Abstract
AeNAT5 (NCBI, ABZ81822), an orphan member of the insect-specific Nutrient Amino acid Transporter subfamily of SoLute Carrier family 6 (NAT-SLC6) and the first representative of a novel eukaryotic methionine-selective transport system (M), was cloned from cDNA of the vector mosquito, Aedes aegypti. It has orphan orthologs throughout several mosquito genomes, but not in Drosophila or outside Diptera. It shows the highest apparent affinity to L-Met (K0.5 = 0.021 mM) and its metabolites Homocysteine and Cysteine (K0.5 = 0.89 and 2.16 mM), but weakly interact with other substrates. It has a Na+ - coupled mechanism (K0.5 Na+ ~ 46 mM) with 1AA:1Na+ stoichiometry that maintains ~ 60% activity in Cl− - free media. In situ hybridization showed accumulation of AeNAT5 transcript in the absorptive and secretory epithelia, as well as in specific peripheral neurons and the central ganglia of mosquito larvae. The labeling pattern is distinct from that of the previously characterized AeNAT1. RNAi of AeNAT5 increases larval mortality during ecdysis and dramatically suppresses adult emergence. Our results showed that in addition to previously characterized broad spectra and aromatic amino acid selective transport systems, the mosquito NAT-SLC6 subfamily evolved a unique mechanism for selective absorption of sulfur-containing substrates. We demonstrated specific patterns of alimentary and neuronal transcription of AeNAT5 in mosquito larvae that is collateral with the indispensable function of this transporter in mosquito development.
Keywords: amino acids transport, methionine, hatching induced RNA interference (hiRNAi), epithelial cell, neurons, mosquito, larvae, SLC6
1. Introduction
L-Methionine and L-Cysteine are sulfur-containing proteinogenic amino acids that form start and disulfide bridge moieties in peptides and proteins, serve as precursors of taurine and glutathione, and comprise essential substrates in a range of fundamental metabolic processes, including: methylation, glutathione-coupled antioxidant defense, biosynthesis of cysteamine and polyamines, and production of coenzyme-A and iron-sulfur clusters (Cooper, 1983). L-Met is synthesized by autotrophs (Bright et al., 1979). However, its expensive and structurally complex synthesis promotes prokaryotes to acquire available environmental L-Met via specialized high-affinity ATPases and high-throughput secondary transporters (Trotschel et al., 2008). Uptake of dietary or symbiotic L-Met is obligatory in Metazoa that lack relevant synthesis cascades (Payne and Loomis, 2006). After metabolic deprivation of its essential L-Met precursor, membrane transport also becomes obligatory for conditionally essential L-Cys. The systemic pool of L-Met/L-Cys can be partly restored via autophagy of intracellular peptides (Mizushima, 2007) or recycling of homocysteine (hCys) accumulated by a conserved methylation process involving the L-Metderived methyl donor, S-Adenosyl-Methionine (Zhao et al., 2012). Obviously, recycling supplies only a fraction of the necessary substrates. Moreover, it involves a complex enzymatic cascade that uses an essential cofactor and metabolic energy. Recycling alone is unable to satisfy the metabolic demand of a metazoan organism that still requires systemic distribution of L-Met, L-Cys and hCys and must compensate for irreversible consumption of these substrates. Hence, the effective alimentary absorption of essential L-Met, as well as systemic distribution of its derivatives L-Cys and hCys, is critical for the metabolic balance of sulfur-containing amino acids in metazoan cells and organisms.
