Abstract
B-precursor acute lymphoblastic leukemia (B-ALL) lymphoblast (blast) internalization of anti-cytokine receptor-like factor 2 (CRLF2) antibody-armored biodegradable nanoparticles (AbBNPs) are investigated. First, AbBNPsaere synthesized by adsorbing anti-CRLF2 antibodies to poly(D,L-lactide- co -glycolide) (PLGA) nanoparticles of various sizes and antibody surface density (Ab/BNP) ratios. Second, AbBNPs are incubated with CRLF2-overexpressing (CRLF2+) or control blasts. Third, internalization of AbBNPs by blasts is evaluated by multicolor flow cytometry as a function of receptor expression, AbBNP size, and Ab/BNP ratio. Results from these experiments are con-firmed by electron microscopy, fluorescence microscopy, and Western blotting. The optimal size and Ab/BNP for internalization of AbBNPs by CRLF2+ blasts is 50 nm with 10 Ab/BNP and 100 nm with 25 Ab/BNP. These studies show that internalization of AbBNPs in childhood B-ALL blasts is AbBNP size-and Ab/BNP ratio-dependent. All AbBNP combinations are non-cytotoxic. It is also shown that CD47 is very slightly up-regulated by blasts exposed to AbBNPs. CD47 is “the marker of self” overexpressed by blasts to escape phagocytosis, or “cellular devouring”, by beneficial macrophages. The results indicate that precise engineering of AbBNPs by size and Ab/BNP ratio may improve the internalization and selectivity of future biodegradable nanoparticles for the treatment of leukemia patients, including drug-resistant minority children and Down’s syndrome patients with CRLF2+B-ALL.
1. Introduction
Acute lymphoblastic leukemia (ALL), despite an ≈ 85% cure rate, remains the second most common cause of cancer-related mortality in children in the U.S.[1] Unfortunately, minority children with ALL have an increased incidence of ALL[2,3] and among the poorest outcome,[4–6] a well-known and significant cancer health disparity. The U.S. Census Bureau predicts that minority children will comprise 62% (vs. 44% in 2008) of the U.S. population by 2050. Given these projections, the potential impact of novel drugs for minority children is profound.
Recent studies show that childhood cancer health disparities are due to both socioeconomic and biological differences. In 2000, Pollock et al. showed that the 5-year relapse-free survival (RFS) rates were 81.9% ± 0.6%, 68.6% ± 2.1%, and 74.9% ± 2.0% for non-Hispanic/ Latino White, African-American, and Hispanic/Latino children.[5] Adjusting for age, lymphoblast (blast) count, sex, era of treatment, and blast ploidy, African American children had a 42% excess mortality rate and Hispanic/ Latino children had a 33% excess mortality rate compared to non-Hispanic/Latino White children. More recent studies are in excellent agreement with this work.[4,6] For example, Batia et al. showed that 5-year RFS rates were the following among patients with high-risk features (age <1 or >10; blast count > 50,000): Asian, 75.1% ± 3.5%; non-Hispanic/ Latino White, 72.8% ± 0.6%; Hispanic/Latino, 65.9% ± 1.5%; and African American, 61.5% ± 2.2%.[6] Adjusting for the same factors as above, Hispanic/Latino children had the worst outcome with a 40% excess mortality rate. In 2009, Willman et al. described a 38-transcript expression classifier predictive of RFS for high-risk B-precursor ALL (B-ALL), where 25% of the 207 residual pre-treatment specimens obtained from Children’s Oncology Group (COG) clinical trial P9906 were from Hispanic/Latino children.[4] Significantly, 13 of these 46 Hispanic/ Latino children (28%) were classified to the highest risk group 8 (29% RFS) by the transcriptomics classifier and flow cytometry measures of minimal residual disease at end-induction day 29. The hazard ratio association of RFS for Hispanic/Latino children (1.6) was second only to that of minimal residual disease (2.8).
Cytokine receptor-like factor 2 (CRLF2), also known as the thymic stromal lymphopoietin receptor, is the most significant cell surface protein in the 38 transcript classifier described above. The CRLF2 receptor is a heterodimeric type I receptor complex for thymic stromal lymphopoietin (TSLP), comprised of CRLF2 and interleukin-7 receptor alpha (IL-7Rα), where the latter is shared with the cytokine receptor common chain γc to form the heterodimeric IL-7 receptor complex for IL-7.[7] Both CRLF2 and IL-7 can activate the transcription factor STAT5, where IL-7Rα binds to JAK1 and the γc binds to JAK3 upon addition of IL-7. Recently, JAK2 has been demonstrated to be involved in STAT5 activation following binding of TSLP to the CRLF2 receptor complex.[7], [8–11]
More than 25% of CRLF2-overexpressing B-ALL patients (CRLF2+B-ALL), particularly minority children, are classified as high-risk for combination chemotherapy treatment failure.[12] High-risk B-ALL is largely defined by pretreatment clinical characteristics (age > 10 years and presenting blast count >50 000/µL) and the absence of genetic abnormalities associated with “low risk” disease [hyperdiploidy, t(12;21)(ETV6-RUNX1)] or “very high risk” disease [hypodiploidy, t(9;22)(BCR-ABL1) (the Philadelphia chromosome)].[4] Strikingly, CRLF2 alterations, including IGH-CRLF2 and P2RY8-CRLF2 fusions, occur in 5–15% of childhood and adult B-ALL patients and in 50–60% of Down’s syndrome B-ALL patients. These patients often harbor “BCR-ABL-like” cytotoxic drug-resistant point mutations/gene expression profiles, activating JAK mutations, and lymphoid transcription factor IKAROS deletions/mutations. Small molecule inhibitors, including the JAK inhibitor INCB018424 (Incyte)[13] and the HSP90 inhibitor AUY922 (Novartis),[14] focus on blocking the stimulation of signaling pathways believed to confer proliferation and survival advantages.[15] However, 225+phosphorylation sites have been discovered in the TSLP signaling pathway,[16] suggesting numerous drug resistance mechanisms for CRLF2+blasts.
