V, Elliott P. Salt intakes around the world: implications for public health. International Journal of Epidemiology. 2009;38:791–813. doi: 10.1093/ije/dyp139. [DOI] [PubMed] [Google Scholar]
- 7.Machnik A, et al. Macrophages regulate salt-dependent volume and blood pressure by a vascular endothelial growth factor-C-dependent buffering mechanism. Nat Med. 2009;15:545–52. doi: 10.1038/nm.1960. [DOI] [PubMed] [Google Scholar]
- 8.Platten M, et al. Blocking angiotensin-converting enzyme induces potent regulatory T cells and modulates TH1- and TH17-mediated autoimmunity. Proc Natl Acad Sci U S A. 2009;106:14948–53. doi: 10.1073/pnas.0903958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stegbauer J, et al. Role of the renin-angiotensin system in autoimmune inflammation of the central nervous system. Proc Natl Acad Sci U S A. 2009;106:14942–7. doi: 10.1073/pnas.0903602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, et al. IL-21 and TGF-beta are required for differentiation of human T(H)17 cells. Nature. 2008;454:350–2. doi: 10.1038/nature07021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junger WG, Liu FC, Loomis WH, Hoyt DB. Hypertonic saline enhances cellular immune function. Circ Shock. 1994;42:190–6. [PubMed] [Google Scholar]
- 12.Zielinski CE, et al. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–8. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L, Littman DR. Transcriptional regulatory networks in Th17 cell differentiation. Curr Opin Immunol. 2009;21:146–52. doi: 10.1016/j.coi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–71. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Codarri L, et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–7. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 16.El-Behi M, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–75. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reboldi A, et al. C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat Immunol. 2009;10:514–23. doi: 10.1038/ni.1716. [DOI] [PubMed] [Google Scholar]
- 18.O’Connell RM, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapiro L, Dinarello CA. Osmotic regulation of cytokine synthesis in vitro. Proc Natl Acad Sci U S A. 1995;92:12230–4. doi: 10.1073/pnas.92.26.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go WY, Liu X, Roti MA, Liu F, Ho SN. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc Natl Acad Sci U S A. 2004;101:10673–8. doi: 10.1073/pnas.0403139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kino T, et al. Brx mediates the response of lymphocytes to osmotic stress through the activation of NFAT5. Sci Signal. 2009;2:ra5. doi: 10.1126/scisignal.2000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, et al. Tonicity-dependent induction of Sgk1 expression has a potential role in dehydration-induced natriuresis in rodents. J Clin Invest. 2009;119:1647–58. doi: 10.1172/JCI35314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortells MC, et al. Transcriptional regulation of gene expression during osmotic stress responses by the mammalian target of rapamycin. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woehrle T, et al. Hypertonic stress regulates T cell function via pannexin-1 hemichannels and P2X receptors. J Leukoc Biol. 2010;88:1181–9. doi: 10.1189/jlb.0410211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waldegger S, Barth P, Raber G, Lang F. Cloning and characterization of a putative human serine/threonine protein kinase transcriptionally modified during anisotonic and isotonic alterations of cell volume. Proc Natl Acad Sci U S A. 1997;94:4440–5. doi: 10.1073/pnas.94.9.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell LM, et al. Hyperosmotic stress stimulates promoter activity and regulates cellular utilization of the serum- and glucocorticoid-inducible protein kinase (Sgk) by a p38 MAPK-dependent pathway. J Biol Chem. 2000;275:25262–72. doi: 10.1074/jbc.M002076200. [DOI] [PubMed] [Google Scholar]
- 27.Waldegger S, et al. h-sgk serine-threonine protein kinase gene as transcriptional target of transforming growth factor beta in human intestine. Gastroenterology. 1999;116:1081–8. doi: 10.1016/s0016-5085(99)70011-9. [DOI] [PubMed] [Google Scholar]
- 28.Webster MK, Goya L, Ge Y, Maiyar AC, Firestone GL. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13:2031–40. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldegger S, Gabrysch S, Barth P, Fillon S, Lang F. h-sgk serine-threonine protein kinase as transcriptional target of p38/MAP kinase pathway in HepG2 human hepatoma cells. Cell Physiol Biochem. 2000;10:203–8. doi: 10.1159/000016351. [DOI] [PubMed] [Google Scholar]
- 30.Sherk AB, et al. Development of a Small-Molecule Serum- and Glucocorticoid-Regulated Kinase-1 Antagonist and Its Evaluation as a Prostate Cancer Therapeutic. Cancer Research. 2008;68:7475–7483. doi: 10.1158/0008-5472.CAN-08-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Astier AL, et al. RNA interference screen in primary human T cells reveals FLT3 as a modulator of IL-10 levels. J Immunol. 2010;184:685–93. doi: 10.4049/jimmunol.0902443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 34.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 35.Wenzel K, et al. Potential relevance of alpha(1)-adrenergic receptor autoantibodies in refractory hypertension. PLoS One. 2008;3:e3742. doi: 10.1371/journal.pone.0003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noubade R, et al. Activation of p38 MAPK in CD4 T cells controls IL-17 production and autoimmune encephalomyelitis. Blood. 2011;118:3290–300. doi: 10.1182/blood-2011-02-336552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engel FB, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–87. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bettelli E, et al. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197:1073–81. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.