Abstract
In recent years RNA-Seq technology has been used not only to quantify differences in gene expression but also to understand the underlying mechanisms that lead to these differences. Nucleotide sequence variation arising through evolution may differentially affect the expression profiles of divergent species. RNA-Seq technology, combined with techniques to differentiate parental alleles and quantify their abundance, have recently become popular methods for allele specific gene expression (ASGE) analyses. However, analysis of gene expression within interspecies hybrids may be difficult when one of the two parental genomes represented in the hybrid does not have robust genomic resources or available transcriptome data. Herein, we compare two strategies for analyzing allele specific expression within interspecies hybrids produced from crossing two Xiphophorus fish species. The first strategy relies upon a robust reference transcriptome assembly from one species followed by identification of SNPs and creation of an in silico reference transcriptome for the second species. The second strategy employs de novo assembly of reference transcriptomes for both parental species followed by identification of homologous transcripts prior to mapping hybrid reads to a combined hybrid reference. Our results show that, although both methods are able to achieve balanced allelic distribution upon read mapping of F1 hybrid fish transcriptomes, the second “de novo” assembly approach is superior for ASGE analyses and leads to results more consistent with those found from quantitative real time PCR assessment of gene expression. In addition, our analysis indicates that indels between the two parental alleles are the major cause of the differences in results observed when employing these two methods.
Keywords: RNA-Seq, Transcriptome, Assembly, Read mapping, Allele specific, Interspecies hybrid
1. Introduction
In recent years a number of studies have assessed ASGE among interspecies hybrids of inbred maize, yeast and Drosophila (Stupar and Springer, 2006; Tirosh et al., 2009; McManus et al., 2010). These studies have provided a better understanding of evolutionary alterations in regulatory elements that may occur upon speciation and lead to modulation of allele expression in the hybrid genetic background. High-throughput methods such as serial analysis of gene expression (SAGE) (Wei et al., 2004), allele-specific microarray (Tirosh et al., 2009) and next generation sequencing (NGS) (Main et al., 2009a) have all been used to assess ASGE on a global genomic scale. Recently, NGS based methods that simultaneously allow identification of polymorphisms and determination of ASGE have emerged as the most widely used method to study species having sparse genomic resources or limited reference transcriptome information.
For well-studied model species with robust genomic resources, markers for divergence among individuals (e.g., in human) or strains/species (e.g., yeast and Drosophila) are ample and well characterized. Public databases for genomic and transcriptomic data simplify the identification of polymorphisms in these systems. However, for species that lack these resources, as with many aquatic species, extra steps are necessary to identify species-specific variants before assessing allele specific expression. A common problem for researchers using genome resource limited species is having genomic or reference transcriptome information available for one species while having experimental necessity dictates the use of a second, related species [e.g.,(Whitehead et al., 2011; Garcia et al., 2012b)]. This situation is similar to the difficulties inherent to having genomic information available for only one species while attempting to assess ASGE within interspecies hybrids where the second species genome in the hybrid makes it necessary to identify allele variants that allow ASGE analysis.
To address ASGE in interspecies hybrids, one strategy identifies species variants by mapping NGS reads sequenced from a second species (the one lacking a transcriptome) to the reference genome/transcriptome (Main et al., 2009b; Rozowsky et al., 2011; Shen et al., 2012). After species variants are identified, hybrids reads can be mapped to these divergent regions for allele specific quantification. However, a drawback of this strategy is the potential to introduce bias due to reduced read mapping efficiency for reads derived from the second (non-reference) parental allele. To overcome the reduced efficiency, an in silico reference transcriptome assembly for the second species may be created by replacing SNPs in a copy of the reference transcriptome with those identified as specific for the second parent. Then, parent-specific transcriptomes (e.g., parent one de novo assembly with parent two in silico assembly) are used for read mapping (Shen et al., 2012). Herein, this method is designated as an “in silico-based” approach. A second strategy is to assume the cost and time burden of obtaining deep transcriptome sequencing reads for the second species and then assembling an independent de novo transcriptome. This method is herein designated as the “de novo-based” approach. With the rise in NGS throughput and corresponding cost reduction, the possibility of obtaining independent de novo transcriptome assemblies for specific experimental projects becomes more practical. For the de novo-based approach, homologous gene pairs from the two parental transcriptome assemblies are identified and compared. Hybrid reads are then mapped to both transcriptomes for quantification of ASGE.