Biodegradable nanoparticles, multifunctional particles with sub-micrometer dimensions conjugated to biomolecules such as antibodies and/or drugs, often exhibit beneficial properties that differ from traditional drugs. For example, the therapeutic index of doxorubicin-loaded, polyethylene glycol-armored, liposomes was dramatically improved over doxorubicin, by increasing intracellular concentrations and preventing cytotoxic drug expulsion by the multi-drug resistance transporter P glycoprotein.[17] In another example, the circulation half-life of L-asparaginase-loaded liposomes was dramatically improved over L-asparaginase, while reducing resistance due to the production of anti-L-asparaginase-antibodies.[18–20] Unlike liposomes, polymeric nanoparticles can be precisely tuned for sustained drug release and other properties because the biodegradation rate depends upon several well-characterized factors, including the ratio of monomers and/or the molecular weight of the polymer.[21–26] For example, poly (D,L-lactide- co -glycolide)-polyethylenimine (PLGA-PEI) with a lactide-to-glycolide ratio of 50:50 and molecular weight of ≈40 000 has been shown to improve the endosomal escape and loading efficiency of small interfering RNA.[27] Akin to antibody-drug conjugates, the therapeutic index can be improved by conjugating to antibodies that selectively target overexpressed cell surface receptors, including: CD20, CD19, CD22 and CD47. Previous work suggests that internalization of antibody-conjugated nanoparticles by receptor-mediated endocytosis and other mechanisms can be receptor expression-, size-, antibody surface density ratio-, and/or time-dependent.[28–33]
CD47, “the marker of self”, is overexpressed by blasts to escape phagocytosis, or “cellular devouring”, by beneficial macrophages (MΦs).[34–42] Chao, Majeti, Weissman et al. showed an ≈ 200% increase in the relative expression of CD47 by blasts and a strong hazard ratio association of RFS for CD47 (1.8) in ALL patients.[43] They exploited this phenomenon to non-selectively accelerate blast phagocytosis by MΦs with anti-CD47 antibodies. Acceleration was FcR-independent and synergized with FcR-dependent acceleration with anti-CD20 antibodies.[44] While these pioneering studies with antibodies are very promising, potential problems with therapeutic index, resistance and toxicity remain. For example, relatively large and frequent doses, particularly difficult for childhood patients, may be needed to overcome ubiquitous CD47 expression in a variety of cell types, including hemat-opoietic stem cells and red blood cells. This may explain why incomplete elimination of ALL in xenotransplant models with bone marrow burden beyond 70% involvement was observed.[43]
Biodegradable nanoparticles might improve outcome for leukemia patients, particularly drug-resistant minority children and Down’s syndrome patients with CRLF2-overexpressing B-ALL. Therefore, we investigated B-ALL blast internalization of anti-CRLF2 antibody-armored biodegradable nanoparticles (AbBNPs). First, AbBNPs were synthesized by adsorbing anti-CRLF2 antibodies to PLGA nanoparticles of various sizes and antibody surface density (Ab/BNP) ratios. Second, AbBNPs were incubated with CRLF2+ or control blasts. Third, internalization was evaluated by multicolor flow cytometry as a function of receptor expression, AbBNP size, and the Ab/BNP ratio. Results from these experiments were confirmed by electron microscopy, fluorescence microscopy and Western blotting.
2. Materials and Methods
2.1. AbBNP Synthesis
AbBNPs were synthesized by adsorbing mouse anti-human CRLF2 monoclonal antibodies (LS-C40791, LifeSpan Biosciences) at 4 °C for 24 h to non-cytotoxic FITC-loaded, 50:50 PLGA nanoparticles of two sizes (50–100 nm) (LGFG100, LGFG50; Phosphorex INC.) and six Ab/BNP ratios (0–200).[30,45] BNPs were dispersed in PBS (pH 7.4) for reactions with 7.34 × 1011 BNPs/mL. AbBNPs were separated from free antibody by centrifugation at 10 000 rpm for 15 min (Eppendorf Centrifuge 5415). The sediment containing purified AbBNPs was washed and re-dispersed with 1 mL of phosphate buffered saline (PBS) prior to storage at 4 °C and characterization.
2.2. Transmission Electron Microscopy
Transmission electron microscopy (TEM) of 50 nm diameter AbBNPs with 10 or 100 Ab/BNP was in manner similar to that described previously.[46] AbBNPs were incubated with 6 nm gold rabbit anti-mouse secondary antibody conjugates (gold conjugates) (Cat: 25104; Electron Microscopy Sciences), where 100 µL of AbBNP (corresponding to 1.8 µg of 10Ab/BNP and 18.8 µg 100Ab/BNP) was incubated 1:1 with 9 × 1010 and 9 × 1011 gold conjugates, respectively, for 1 h. These were washed 2× with PBS for 5 min after centrifugation at 10 000rpm for 10 min.[47] Next, 10 µL of AbBNP-gold conjugates were deposited on holey carbon film 300 mesh copper grids (HC300-Cu; Electron Microscopy Sciences). High-resolution TEM (HRTEM) characterization was performed with JEOL 2010F operated at an accelerating voltage of 200 kV, to confirm the size distribution of AbBNPs and number of gold conjugates that correspond to various Ab/BNP ratios.
2.3. Western Blotting
A 20 µL aliquot of each AbBNP was resolved on a non-denaturing 4–20% SDS-PAGE gel (161–1159; BIO-RAD) at 90 V for 2 h prior to transfer to a nitrocellulose membrane (162–0095; BIO-RAD) at 90 V for 4 h at 4 °C. After blocking with 5% nonfat dry milk and 3 washes in PBS, blots were incubated with 1:4000 dilution of secondary anti-mouse-HRP antibody (sc-2005; Santa Cruz Biotechnologies) for 1 h. After 3 washes with PBS, blots were developed for chemiluminescence imaging (sc-2048, Santa Cruz Biotechnologies). A 64bit image was obtained (Typhoon, BioRad) for densitometry analysis using the photoanalyzer tool in Image J (NIH) to estimate the yield of AbBNP synthesis for each Ab/BNP ratio.
2.4. Blast Culture
Commercial childhood B-ALL blasts lines were employed for these studies, including CRLF2-overexpressng (CRLF2+) MHH-CALL4[48] (DSMZ, Braunschweig, Germany) and control GM04154 (Corielle Cell Repositories, New Jersey) cell lines.[49,50] Blasts were grown in RPMI1640 medium (ATCC) with 20% FBS (GibCo)/5% Penicillin-Streptomycin (MPBiomedicals) in 75 cm2 T- flasks at 37 °C, 5psi CO 2 and doubling times of 4 and 2 days, respectively. AbBNPs were incubated with 5 × 106 blasts at 0.1 µg antibody equivalents for a range of times (5 min to 24 h) at 37 °C.
2.5. Fluorescence Microscopy
Laser confocal microscopy was performed on blasts incubated with 100 nm diameter AbBNPs with 10 or 100 Ab/BNP for 24 h at 37 °C in glass microscope chamber slides with a poly-L-lysine mimetic (Labtek) to adhere suspended blasts. After 2 washes with PBS, blasts were fixed with 4% paraformalde-hyde. Primary polyclonal anti-CRLF2 antibodies (sc83871, Santa Cruz) and secondary FITC-labeled anti-CRLF2 antibodies were used to stain blasts not incubated with AbBNPs (but not blasts incubated with AbBNPs since they are FITC-loaded). Mounting media containing DAPI (sc-24941) was added before sealing the slides. Fluorescence images were collected with a laser confocal microscope (Zeiss LSM-510) and voxels were analyzed (Imaris) for co-localization of green FITC-loaded AbBNPs and blue DAPI-stained nuclei to estimate blast internalization of AbBNPs.
2.6. Flow Cytometry
Receptor expression, internalization and cytotoxicity were evaluated simultaneously by multicolor flow cytometry in co-cultures of MHH-CALL4 and GM04154 blasts mixed 1:1 by cell count (5 × 106 /mL). Single cell suspensions of blasts incubated with AbBNPs were first blocked with human Fc Blocker (16–9161–73, BD Pharmingen) on ice before washing twice with PBS and staining with 0.5–1.0 µg of fluorescent-dye-labeled antibodies for CRLF2, CD19 and CD47 for 45 min [(PerCP, 46–5499, eBiosciences); (Alexa Fluor 700, 561031, BD-Pharmingen); (V405, 561594, BD-Pharmingen)]. After surface staining, apoptosis was measured by staining with PE-Annexin V (5165875X, BD-Pharmingen) and 7AAD (51–68981, BD Pharmingen) for 30 min. After adjusting the samples to a final volume of 400 µL in flow buffer (ddPBS), analysis was performed with a flow cytometer (FACSAria II, BD Biosciences) set to collect 20 000 events. Receptor expression, internalization and cytotoxicity were calculated from mean fluorescence intensities according to the following equations (1–4, respectively).[51] The number of blasts in the parent population (P) and autofluorescence from unstained controls were also accounted for.