Several transport systems that accept L-Met and may potentially aid in its absorption have been recorded in vertebrate and invertebrate tissues. However, the contribution of broad spectra systems remains arguable as they are considered insufficiently selective, while the molecular identity of a selective transport system for L-Met remains unknown. L-Met is a substrate of several Na+ - dependent (Bo) and Na+ - independent (bo,+, y+L and L) amino acid transport systems. Molecular mechanisms with similar properties have been identified among phylogenetically related secondary transporters of the SoLute Carrier (SLC) families 6 and 7 (tabulated in (Boudko, 2010; Boudko, 2012)). To date, a few characterized mammalian and insect members of these families can aid in primary apical/basolateral transport of sulfurcontaining amino acids across alimentary epithelia and absorb these substrates through plasma membranes of neighboring cells. Specifically, B0-like SLC6 transporters (Chen et al., 2004) contribute transport mechanisms that use the generic electrochemical gradients of alkali cations (mainly Na+) for active translocation of amino acids against unfavorable chemical gradients. In addition, b0,+, y+L and L transporters of the SLC7 family (Verrey et al., 2004) facilitate passive transmembrane diffusion and/or exchange of essential and metabolic amino acids at the cellular membrane, providing mechanisms sufficient for selective substrate translocation toward favorable (permeases) or against small unfavorable (exchangers) gradients. Several characterized mammalian SLC6 members share broad spectra substrate selectivity profiles that include large aliphatic amino acids as well as sulfur-containing L-Met and L-Cys. Similarly, previously characterized insect transporters: caterpillar Manduca Sexta MsKAAT1 (Castagna et al., 1998), MsCATCH1(Feldman et al., 2000), mosquito A. aegypti AaNAT1 (Boudko et al., 2005a) and fly D. melanogaster DmNAT1(Miller et al., 2008) share relatively broad spectra mechanisms. Except for DmNAT1, all characterized insect transporters exhibit low relative affinity and velocity as L-Met transporters. Moreover, due to substrate competition, the B0-like mechanism is ineffective for selective absorption of underrepresented substrates, and may be insufficient under conditions with limited availability of sulfur containing amino acids.
Previously, we identified two mosquito transporters with unique adaptations for selective transport of either indole-branched L-Tryptophan and L-5-HydroxyTryPtophan (L5-HTP)(Meleshkevitch et al., 2009) or phenol-branched L-Phenylalanine, L-Tyrosine, and L-3,4-DihydroxyPhenylAlanine (L-DOPA)(Meleshkevitch et al., 2006). High affinity and L-Met selective transport systems have been reported in earlier studies of amino acid absorption in insect tissues (Neal et al., 1996; Wolfersberger, 2000) as well as in other invertebrate (Gache and Vacquier, 1983) and vertebrate organisms (Soriano-Garcia et al., 1998). However, the molecular identities of these mechanisms remain enigmatic. In this work we characterized the first known eukaryotic Methionine selective transporter. It was cloned from the larval gut of the vector mosquito, Ae. aegypti. We also determined the spatial expression profile of this transporter in the larval alimentary canal and neuronal tissues using whole mount in situ hybridization, and demonstrated the essential contribution of this mechanism to mosquito development.
2. Materials and Methods
2.1. Experimental organism
The eggs of the Aedes aegypti Costa Rica strain used in all experiments were acquired from the Malaria Research and Reference Reagent Resource Center (MR4, Cat # MRA-726). Mosquito eggs were hatched in deionized water (DW), except for those in the RNAi experiments (see below). Groups of 20 larvae were maintained in plastic containers with ~ 100 ml DW at 25 °C and ½ daylight cycles. Larvae were fed every two days with approximately 20 mg of a mixture containing one part yeast and three parts Wardley® Tetramin fish flakes. The rearing medium was refreshed every 2 days by replacing ~ 90% of the media with DW. Third to early fourth instar developmental stages of mosquito were used for preparation of the cDNA collection and for whole-mount in situ hybridization.
2.2. Bioinformatics
Protein sequences of selected SLC6 transporters, derived from the best reciprocal matches to the earlier cloned AeAAT1, were acquired from the NCBI database using protein BLAST (Altschul et al., 1990). The phylogenomic analysis was completed using Mega 5 software and an initial alignment of SLC6 family members from the completed genomes of selected organisms. PROMALS3D (Pei et al., 2008) and Clustal X(Larkin et al., 2007) implementations within the Geneious Pro 4.8 (Biomatters Ltd, New Zealand) software, structural alignment of LeuT and a 3D model of AeNAT5 with Yasara Structure(Krieger et al., 2002), and an implementation of the MUSTANG algorithm (Konagurthu et al., 2006) for structure superposition were used to reconstruct and tune the final alignment of selected NATs and structural motifs.