Herein we compare these two strategies (in silico-based and de novo-based) using the same set of RNA-Seq reads produced from two inbred Xiphophorus parental species and from F1 interspecies hybrids produced from crossing them. We provide detailed pros and cons of the two methods that may provide guidance for similar ASGE studies in species having little or no publicly available genomic and/or transcriptomic information.
2. Materials and methods
2.1. RNA isolation and sequencing
All fishes were supplied by the Xiphophorus Genetic Stock Center (see http://www.xiphophorus.txstate.edu). The X. maculatus Jp 163 A parental line in its 107th generation of inbreeding produced a brood of four fish (0.73 g) sacrificed for RNA as immature, but sexually dimorphic, fry (45 do.). X. hellerii (Rio Sarabia line, pedigree 11479) produced a brood of 4 fry (0.88 g) that were utilized for RNA isolation (31 do). X. maculatus Jp 163 A (x) X. hellerii F1 interspecies hybrid (pedigree 11470) RNA was isolated from a brood of 5 fry (0.84 g, 45 do). In this interspecies cross the female X. maculatus parent was from the 106th generation, and the male X. hellerii was from pedigree 11103. The total RNA was isolated after maceration of liquid nitrogen frozen whole fry using a pestle followed by re-suspension in Trizol (Invitrogen, Carlsbad, CA, USA). RNA was further purified using an RNAeasy mini RNA isolation kit (Qiagen, Valencia, CA, USA). Residual DNA was eliminated by column DNase digestion at 37 °C (30 min). The integrity of RNA was assessed by gel electrophoresis (2% agarose in TAE running buffer) and the concentration was determined using a spectrophotometer (Nano Drop Technologies, Willmington, DE, USA).
At least 20 μg total RNA was used for library construction and subjected to ABI-SOLiD-3 sequencing at Cofactor Genomics Inc. (St. Louis, MO, USA). Two barcodes were run for each sample, yielding 63, 45, and 40 million reads of 50 bp single end sequences for X. maculatus, X. hellerii, and the F1 interspecies hybrid RNAs, respectively.
2.2. Assembly of the X. maculatus and X. hellerii reference transcriptomes
For X. maculatus, RNA samples isolated from the brain, heart, and liver tissues of mature individuals were sequenced using the Illumina GAIIx platform as 60 bp, paired end reads (Expression Analysis® Inc. Durham, NC, USA). After custom designed filtration, [to check uncalled base, ‘B’ flags, and low quality regions, [see (Garcia et al., 2012a)]], 200 million paired reads and 6 million singletons remained in a combined read set and were used for transcriptome assembly. For X. hellerii, RNA samples were extracted from1month old whole fry, and the brain, liver, ovaries and testes of mature adults, then sequenced and processed as described above. After custom filtration, 173 million paired reads and 22 million singletons were used for transcriptome development.
We employed VELVET (Zerbino and Birney, 2008) to guide the assembly using combined paired-end and singleton reads. We first used k-mer sizes from 21 bases to 49 bases and compared assemblies produced from different k-mer sizes to identify the assembly with the longest N50 length. For the final assembly, we used a k-mer size of 43 and 35 bases for X. maculatus and X. hellerii, respectively (Supplementary data Fig. 1). We employed Oases (http://www.ebi.ac.uk/∼zerbino/oases/) to perform the final assembly and reporting of putative transcripts and splice variants using a coverage cutoff of 4, insert length estimate of 120, and other parameters at default values. We filtered out sequences smaller than 500 bp. The final X. maculatus assembly contained 110,604 transcripts with an N50 of 3922 bp, an average length of 2197 bp, and a total size of 243 Mbp. For X. hellerii, the final assembly contained 242,675 transcripts with an N50 of 3280 bp, average length of 1991 bp and a total size of 483 Mbp. We did not filter out sequences in the X. hellerii transcriptome based on length in order to maintain a deeper coverage for transcriptome alignment.