| (1) |
| (2) |
| (3) |
| (4) |
3. Results and Discussion
3.1. Synthesis and Characterization of AbBNPs with Various Sizes and Antibody Surface Density (Ab/BNP) Ratios
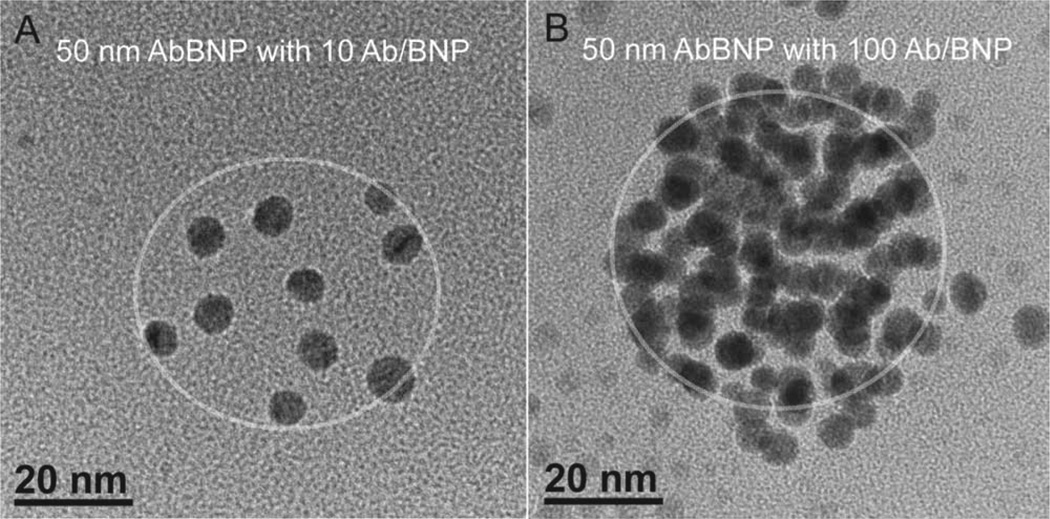
AbBNPs were synthesized by adsorbing anti-CRLF2 antibodies to PLGA nanoparticles of various sizes and antibody surface density (Ab/BNP) ratios. Shown in Figure 1 shows TEM images of 50 nm diameter AbBNPs with antibody surface density (Ab/BNP) ratios of 10 (A) or 100 (B), respectively. A circle was superimposed on these images to show the relative size of the naked nanoparticles. We observed a clear difference in the Ab/ BNP ratio, confirming a ten-fold change in antibody equivalents during AbBNP synthesis. Western blotting provided additional evidence for the synthesis of all AbBNP combinations, including two different AbBNP sizes (50 and 100 nm diameter) and six Ab/BNP ratios (0–200) (Supporting Information Figure 1).
Figure 1.
TEM images of 50 nm diameter AbBNPs with antibody surface density (Ab/BNP) ratios of 10 (A) or 100 (B), respectively. A circle was superimposed on these images to show the relative size of the naked nanoparticles. The scale bar is 20 nm.
3.2. Internalization of AbBNPs by Blasts is Size- and Ab/BNP Ratio-Dependent
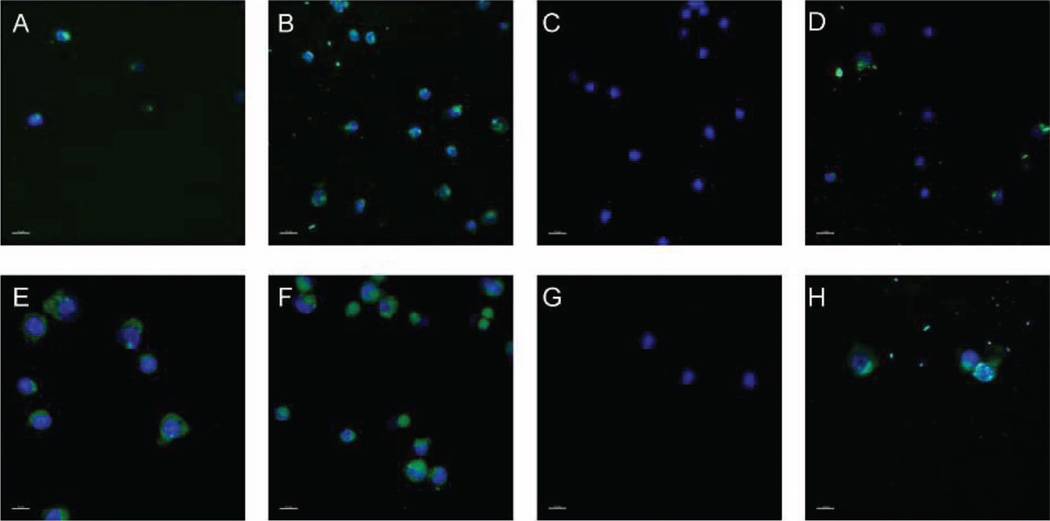
AbBNPs were incubated with CRLF2+ or control blasts. Shown in Figure 2 are fluorescence microscopy images of CRLF2-overexpressing (CRLF2+) (A–D) or control (E–H) B-ALL blasts incubated with 100 nm diameter AbBNPs with Ab/BNP ratios of 10 or 100. AbBNPs (FITC-loaded) co-localized with DAPI-stained nuclei (blue) (A, B, E, F). Three-dimensional co-localization analysis of these images, as performed in this work, is a semi-quantitative method for studying internalization and receptor expression by the non-adherent blasts studied herein. The percent of FITC signals that co-localized with DAPI signals were: 6% (A); 34% (B); 0% (C); 41% (D); 48% (E); 67% (F); 0% (G); 61% (H). Blasts incubated with 100 nm diameter AbBNPs with Ab/BNP ratios of 10 (A,E) or 100 (B,F) clearly show that both CRLF2+ and control blasts internalized AbBNPs. AbBNPs of 100 nm with 100 Ab/BNP were internalized by CRLF2+ and control blasts (B,F) more than those with 10 Ab/BNP (A,E). Controls included blasts that were not incubated with AbBNPs but stained with FITC-labeled anti-CRLF2 secondary antibodies (D,H) and DAPI-stained nuclei of blasts alone (C,G).
Figure 2.
Fluorescence microscopy images of CRLF2-overexpressing (CRLF2+) (A–D) or control (E–H) B-ALL lymphoblasts (blasts) incubated with 100 nm diameter AbBNPs with Ab/BNP ratios of 10 or 100. AbBNPs (FITC-loaded) co-localized with DAPI-stained nuclei (blue) (A,B,E,F). 3D co-localization analysis of FITC and DAPI in these images, as performed here, is a semiquantitative method for studying internalization and CLRF2 expression in non-adherent blasts. Co-localization values were: 6% (A); 34% (B); 0% (C); 41% (D); 48% (E); 67% (F); 0% (G); 61% (H). Blasts incubated with 100 nm diameter AbBNPs with Ab/BNP ratios of 10 (A,E) or 100 (B,F) show that both CRLF2+ and control blasts internalized AbBNPs. Controls included blasts that were not incubated with AbBNPs but stained with FITC-labeled anti-CRLF2 secondary antibodies (D,H) and DAPI-stained nuclei of blasts alone (C,G). The scale bar is 15 µm.