2.3. Molecular cloning
To ensure correctness of the AeNAT5 Open Reading Frame (ORF), a homology-cloning approach based on a combination of nested PCR with degenerate primers and rapid amplification of cDNA ends (RACE) techniques was employed. The set of standard degenerate primers was designed based on conserved contiguous blocks of the SLC6 protein alignment using CODEHOP software (Rose et al., 2003) and optimized to target a conserved peptide motif among mosquito members of the NAT-SLC6 subfamily (Table 1). The primers were used to screen a cDNA collection from the alimentary canal of A. aegypti larvae that was prepared as previously described (Matz, 2002). Touchdown hot-start PCR was performed with TaqDNA Polymerase (Roche Molecular Biochemicals) in 1 mM MgCl2 solution. The PCR cycling conditions were as follows: 35 cycles of denaturation at 94 °C , 1 min; annealing at 58 °C , 2min; exten sion at 72 °C , 2 min. A final 10 min extension step at 7 2 °C was performed.
Table 1.
Degenerate, cloning, sequencing, and dsRNA probe primers used in this work.
| Name | Sequence |
|---|---|
| iNAT3F | TGGGATTCGGTAACGTCTGGMRNTTYCC |
| iNAT4F | GGTCGGACGGCCGATCTAYTAYHTNGA |
| iNAT5R | CGATTCCCAGCATGAACAACATNRVRAARAA |
| iNAT6R | GCACAACCGGTCCACTCCRTANANCCA |
| AeNAT5-195R | CACGCCAATGCCCCTCAT |
| AeNAT5-294R | TGGCAACTCGGGGCTAAAT |
| AeNAT5-378R | AACCTTTGTGCCATTTCCCAT |
| AeNAT5-939F | TGTGATTGCCAAGTTCGATTTC |
| AeNAT5-1119F | CTGCGTTGGATTGGTGTATTTG |
| AeNAT5-1203F | CACGCTGGCTGTCTTTGAACT |
| TRsa | CGCAGTCGGTACTTTTTTTTTTTTT |
| LU4 | CGACGTGGACTATCCATGAACGCA |
| AeNAT5_5’_Hindlll | CCCAAGCTTCACGCCAATGCCCCTCAT |
| AeNAT5_3’_Notl | ATAGTTTAGCGGCCGCCTAATGGTAGGTGGGCGG |
| Ae18SrRNA1530F | AGCGATAACAGGTCCGTGATG |
| Ae18SrRNA1598r | CGTTGCTGCGCACATTGTA |
| AgNAT5-109F | CATCACACTACGGTCGATCTGAA |
| AgNAT5-180R | CTCCCGAAGTGGTGGCTTT |
The resulting ~1kb PCR fragments were TA-cloned into the TOPO vector (Invitrogen, Carlsbad, CA, USA) and sequenced. To define the complete ORF of the target transporters, a second set of nested PCR primers was designed based on the unique sequence of fragments from the degenerate screening and used in combination with cDNA-flanked TRsa and LU4 terminal adapters (Matz, 2002) for Rapid Amplification of CDNA Ends (RACE). SEQMAN II software (DNASTAR, Madison, WI) was used to assemble the contiguous transcripts and determine the exact ORFs of the targeted transporters. NCBI BLAST was used to confirm that the ORFs included a comprehensive set of SLC6 motifs. To obtain an expression construct of AeNAT5, an ORF-flanking pair of primers with non-redundant HindIII and NotI restriction sites was designed for the 5’ and 3’ ends of the transporter (Table 1). The cDNA collection was used again as a template to obtain the ORF of AeNAT5-flanked by the specified restriction sites. The PCR cycling conditions were as follows: 35 cycles of denaturation at 94 °C , 40 s; annealing at 57 °C , 1 mi n; extension at 72 °C , 2 min 30 s. The PCR product was purified with a Qiagen PCR Purification Kit and cut with HindIII and NotI restriction enzymes. The product was run on a 1% agarose gel, and the band was excised and cleaned with a Gel Extraction Kit (Qiagen, Chatsworth, CA). The resulting product was ligated into the expression vector pXOOM (Jespersen et al., 2002) . The integrities of the cloned products were confirmed by sequencing. The RNA for oocyte injection was produced from XbaI - linearized AeNAT5-pXOOM construct using mMessage mMachine, a high-yield capped RNA transcription Kit (Ambion Inc. Austin, TX), according to the manufacturer’s instruction. The integrity of the transcript was confirmed by agarose gel electrophoresis, and cRNA was stored at − 80° C until injection.