2.3. Allele-specific expression analysis
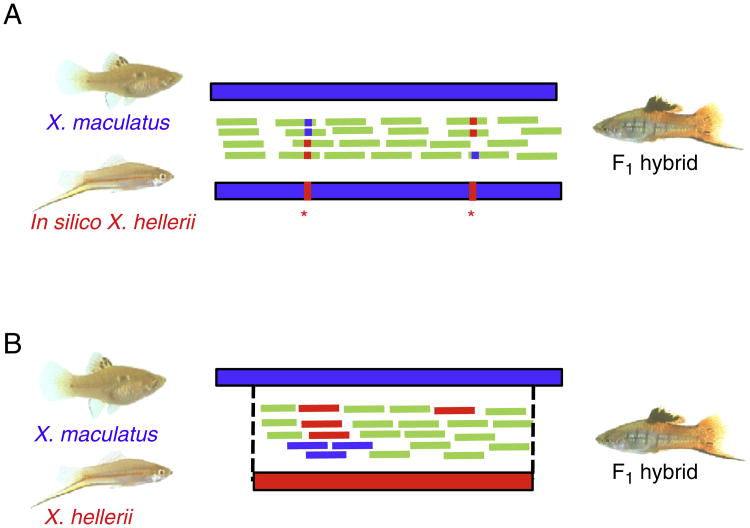
We used two methods to normalize the read count data and quantify allele-specific expression. The in silico-based method was performed as previously detailed in a study of X. maculatus and X. couchianus interspecies hybrids (Shen et al., 2012). Briefly, X. hellerii reads were mapped to the X. maculatus reference transcriptome by Bowtie and used to identify 127,585 species-specific SNP variants between two species using Samtools (Langmead et al., 2009; Li et al., 2009). We then used these SNPs to create an in silico X. hellerii reference transcriptome by replacing each nucleotide base in a copy of the X. maculatus transcriptome with the appropriate X. hellerii consensus SNP base (Fig. 1). Reads in the F1 interspecies hybrid were then mapped to both de novo X. maculatus and in silico X. hellerii transcriptomes using PerM (Chen et al., 2009) to quantify allele-specific expression.
Fig. 1.
Overview of two allele-specific expression profiling methods. A). In silico-based method. An in silico X. hellerii transcriptome, which was made by replacing variants (in red and marked by asterisks) in a copy of the X. maculatus transcriptome, was used along with the unaltered (de novo) X. maculatus transcriptome as targets for read mapping. Reads were counted based on the species-specific nucleotide variants at locations previously identified (marked as red or blue). B). The homologous regions of parental transcripts were identified by comparative alignments. F1 hybrid reads were mapped to both transcriptomes and only those reads mapped to one transcript were used for expression profiling. Reads that did not map to any transcript (not shown) or mapped to both transcripts (green) were not used in profiling.
For the de novo-based method, the same X. maculatus assembly was used, but a new transcriptome for X. hellerii was assembled de novo. These two de novo parental transcriptomes were aligned with each other using BLASTN (E value <10−06) and aligned regions were extracted using a custom written Perl script. There were 75,164 reciprocal best-hit pairs in the alignment representing 145Mbp of total aligned sequences. Only aligned sequences from reciprocal best hits were used for the ensuing read mapping analyses. A Perl script was used to count the allele specific reads in the hybrid. Reads in the F1 interspecies hybrid were then mapped to both transcriptomes using PerM (Chen et al., 2009) to quantify allele-specific expression.