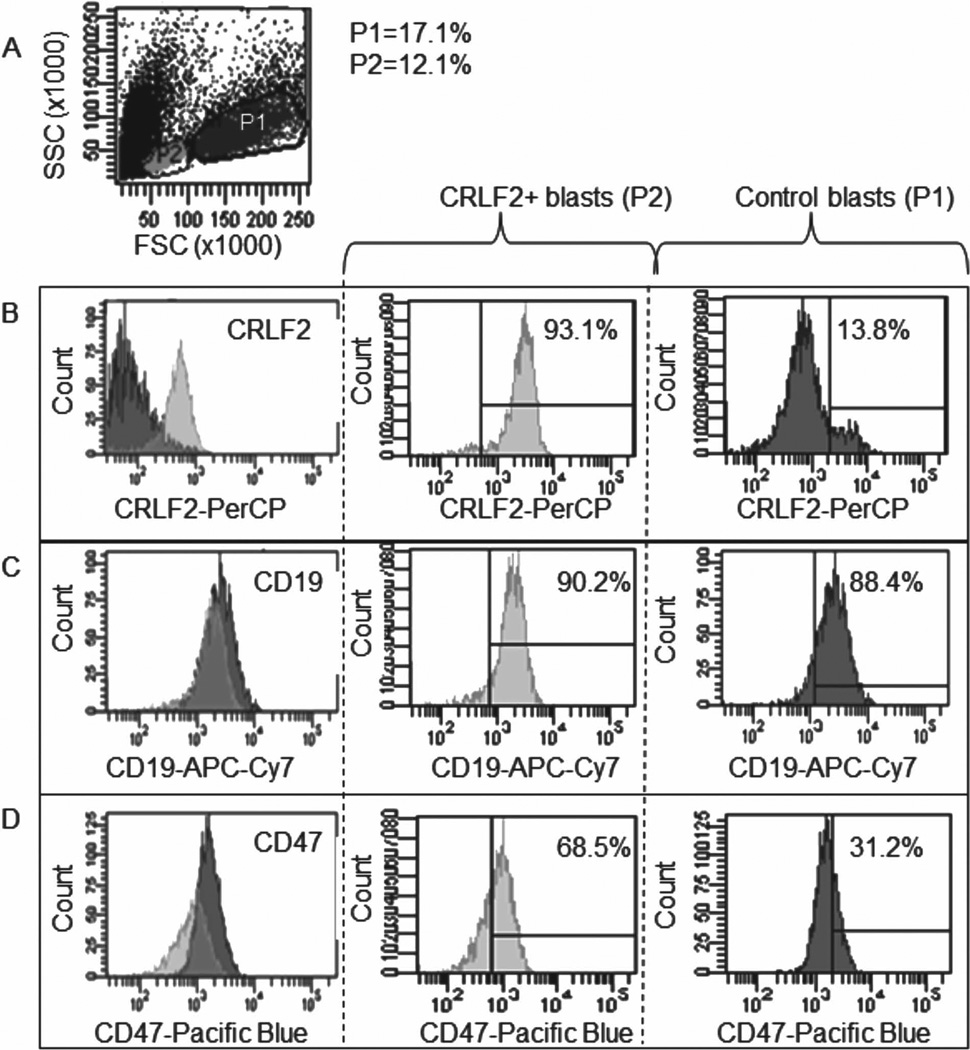
Internalization of AbBNPs by blasts was quantified by multicolor flow cytometry as a function of receptor expression, AbBNP size, and Ab/BNP ratio. Shown in Figure 3 are flow cytometry plots of receptor expression for co-cultures of CRLF2+ (P2) and control (P1) blasts. The relative expression for CRLF2 (B), CD19 (C), and CD47 (D) by CRLF2+ blasts, compared to control blasts, was 14.6 ± 0.8, 1.0 ± 0.7, and 2.2 ± 0.5, respectively. These values were calculated from mean fluorescence intensities as described in the methods section. The number of blasts in the parent population (P) and autofluores-cence from unstained controls were also accounted for. In other words, CRLF2+ blasts express more CRLF2 and CD47 than control blasts, as expected. These results are in excellent agreement with previous studies.[52]
Figure 3.
Flow cytometry plots of receptor expression for co-cultures of CRLF2+ (P2) and control (P1) blasts (A). The relative expression for CRLF2 (B), CD19 (C), and CD47 (D) by CRLF2+ blasts, compared to controls, was 14.6 ± 0.8, 1.0 ± 0.7, and 2.2 ± 0.5, respectively.
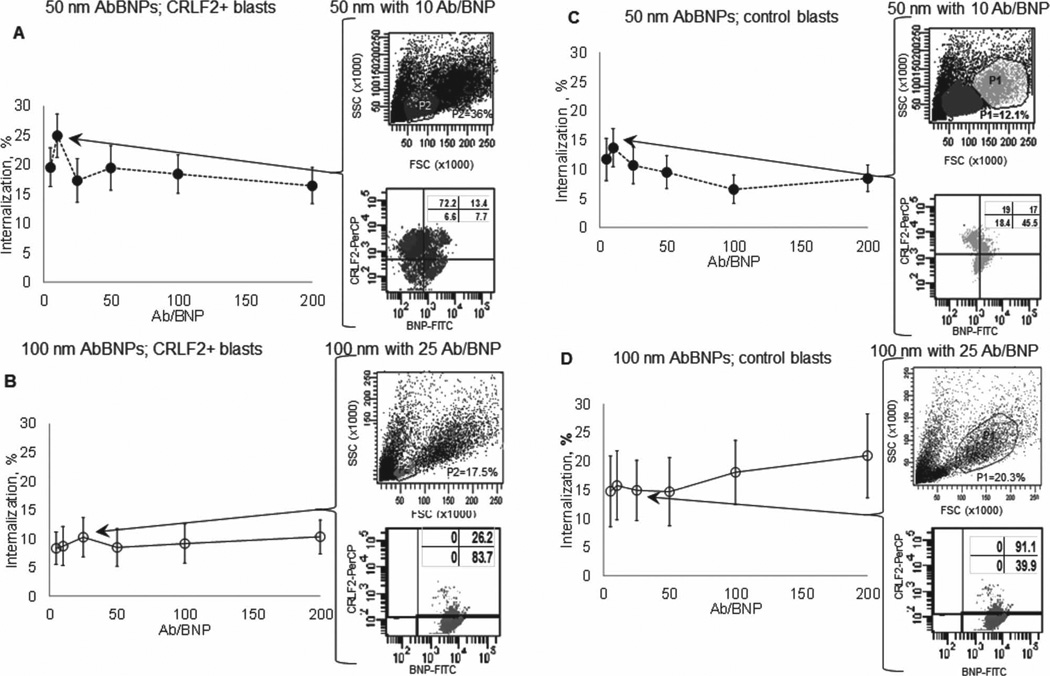
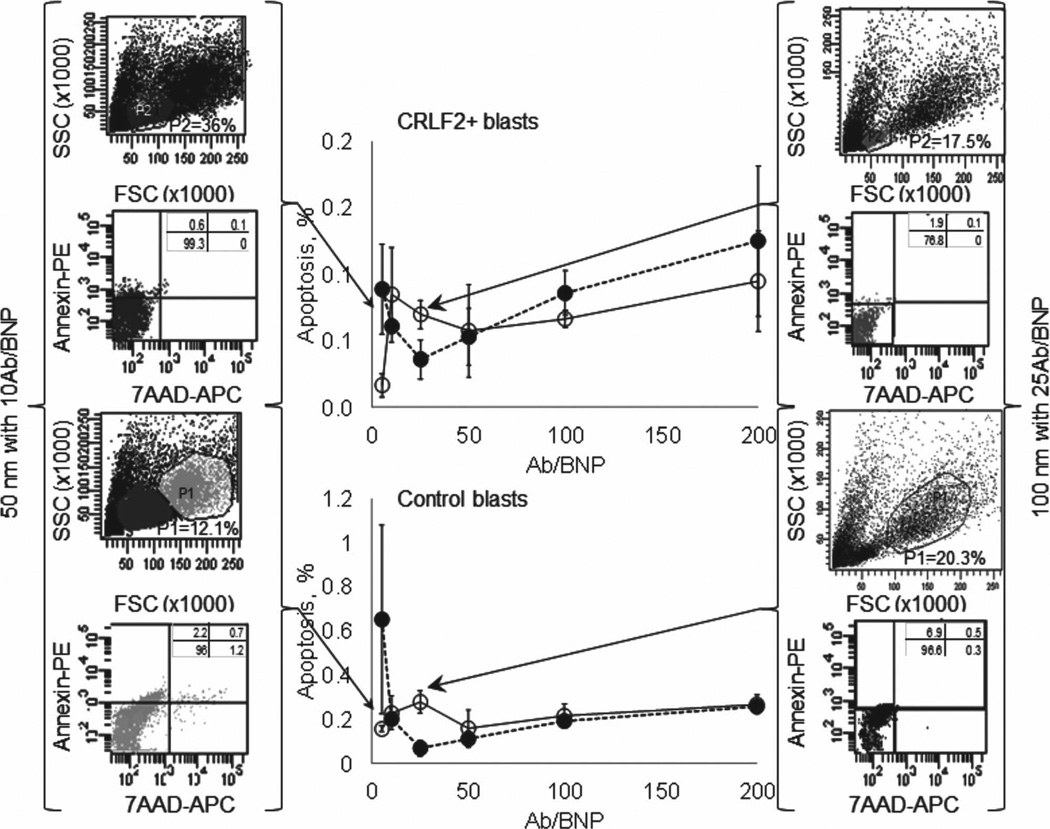
Shown in Figure 4 is blast internalization of AbBNPs as a function of AbBNP size (· = 50 nm; ο = 100 nm) and Ab/BNP ratio (Ab/BNP = 0–200) for CRLF2+ (A,B) and control (C,D) blasts (n = 7). The optimal AbBNP sizes and Ab/BNP ratios for internalization by CRLF2+ blasts was 50 nm with 10 Ab/BNP (A) and 100 nm with 25 Ab/BNP (B), and this was reached in less than 1 h (Supporting Information Figure 2). The optimal AbBNP sizes and Ab/BNP ratios for internalization by control blasts were similar (C,D). Representative flow cytometry plots are shown for optimal AbBNP sizes and Ab/BNP ratios. These measurements by fluorescence microscopy and flow cytometry strongly suggest internalization of AbBNPs by blasts is size- and Ab/BNP ratio-dependent. First, we discovered that CRLF2+ blasts most efficiently internalized 50 nm AbBNPs with 10 Ab/BNP and 100 nm AbBNPs with 25 Ab/ BNP. This is in excellent agreement with the 34 anti-trans-ferrin receptor antibodies (Ox26) per conjugated 100 nm diameter polymersome that were most efficiently internalized.[53] Ab/BNP ratio effects on internalization, albeit subtle, were also observed for anti-JC virus T-antigen antibodies conjugated to iron oxide nanoparticles, where a narrow range of 2 to 8 Ab/NP was investigated due to aggregation problems.[54] Second, we discovered that CRLF2+ blasts internalized 50 nm AbBNPs more efficiently than 100 nm AbBNPs is in excellent agreement with previous work showing that 25–50 nm anti-human epidermal growth factor receptor 2 (HER2) antibody-conjugated colloidal gold nanoparticles were most efficiently internalized.[32]
Figure 4.
Blast internalization of AbBNPs as a function of AbBNP size (· = 50 nm; ο = 100 nm) and antibody surface density ratio (Ab/BNP = 0–200) for CRLF2+ (A,B) and control (C,D) blasts ( n = 7). The optimal AbBNP sizes and Ab/BNP ratios for internalization by CRLF2+ blasts was 50 nm with 10 Ab/BNP (A) and 100 nm with 25 Ab/BNP (B), and this was reached in less than 1 h (Supporting Information Figure 2). The optimal AbBNP sizes and Ab/BNP ratios for internalization by control blasts were similar (C,D). Representative flow cytometry plots are shown for optimal AbBNP sizes and Ab/BNP ratios.
3.3. Factors Contributing to Selective Internalization of AbBNPs
Selectivity is the ratio of internalization of AbBNPs by CRLF2+ blasts to that by control blasts. Differences in AbBNP sizes and Ab/BNP ratios can lead to seemingly counterintuitive differences in selectivity. We observed striking changes in selectivity for different combinations of AbBNP sizes and Ab/BNP ratios. For example, Figure 4 shows that 50 nm AbBNPs with 10 Ab/ BNP were internalized more by CRLF2+ blasts (A) than control blasts (C). In contrast, 100 nm AbBNPs with 25 Ab/BNP were internalized less by CRLF2+ blasts (B) than control blasts (D). The relative expression of CRLF2 is greater in CRLF2+ blasts than control blasts. Therefore, selectivity is not necessarily dependent on receptor expression. CRLF2+ blasts are also significantly smaller than control blasts (Figure 2). This suggests that internalization of AbBNPs may be blast size-dependent, in addition to being AbBNP size- and Ab/BNP ratio-dependent. Another explanation may be that non-selective internalization by receptor mediated endocytosis through Fc receptors, instead of CRLF2, is less pronounced in CRLF2+ blasts than control blasts. These conclusions are in excellent agreement with previous work with anti-CD22 and anti-CD33 antibody drug conjugates, where cell- and receptor specific-differences for saturation of receptors, internalization and receptor recycling were observed.[55]
3.4. All AbBNP Combinations are Non-Cytotoxic
Flow cytometry measurements of apoptosis show that all AbBNP combinations are non-cytotoxic irrespective of AbBNP size or Ab/BNP ratio. Shown in Figure 5 is apoptosis as a function of AbBNP size (· = 50 nm; o = 100 nm) and antibody surface density ratio (Ab/BNP = 0–200) for CRLF2+ (top) and control (bottom) blasts ( n = 3). Cytotoxicity was less than ≈1% for all AbBNP combinations. Representative flow cytometry plots are shown for optimal AbBNP sizes and Ab/BNP ratios. This confirms previous reports establishing PLGA as a FDA-approved non-cytotoxic biodegradable material for a variety of medical devices.
Figure 5.
Apoptosis as a function of AbBNP size (· = 50 nm; ο = 100 nm) and antibody surface density ratio (Ab/BNP = 0–200) for CRLF2+ (top) and control (bottom) blasts (n = 3). Cytotoxicity was less than ≈ 1% for all AbBNP combinations. Representative flow cytometry plots are shown for optimal AbBNP sizes and Ab/BNP ratios.