2.4. Real-time PCR
Results were verified for selected genes by qualitative RT-PCR analysis and SYBR Green-based detection with an Applied Biosystems 7500Fast system (Applied Bioscience, Carlsbad, CA, USA). Initially, each set of designed primers was tested for allele specificity in a 20 μL reaction consisting of 1 μL of cDNA, 0.5 μM of each primer, and 10 μL SYBR Green ready mix. Each reaction was subjected to 40 cycles each at 95 °C for 20 s, 95 °C for 15 s, and 60 °C for 30 s, before being subjected to melting curve analysis. Amplified products were also analyzed for size by agarose gel electrophoresis (2% agarose in TAE running buffer). The 18S gene was selected for normalization of all samples. The mean CT values from triplicate runs were used to calculate relative expression levels. The allele specific primers were then used to determine efficiency following the same procedure outlined above but using a series of dilutions of cDNA to establish a standard curve (100 ng, 10 ng, 1 ng, and 0.1 ng, respectively). Genes were selected based on the following criteria. First, in the hybrids, genes must have more than 20 allele specific reads in both de novo and in silico strategies. Second, the alignment region between two Xiphophorus homologous transcripts must be longer than 200 bp and have enough divergence between sequences to design allele specific primers. Third, only primer sets with an efficiency percentage of ±10% of one another were utilized. Once the primer efficiency and allele specificity were established, the primers were used to test relative expression of each allele in immature X. maculatus (pedigree 109a), X. hellerii (pedigree 11479) and F1 hybrid (pedigree 11470) fish using eight technical repeats each. Primer sequences and real time PCR results are presented in Supplementary data 3.
3. Results and discussion
3.1. ASGE analyses using in silico-based method
The interspecies hybrid model represents two parental genomes, presenting the challenge of assigning reads to their corresponding parental alleles in an unbiased fashion. Thus, we first created an in silico X. hellerii transcriptome based on the X. maculatus de novo transcriptome by replacing X. maculatus bases with identified X. hellerii (Fig. 1). A total of 127,585 SNPs was initially called by Samtools (Li et al., 2009). Subsequent quality filters (Garcia et al., 2012a; Shen et al., 2012) left 32,236 SNPs that had at least 20 reads supporting the SNP call. Only these SNPs were used for the ensuing analyses and were able to be unambiguously assigned to 8132 transcripts. Thus, in the transcriptomes of these two species, one SNP occurred, on average, every 7.5 kb.
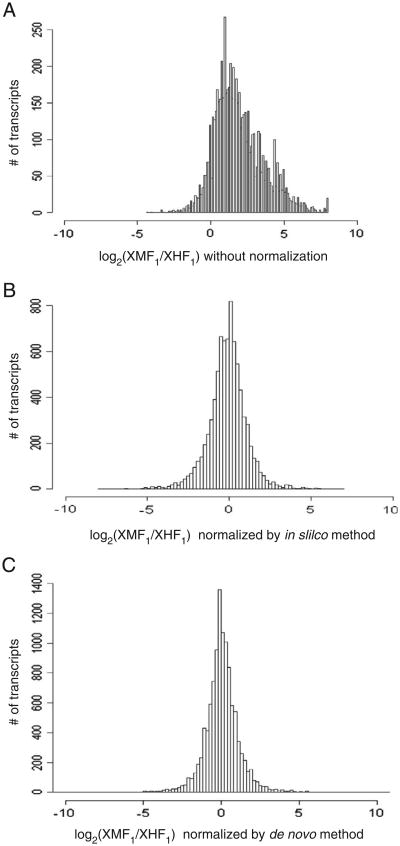
Once we created the in silico X. hellerii transcriptome, both parental transcriptomes were fused to produce a hybrid transcriptome. RNA-Seq reads from the F1 hybrid were mapped to the hybrid (fused) transcriptome and the expression levels of X. maculatus and X. hellerii alleles were calculated from the mapping results. Fig. 2 presents a histogram of the percentage of X. hellerii specific allele reads mapped to each transcript in the F1 hybrid background for various transcriptomes. In Fig. 2A, only unaltered X. maculatus transcripts were used for mapping and they show a bias towards the X. maculatus alleles (shift to the right). In contrast, the in silico-based transcriptome (Fig. 2B) exhibits balanced allele distribution for the F1 hybrid reads against the X. hellerii transcriptome. Both X. maculatus and X. hellerii alleles exhibit a near normal distribution of allele expression in the hybrid genetic background (with a mean of 52% and standard deviation of 17%) when mapped to the fused transcriptome.