3.5. AbBNPs Very Slightly Up-Regulate CD47 Expression by Blasts
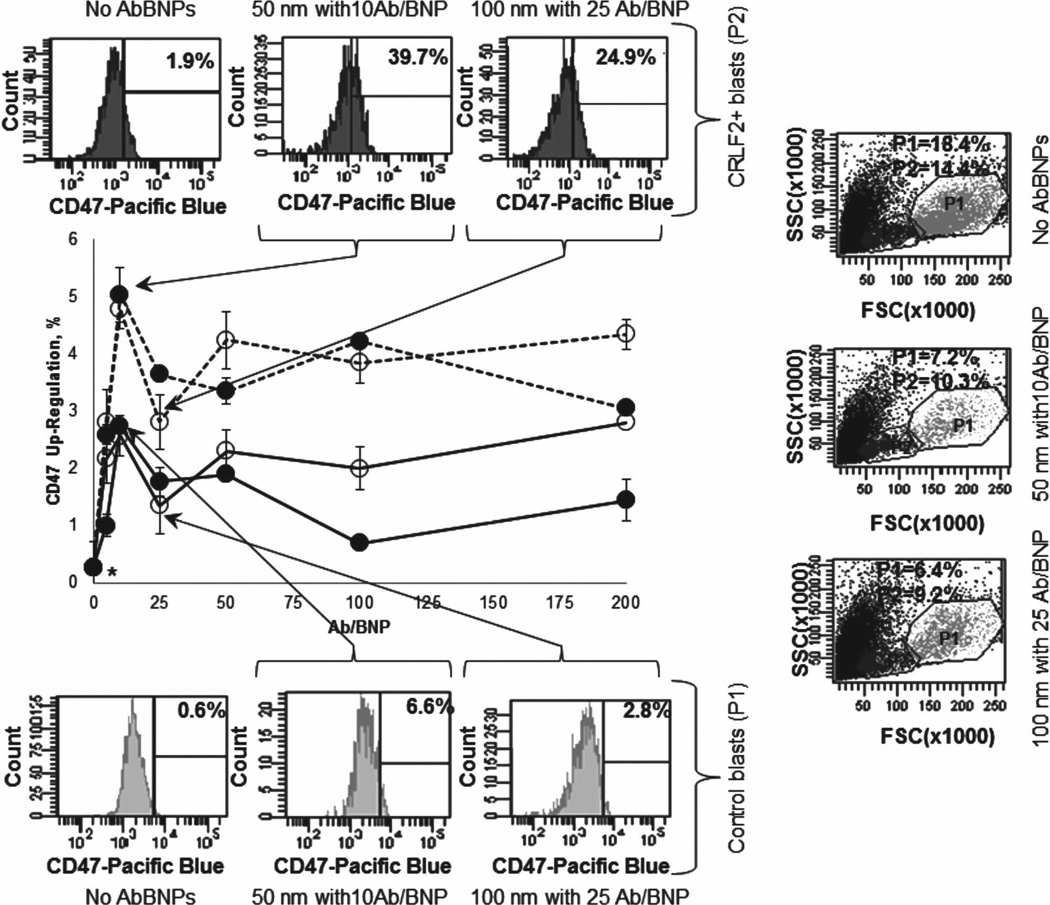
While up-regulation of CD47 by blasts to escape phagocytosis by macrophages (MOs) has been previously reported,[34–42] this study shows that AbBNPs very slightly up-regulate CD47 expression by blasts (Figure 6). Up-regulation of CD47 expression by CRLF2+ blasts incubated with AbBNPs was 4.8% ± 0.3% (P = 0.011; · = 50 nm; 10 Ab/BNP) and 2.8% ± 0.5% (P = 0.096; o = 100 nm; 25 Ab/BNP). Likewise, up-regulation of CD47 expression by control blasts incubated with AbBNPs was 2.7% ± 0.4% (P = 0.036; · = 50 nm; 10Ab/BNP) and 1.3% ± 0.3% (P = 0.018; ο = 100 nm; 25 Ab/BNP). In contrast, up-regulation of CD47 expression in CRLF2+ or control blasts not incubated with AbBNPs was only 0.27 % ± 0.0% (asterisk). Representative flow cytometry plots are shown for optimal AbBNP sizes and Ab/BNP ratios. Akin to the selective internalization of AbBNPs by blasts, up-regulation of CD47 by blasts incubated with AbBNPs is AbBNP size- and Ab/BNP ratio-dependent. The relative up-regulation of CD47 expression by CRLF2+blasts, compared to control blasts, was ≈ 2.0 for both combinations of AbBNPs (1.8 (= 4.8%/2.7%) for · =50 nm; 10 Ab/BNP) and 2.2 (= 2.8%/1.3%) for ο =100 nm; 25 Ab/ BNP)). The reason for this is unknown. We posit that up-regulation of CD47 expression by blasts incubated with AbBNPs may also be affected by cell-specific-differences in the saturation of receptors, internalization and receptor recycling.[55] Nonetheless, our protein expression results are in agreement with previous studies by Weinstock et al. where up-regulation of CD47 messenger RNA by CRLF2+ blasts exposed to JAK/ HSP90 inhibitors was ≈280% compared to controls (Supporting Information).[56]
Figure 6.
CD47 expression is very slightly up-regulated by CRLF2+ (dashed lines) and control (solid lines) blasts incubated with AbBNPs, respectively (n = 4). Up-regulation of CD47 expression by CRLF2+ blasts incubated with AbBNPs was 4.8% ± 0.3% (P = 0.011 ; · = 50 nm; 10 Ab/BNP) and 2.8% ± 0.5% (P = 0.09 6; ο = 100 nm; 25 Ab/BNP). Likewise, up-regulation of CD47 expression by control blasts incubated with AbBNPs was 2.7% ± 0.4% (P = 0.036 ; · = 50 nm; 10Ab/BNP) and 1.3% ± 0.3% (P = 0.018; ο = 100 nm; 25 Ab/BNP). In contrast, up-regulation of CD47 expression in CRLF2+ or control blasts not incubated with AbBNPs was only 0.27 % ± 0.0% (asterisk). Representative flow cytometry plots are shown for optimal AbBNP sizes and Ab/BNP ratios.
3.6. Future Work
We propose that precise engineering of AbBNPs by size and Ab/BNP ratio may improve the internalization and selectivity of future biodegradable nanoparticles for the treatment of leukemia patients, including drug-resistant minority children and Down’s syndrome patients with CRLF2+B-ALL. For example, since AbBNPs very slightly up-regulate CD47 expression, nanoparticles might be engineered to render CRLF2+ blasts more susceptible to non-selective acceleration of phagocytosis by MΦs with anti-CD47 antibodies; albeit “stealth” modification of nanoparticles with methoxypolyethylene glycol (mPEG) may be necessary to block their phagocytosis by MΦs and improve circulation half-life.[57] Finally, the successes of doxorubicin- and L-asparaginase-loaded liposomes suggest that drug-loaded AbBNPs may help overcome potential problems with therapeutic index, resistance and toxicity.
4. Conclusions
This study shows that internalization of AbBNPs in childhood B-ALL blasts is AbBNP size- and Ab/BNP ratio-dependent. All combinations of AbBNPs were non-cytotoxic. We also show that CD47 is very slightly up-regulated by blasts exposed to AbBNPs. CD47 is the marker of self overexpressed by blasts to escape phagocytosis, or cellular devouring, by MΦs. Based on our results, we propose that precise engineering of AbBNPs by size and Ab/BNP ratio may improve the internalization and selectivity of future biodegradable nanoparticles for the treatment of leukemia patients, including drug-resistant minority children and Down’s syndrome patients with CRLF2+B-ALL.