Fig. 2.
Allele distribution in F1 hybrid genetic background. The x-axis is the log2 ratio of the number of reads carrying the X. maculatus allele vs. the number of reads carrying the X. hellerii allele. A value of zero indicates an equal distribution of the two alleles while a positive value indicates stronger X. maculatus allele expression than the X. hellerii allele; the reverse balance in expression is indicated by a negative value. (A) F1 hybrid reads are mapped to only X. maculatus reference transcriptome. (B) F1 hybrid reads are mapped to X. maculatus and in silico X. hellerii transcriptomes allowing five mismatches. (C) F1 hybrid reads are mapped to de novo fused assemblies of both X. maculatus and X. hellerii transcriptomes allowing no mismatches.
3.2. ASGE analyses using de novo based transcriptome comparison method
The second, de novo-based approach, involved creating a de novo X. hellerii transcriptome assembly, identifying homologous sequences between the two transcriptomes, and mapping reads to the combined set of transcripts. One consideration when using two independent assembliesis that many assembled sequences (gene or partial transcripts) may appear exclusively in one of the assemblies and this could cause overestimation of one allele over the other. To eliminate differences between the two reference transcriptomes, reciprocal BLAST best hits were used to identify the homologous sequences present in both transcriptomes and only the aligned sequences of these best hits were used for ASGE analyses. There were two major differences between this approach and the in silico approach were used in mapping the hybrid RNA-Seq reads to the de novo transcriptomes; (1) no mismatches were allowed between a read and a transcript, while five mismatches were allowed for the in silico approach, and (2) reads were mapped to both parental transcriptomes in a combined set, but the mapped reads were only counted if they mapped uniquely to one transcript (and not the homologous transcript from the other parent). This approach allowed us to measure only reads that mapped to species-specific variable regions in the transcripts and thus, obtain an accurate estimation of ASGE. A total of 11,029 transcripts was identified that exhibited more than 20 allele-specific mapped reads and only these transcripts were used in ensuing analyses. This method also produced a balanced distribution of allele preference (mean of 49% and a standard deviation of 16%, Fig. 2C).
3.3. Similarities of in silico-based and de novo-based methods
Of 110,604 transcripts in the X. maculatus reference transcriptome, both in silico-based and de novo-based methods provided estimations of ASGE for a similar number of transcripts (8132 vs. 11,029, respectively). We can only study ASGE in transcripts with divergent sequences between the two species donating genomes to the interspecies hybrid. Thus, only a relatively low percentage of transcripts (∼10% of total transcripts in the transcriptomes) could be assessed, suggesting that most mRNA sequences between the two parental species are well conserved. A SNP call frequency of 7.5/kb suggests that the vast majority of assembled transcripts (with an average length of 2197 bp in the X. maculatus transcriptome) were identical in sequence between these two species. Initially, 19,337 and 18,945 transcripts were identified that had sequence divergence in the in silico and de novo based methods, respectively. The number of transcripts that yielded ASGE information is further reduced by our requirement for at least 20 species-specific reads to ensure reliability of the ASGE estimation. The ASGE results presented here are based on 40 million 50 bp single-ended RNA-Seq reads from F1 hybrid fish. With increased read length and read depth that would be provided by continuously improving sequencing technology, ASGE analysis is expected to improve and gain sensitivity for lower-coverage transcripts.