Supplementary Material
Acknowledgements
This work was supported by grants NIH5G12RR013646–12 and NIH5U54RR022762–05 from the National Institute of Health. The authors thank the RCMI program and facilities and Dr. Kelly Nash, Dr. Niannian Ji, Dr. Fidel Santamaria, Dr- Lorenzo Brancaleon (UTSA) and Dr. Robyn Dennis, Dr. Terzah Horton and Dr. Karen Rabin (Texas Children’s Hospital) for technical assistance and insightful discussion. The authors also acknowledge the support of the Cancer Therapy and Research Center at the University of Texas Health Science Center San Antonio, a National Cancer Institute-designated Cancer Center (NIHP30CA054174). The authors thank H.L.H. for inspiration.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Rekha Raghunathan, Pediatric Biochemistry Laboratory Department of Chemistry-BSE 3.108A University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA.
Swetha Mahesula, Pediatric Biochemistry Laboratory Department of Chemistry-BSE 3.108A University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA.
Kranthi Kancharla, Pediatric Biochemistry Laboratory Department of Chemistry-BSE 3.108A University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA.
Preethi Janardhanan, Department of Biology University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA.
Yeshwant L. A. Jadhav, Pediatric Biochemistry Laboratory Department of Chemistry-BSE 3.108A University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA
Robert Nadeau, Pediatric Biochemistry Laboratory Department of Chemistry-BSE 3.108A University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA.
German P. Villa, Department of Physics University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA
Robert L. Cook, Department of Physics University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA
Colleen M. Witt, Department of Physics University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA
Jonathan A. L. Gelfond, Department of Epidemiology & Biostatistics University of Texas Health Science Center at San Antonio San Antonio, TX, 78229, USA
Thomas G. Forsthuber, Department of Biology University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA
William E. Haskins, Email: WEH.scholar@gmail.com, Pediatric Biochemistry Laboratory Department of Chemistry-BSE 3.108A University of Texas at San Antonio One UTSA Circle, San Antonio, TX 78249, USA.
References
- 1.Pui CH, Jeha S. Nat. Rev. Drug Discovery. 2007;6:149. doi: 10.1038/nrd2240. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson JD, Gonzalez A, Wohler-Torres B, Fleming LE, MacKinnon J, Trapido E, Button J, Peace S. Rev. Panam. Salud Publica. 2005;18:5. doi: 10.1590/s1020-49892005000600002. [DOI] [PubMed] [Google Scholar]
- 3.Howe H, Wu X, Ries L, Cokkinides V, Ahmed F, Jemal A, Miller B, Williams M, Ward E, Wingo P, Ramirez A, Edwards B. Cancer. 2006;107:1711. doi: 10.1002/cncr.22193. [DOI] [PubMed] [Google Scholar]
- 4.Kang H, Chen IM, Wilson CS, Bedrick EJ, Harvey RC, Atlas SR, Devidas M, Mullighan CG, Wang X, Murphy M, Ar K, Wharton W, Borowitz MJ, Bowman WP, Bhojwani D, Carroll WL, Camitta BM, Reaman GH, Smith MA, Downing JR, Hunger SP, Willman CL. Blood. 2010;115:7. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollock BH, DeBaun MR, Camitta BM, Shuster JJ, Ravindranath Y, Pullen DJ, Land VJ, Mahoney GH, Jr, Lauer SJ, Murphy SB. J. Clin. Oncol. 2000;18:813. doi: 10.1200/JCO.2000.18.4.813. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia S, Sather HN, Heerema NA, Trigg ME, Gaynon PS, Robison LL. Blood. 2002;100:1957. doi: 10.1182/blood-2002-02-0395. [DOI] [PubMed] [Google Scholar]
- 7.Rochman Y, Kashyap M, Robinson GW, Sakamoto K, Gomez-Rodriguez J, Wagner KU, Leonard WJ. Proc. Natl. Acad. Sci. USA. 2010;107:19455. doi: 10.1073/pnas.1008271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roll JD, Reuther GW. Cancer Res. 2010;70:7347. doi: 10.1158/0008-5472.CAN-10-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertzberg L, Vendramini E, Ganmore I, Cazzaniga G, Schmitz M, Chalker J, Shiloh R, Iacobucci I, Shochat C, Zeligson S, Cario G, Stanulla M, Strehl S, Russell LJ, Harrison CJ, Bornhauser B, Yoda A, Rechavi G, Bercovich D, Borkhardt A, Kempski H, te Kronnie G, Bourquin JP, Domany E, Izraeli S. Blood. 2010;115:1006. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 10.Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, Kang H, Liu W, Dobbin KK, Smith MA, Carroll WL, Devidas M, Bowman WP, Camitta BM, Reaman GH, Hunger SP, Downing JR, Willman CL. Blood. 2010;115:5312. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, Devidas M, Atlas SR, Chen IM, Clifford RJ, Gerhard DS, Carroll WL, Reaman GH, Smith M, Downing JR, Hunger SP, Willman CL. Proc. Natl. Acad. Sci. USA. 2009;106:9414. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasian S, Loh ML. Crit. Rev. Oncog. 2011;16:13. doi: 10.1615/critrevoncog.v16.i1-2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, Estrov Z, Fridman JS, Bradley EC, Erickson-Viitanen S, Vaddi K, Levy R, Tefferi A. New Engl. J. Med. 2010;363:1117. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigert O, Lane AA, Bird L, Kopp N, Chapuy B, van Bodegom D, Toms AV, Marubayashi S, Christie AL, McKeown M, Paranal RM, Bradner JE, Yoda A, Gaul C, Vangrevelinghe E, Romanet V, Murakami M, Tiedt R, Ebel N, Evrot De A, Pover E, Regnier CH, Erdmann D, Hofmann F, Eck MJ, Sallan SE, Levine RL, Kung AL, Baffert F, Radimerski T, Weinstock DM. J. Exp. Med. 2012;209:259. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiskus W, Verstovsek S, Manshouri T, Rao R, Balusu R, Venkannagari S, Rao NN, Ha K, Smith JE, Hembruff SL, Abhyankar S, McGuirk J, Bhalla KN. Clin. Cancer Res. 2011;17:7347. doi: 10.1158/1078-0432.CCR-11-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong J, Kim MS, Chaerkady R, Wu X, Huang TC, Getnet D, Mitchell CJ, Palapetta SM, Sharma J, O’Meally RN, Cole RN, Yoda A, Moritz A, Loriaux MM, Rush J, Weinstock DM, Tyner JW, Pandey A. Mol. Cell Proteomics. 2012;11:M112017764. doi: 10.1074/mcp.M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vail DM, Amantea MA, Colbern GT, Martin FJ, Hilger RA, Working PK. Semin. Oncol. 2004;31:16. doi: 10.1053/j.seminoncol.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Ho DH, Brown NS, Yen A, Holmes R, Keating M, Abuchowski A, Newman RA, Krakoff IH. Drug Metab. Dispos. 1986;14:349. [PubMed] [Google Scholar]
- 19.Jorge JC, Perez-Soler R, Morais JG, Cruz ME. Cancer Chemother. Pharmacol. 1994;34:230. doi: 10.1007/BF00685082. [DOI] [PubMed] [Google Scholar]