Both in silico-based and de novo-based methods produced similar patterns in terms of defining the relative use of parental alleles (Fig. 2B and C). In the hybrid genetic background, near normal distributions are shown for both methods. In addition, both distributions have similar standard deviations (16% vs. 17%) of allele usage in the F1, further indicating that both methods provide similar estimations of the global characteristics of ASGE in the F1 hybrid.
3.4. Differences between in silico-based and de novo-based methods
Although both read mapping methods use the same principle to identify divergent markers between the parental species, there are major differences between them. First, the de novo-based method requires sequencing and assembling a second transcriptome with at least comparable depth to the first transcriptome. In this work, six lanes of sequencing data from Illumina GAIIx were required in order to build the second X. hellerii transcriptome with a cost of more than $2000 per lane. In addition, a computer with at least 200 GB of RAM is recommended in order to use the best currently available transcriptome assembler software packages. Thus, methods, cost, and equipment may be prohibitive for many investigators with this method. The in silico-based method, at the same time, requires fewer reads; we were able to use only 45 million X. hellerii single end reads to achieve ASGE data for a comparable numbers of transcripts. In addition, if one needed to examine more than two species, data interpretation would be easier if all divergence is determined from a single reference transcriptome.
In addition, the two methods are prone to different types of errors based on how reads are mapped and counted. The in silico-based method focuses on single nucleotide divergence while other types of divergence, such as insertions or deletions (indels), between two sequences are omitted. Bowtie1 (Langmead et al., 2009), the read mapping tool used herein, does not support gap alignment and is therefore insensitive to indels. This problem can be partially solved using mapping tools that support gap alignment [e.g., BWA (Li and Durbin, 2009)] or only using coding sequences where lower indel frequencies are expected (Shabalina and Spiridonov, 2004). However these steps would further increase the complexity of analysis. For the de novo-based method, single nucleotide divergence and small indels are allowed by the BLAST alignment and thus, reads that span indels can be mapped to their corresponding transcripts. This might explain why more total reads are mapped in the de novo-based method than the in silico-based method (824,008 vs. 522,326, respectively, for F1 hybrid reads). However, the de novo-based method allowed no mismatches during read mapping in order to differentiate alleles. Therefore, reads with sequencing errors cannot be mapped or counted. In contrast, the in silico-based method can tolerate up to five mismatches from sequencing errors (estimated to be 0.1% in SOLiD and 1% in Illumina) or from naturally occurring divergence. The 50 bp SOLiD reads we used have an estimated 0.1% rate of error (McKernan et al., 2009) or approximately 1 bp in every 20 reads. However, with improved technology yielding longer NGS reads, sequencing errors may have a larger impact when using de novo-based methods for ASGE. A combination of the two methods might be required to establish the most accurate estimation of the ASGE.
Finally, although similar numbers of transcripts were identified for ASGE by the two methods tested, only half of them are present in both lists. Of 8132 (in silico-based) and 11,029 (de novo-based) transcripts with ASGE data, only 4136 have at least 20 reads in both results (Supplementary data Table 2). A possible explanation for the transcripts unique to each method that do not appear in the other is that indels between two mRNA sequences preclude them from inclusion in the list generated by the in silico-based method. In turn, the de novo-based method relies upon direct matching of independent transcriptome assemblies (e.g., reciprocal best hits) that produces higher numbers of decision points as the assembly process runs its course. How closely these decision points mirror one another, or conversely, how often they become skewed and shift the final result remains to be studied.
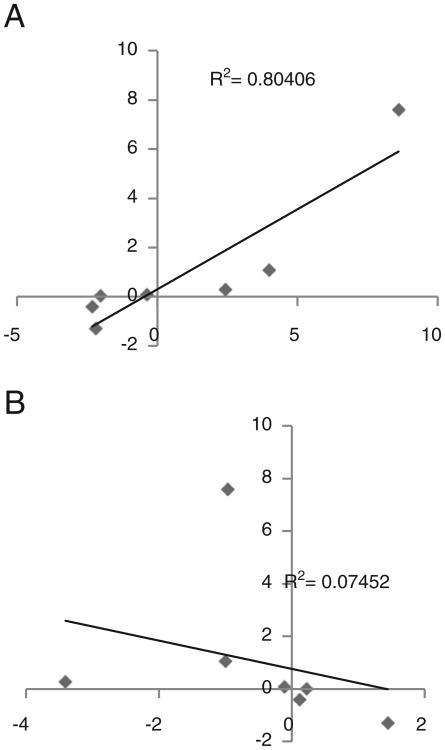
To examine which method is better in representing the true ASGE pattern in hybrid Xiphophorus, we compared relative expression levels obtained by allele specific real time-PCR for seven transcripts (Supplementary data 3) with read counting data from both in silico and de novo based methods. For each transcript, a ratio of the expression of the two alleles (X. maculatus/X. hellerii) in the hybrid was assessed by real time PCR and compared with the same ratios obtained by de novo and in silico methods, respectively. Linear regression was used to model the relationship between real time PCR results and read count data (Fig. 3). A coefficient of determination R2=0.804 was observed between real time PCR and the de novo method, suggesting that the de novo method produced a reasonably accurate estimate of allele abundance in the hybrid. However, the in silico method produced a very weak correlation (R2=0.07) with our real time PCR results. This suggests that the in silico method might not be reliable for specific expression analyses. It should be mentioned that we do not have biological replicates in the hybrid RNA-Seq sample. So another possible way to improve the correlation between experimental data and reads count might be to introduce multiple biological replicates to reduce noise in the read counting data.
Fig. 3.
Comparison of allele specific gene expression detected by real time-PCR and RNA-Seq. A comparison of RNA-Seq read counts and real-time-PCR relative expression. Log2-transformed ratios of two species-specific alleles in the F1 hybrid were compared between RNA-Seq data (x-axis) and real time-PCR data (y-axis) (A) real time-PCR vs. the de novo method. (B) real time-PCR vs. the in silico method.
We further looked at several examples in the ASGE results where the two methods did not agree with each other and found that most of them were due to indels or a close clustering of SNPs at a discrete location. For example, Ribosomal Protein L41 (Locus_228710) has a 6 nt indel and several SNPs. The in silico method identified reads mapped to SNPs but not those reads that spanned an indel, thus producing a biased estimation of the abundance of the two alleles.
4. Conclusions
Quantitative allele specific gene expression analysis in hybrids is an important application of next generation sequencing technology. This study compared two strategies for analyzing allele specific expression within interspecies hybrids produced from crossing two Xiphophorus fish species. The results show that global allele distribution patterns obtained by both methods are very similar and balanced allelic distribution is found in F1 hybrid fish transcriptomes. Disagreements between two methods mostly occurred in genes with indels between the two parental alleles and we found the “de novo” approach superior in handling these variations. In addition, real time PCR results further confirmed that the “de novo” approach is more reliable at estimating allele abundance than the “in silico” approach. Overall, we believe a second “de novo” transcriptome should be used whenever possible to improve the precision of allele specific analyses. When a second transcriptome is not available, alignment tools supporting indels (such as BWA) may be tried, in conjunction with “in silico” approaches to attempt to reduce bias. However, the effect of such alignment tools must be empirically determined.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.cbd.2012.10.006.
Supplementary Material
Acknowledgments
The authors would like to thank Markita Savage and the other employees of the Xiphophorus Genetic Stock Center, Texas State University, for maintaining the pedigreed fish lines, performing interspecies crosses, and caring for the hybrid animals used in this study. This work was supported by the Texas State University and the National Institutes of Health, Division of Comparative Medicine grants R24OD011199 (WAC) and R24OD011120 (RBW), including an American Recovery and Reinvestment Act supplement to this award.
Contributor Information
Yingjia Shen, Email: ys14@txstate.edu.
Tzintzuni Garcia, Email: tzintzuni@gmail.com.