- 20.Gaspar MM, Perez-Soler R, Cruz ME. Cancer Chemother.Pharmacol. 1996;38:373. doi: 10.1007/s002800050497. [DOI] [PubMed] [Google Scholar]
- 21.Mahapatro A, Singh DK. J. Nanobiotechnol. 2011;9:55. doi: 10.1186/1477-3155-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elsabahy M, Wooley KL. Chem. Soc. Rev. 2012;41:2545. doi: 10.1039/c2cs15327k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasper MM, Blanco D, Cruz ME, Alonso MJ. J. Controlled Release. 1998;52:53. doi: 10.1016/s0168-3659(97)00196-x. [DOI] [PubMed] [Google Scholar]
- 24.Wolf M, Wirth M, Pittner F, Gabor F. Int. J. Pharm. 2003;256:141. doi: 10.1016/s0378-5173(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 25.Cun D, Jensen DK, Maltesen MJ, Bunker M, Whiteside P, Scurr D, Foged C, Nielsen HM. Eur. J. Pharm. Biopharm. 2011;77:26. doi: 10.1016/j.ejpb.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, Lovelace P, Scheeren FA, Chao MP, Weiskopf K, Tang C, Volkmer AK, Naik TJ, Storm TA, Mosley AR, Edris B, Schmid SM, Sun CK, Chua MS, Murillo O, Rajendran P, Cha AC, Chin RK, Kim D, Adorno M, Raveh T, Tseng D, Jaiswal S, Enger PO, Steinberg GK, Li G, So SK, Majeti R, Harsh GR, van de Rijn M, Teng NN, Sunwoo JB, Alizadeh AA, Clarke MF, Weissman IL. Proc. Natl. Acad. Sci. USA. 2012;109:6662. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil Y, Panyam J. Int. J. Pharm. 2009;367:195. doi: 10.1016/j.ijpharm.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qaddoumi MG, Gukasyan HJ, Davda J, Labhasetwar V, Kim KJ, Lee VH. Mol. Vis. 2003;9:559. [PubMed] [Google Scholar]
- 29.Jiang W, Kim BY, Rutka JT, Chan WC. Nat. Nanotechnol. 2008;3:145. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 30.Pang Z, Lu W, Gao H, Hu K, Chen J, Zhang C, Gao X, Jiang X, Zhu C. J. Controlled Release. 2008;128:120. doi: 10.1016/j.jconrel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Biochem. J. 2004;377:159. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang W, Kim BY, Rutka JT, Chan WC. Nat. Nanotechnol. 2008;3:145. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 33.Charrois GJ, Allen TM. Biochim. Biophys .Acta. 2003;1609:102. doi: 10.1016/s0005-2736(02)00661-2. [DOI] [PubMed] [Google Scholar]
- 34.Tsai RK, Rodriguez PL, Discher DE. Blood Cells Mol. Dis. 2010;45:67. doi: 10.1016/j.bcmd.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsson M, Oldenborg PA. Blood. 2008;112:4259. doi: 10.1182/blood-2008-03-143008. [DOI] [PubMed] [Google Scholar]
- 36.Bouguermouh S, Van VQ, Martel J, Gautier P, Rubio M, Sarfati M. J. Immunol. 2008;180:8073. doi: 10.4049/jimmunol.180.12.8073. [DOI] [PubMed] [Google Scholar]
- 37.van den, Berg TK, van der Schoot CE. Trends Immunol. 2008;29:203. doi: 10.1016/j.it.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Van VQ, Lesage S, Bouguermouh S, Gautier P, Rubio M, Levesque M, Nguyen S, Galibert L, Sarfati M. EMBO J. 2006;25:5560. doi: 10.1038/sj.emboj.7601415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian S, Parthasarathy R, Sen S, Boder ET, Discher DE. Blood. 2006;107:2548. doi: 10.1182/blood-2005-04-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oldenborg PA. Leuk. Lymphoma. 2004;45:1319. doi: 10.1080/1042819042000201989. [DOI] [PubMed] [Google Scholar]
- 41.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Science. 2000;288:2051. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 42.Olivier JC. NeuroRx. 2005;2:108. doi: 10.1602/neurorx.2.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao MP, Alizadeh AA, Tang C, Jan M, Weissman-Tsukamoto R, Zhao F, Park CY, Weissman IL, Majeti R. Cancer Res. 2011;71:1374. doi: 10.1158/0008-5472.CAN-10-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao MP, Alizadeh AA, Tang C, Myklebust JH, Varghese B, Gill S, Jan M, Cha AC, Chan CK, Tan BT, Park CY, Zhao F, Kohrt HE, Malumbres R, Briones J, Gascoyne RD, Lossos IS, Levy R, Weissman IL, Majeti R. Cell. 2010;142:699. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kocbek P, Obermajer N, Cegnar M, Kos J, Kristl J. J. Controlled Release. 2007;120:18. doi: 10.1016/j.jconrel.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Arora G, Shukla J, Ghosh S, Maulik SK, Malhotra A, Bandopadhyaya G. PLoS One. 2012;7:e34019. doi: 10.1371/journal.pone.0034019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olivier JC, Huertas R, Lee HJ, Calon F, Pardridge WM. Pharm. Res. 2002;19:1137–1143. doi: 10.1023/a:1019842024814. [DOI] [PubMed] [Google Scholar]
- 48.Tomeczkowski J, Yakisan E, Wieland B, Reiter A, Welte K, Sykora KW. Br. J. Haematol. 1995;89:771. doi: 10.1111/j.1365-2141.1995.tb08414.x. [DOI] [PubMed] [Google Scholar]
- 49.Shipp MA, Schwartz BD, Kannapell CC, Griffith RC, Scott MG, Ahmed P, Davie JM, Nahm MH. J. Immuno.l. 1983;131:2458. [PubMed] [Google Scholar]
- 50.Lazarus H, Barell EF, Oppenheim S, Krishan A. In Vitro. 1974;9:303. [PubMed] [Google Scholar]
- 51.Antczak AJ, Vieth JA, Singh N, Worth RG. Clin. Vaccine Immunol. 2011;18:210. doi: 10.1128/CVI.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, Chandrasekaran T, Chapiro E, Gesk S, Griffiths M, Guttery DS, Haferlach C, Harder L, Heidenreich O, Irving J, Kearney L, Nguyen-Khac F, Machado L, Minto L, Majid A, Moorman AV, Morrison H, Rand V, Strefford JC, Schwab C, Tonnies H, Dyer MJ, Siebert R, Harrison CJ. Blood. 2009;114:2688. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- 53.Pang Z, Lu W, Gao H, Hu K, Chen J, Zhang C, Gao X, Jiang X, Zhu C. J. Controlled Release. 2008;128:120. doi: 10.1016/j.jconrel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Knight LC, Romano JE, Krynska B, Faro S, Mohamed FB, Gordon J. J. Mol. Biomark Diagn. 2010;1:1000102. doi: 10.4172/2155-9929.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Vries JF, Zwaan CM, De Bie M, Voerman JS, den Boer ML, van Dongen JJ, van der, Velden VH. Leukemia. 2012;26:255. doi: 10.1038/leu.2011.206. [DOI] [PubMed] [Google Scholar]
- 56.Weigert O, Lane AA, Bird L, Kopp N, Chapuy B, van Bodegom D, Toms AV, Marubayashi S, Christie AL, McKeown M, Paranal RM, Bradner JE, Yoda A, Gaul C, Vangrevelinghe E, Romanet V, Murakami M, Tiedt R, Ebel N, Evrot E, De Pover A, Regnier CH, Erdmann D, Hofmann F, Eck MJ, Sallan SE, Levine RL, Kung AL, Baffert F, Radimerski T, Weinstock DM. J. Exp. Med. 2012;209:259. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bazile D, Prud’homme C, Bassoullet MT, Marlard M, Spenlehauer G, Veillard M. J. Pharm. Sci. 1995;84:493. doi: 10.1002/jps.2600840420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.