Vagmita Pabuwal, Email: v_p48@txstate.edu.
Mikki Boswell, Email: mboswell@txstate.edu.
Amanda Pasquali, Email: ap1392@txstate.edu.
Ion Beldorth, Email: ib12@txstate.edu.
Wes Warren, Email: wwarren@genome.wustl.edu.
Manfred Schartl, Email: phch1@biozentrum.uni-wuerzburg.de.
William A. Cresko, Email: wcresko@uoregon.edu.
Ronald B. Walter, Email: RWalter@txstate.edu.
References
- Chen Y, Souaiaia T, Chen T. PerM: efficient mapping of short sequencing reads with periodic full sensitive spaced seeds. Bioinformatics. 2009;25:2514–2521. doi: 10.1093/bioinformatics/btp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia TI, Shen Y, Catchen J, Amores A, Schartl M, Postlethwait J, Walter RB. Effects of short read quality and quantity on a de novo vertebrate transcriptome assembly. Comp Biochem Physiol C Toxicol Pharmacol. 2012a;155:95–101. doi: 10.1016/j.cbpc.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia TI, Shen Y, Crawford D, Oleksiak MF, Whitehead A, Walter RB. RNA-Seq reveals complex genetic response to deepwater horizon oil release in Fundulus grandis. BMC Genomics. 2012b;13:474. doi: 10.1186/1471-2164-13-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Bickel RD, McIntyre LM, Graze RM, Calabrese PP, Nuzhdin SV. Allele-specific expression assays using Solexa. BMC Genomics. 2009a;10 doi: 10.1186/1471-2164-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Bickel RD, McIntyre LM, Graze RM, Calabrese PP, Nuzhdin SV. Allele-specific expression assays using Solexa. BMC Genomics. 2009b;10:422. doi: 10.1186/1471-2164-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, Zhang Z, Ranade SS, Dimalanta ET, Hyland FC, Sokolsky TD, Zhang L, Sheridan A, Fu H, Hendrickson CL, Li B, Kotler L, Stuart JR, Malek JA, Manning JM, Antipova AA, Perez DS, Moore MP, Hayashibara KC, Lyons MR, Beaudoin RE, Coleman BE, Laptewicz MW, Sannicandro AE, Rhodes MD, Gottimukkala RK, Yang S, Bafna V, Bashir A, MacBride A, Alkan C, Kidd JM, Eichler EE, Reese MG, De La Vega FM, Blanchard AP. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010;20:816–825. doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Abyzov A, Wang J, Alves P, Raha D, Harmanci A, Leng J, Bjornson R, Kong Y, Kitabayashi N, Bhardwaj N, Rubin M, Snyder M, Gerstein M. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5:105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Catchen J, Garcia T, Amores A, Beldorth I, Wagner J, Zhang Z, Postlethwait J, Warren W, Schartl M, Walter RB. Identification of transcriptome SNPs between Xiphophorus lines and species for assessing allele specific gene expression within F(1) interspecies hybrids. Comp Biochem Physiol C Toxicol Pharmacol. 2012;155:102–108. doi: 10.1016/j.cbpc.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Springer NM. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173:2199–2210. doi: 10.1534/genetics.106.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- Wei CL, Ng P, Chiu KP, Wong CH, Ang CC, Lipovich L, Liu ET, Ruan Y. 5′ Long serial analysis of gene expression (LongSAGE) and 3′ LongSage for transcriptome characterization and genome annotation. Proc Natl Acad Sci U S A. 2004;101:11701–11706. doi: 10.1073/pnas.0403514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Dubansky B, Bodinier C, Garcia TI, Miles S, Pilley C, Raghunathan V, Roach JL, Walker N, Walter RB, Rice CD, Galvez F. Genomic and physiological footprint of the Deepwater Horizon oil spill on resident marsh fishes. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1109545108. http://dx.doi.org/10.1073/pnas.1109545108. [DOI] [PMC free article] [PubMed]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.