2.4. Heterologous expression and electrophysiological analysis
Surgically isolated and collagenase-treated stage V-VI Xenopus laevis oocytes (Nasco, Fort Atkinson, WI) were injected with ~ 40 ng of AeNAT5 cRNA and incubated for 3–7 days at 17°C in ste rile oocyte media (98.0 mM NaCl; 2.0 mM KCl; 1.0 mM MgCl2; 1.8 mM CaCl2; 2.5 mM Na Pyruvate; 100 units/ml penicillin; 100 µg/ml streptomycin; 5% horse serum; 10 mM Hepes, pH 7.4). Oocyte recordings were performed in a 50-µl constant-flow perfusion chamber. An OC-725C Oocyte Clamp amplifier (Warner Instruments, Hamden, CT, USA) and saturated KCl - filled glass microelectrodes with 0.5 – 1 MΩ resistance were used for recordings. Current/voltage (IV) signals were acquired and analyzed using parallel DigiData 1322A/MiniDigi acquisition devices and pCLAMP software (Molecular Devices, Sunnyvale, CA, USA). ClampFit (Molecular Devices, Sunnyvale, CA) and SigmaPlot 11 (Systat Software, Inc., San Jose, CA, USA) were used for preparing final IV plots and statistical analysis of the data. The composition of solutions used for ion substitution assays has been described earlier (Boudko et al., 2005a). The saturation profiles and constants were derived by curve fitting of normalized data sets with a three parameter Hill approximation.
2.5. Whole mount in situ hybridization
The relative spatial expression profile of AeNAT5 was visualized by in situ hybridization of AeNAT5 transcript in whole mount preparations of 4th instar Aedes aegypti larvae. The procedures for preparing whole mount mosquito larvae and NAT-specific digoxigenin labeled probes have been previously described (Boudko et al., 2005a; Meleshkevitch et al., 2006; Meleshkevitch et al., 2009). In this study we used full-length open reading frames of AeNAT5 and EGFP clones to generate the experimental and control antisense probes. To prepare probes, an in situ hybridization kit Riboprobe® T7 was used per the manufacturer’s protocol (Promega, USA).
2.6. RNAi experiments
ORF coding cDNAs for AeNAT5 or EGFP (control) were cloned into the pLitmus 28i vector between two T7 promoters. dsRNA RNAi probes were synthesized in vitro using a Hiscribe T7 RNA polymerase Transcription Kit (New England Biolabs, Beverly, MA, USA). Previously described abdominal injectio-ninduced RNAi (Blitzer et al., 2005) was found to be damaging for earlier stage mosquito larvae. This technique was replaced by hatching induced dsRNA delivery, which yields more consistent results, reduces nonspecific mortality, and induces detectable silencing of the targeted transcript in earlier instar larvae. Specifically, ~ 50 competent mosquito eggs were submerged into a 40 µl drop of 0.05, 0.5 and 5 µg/µl dsRNA DW solution inside a Petri dish, which was subsequently maintained in a humidified chamber. After ~10 hours of incubation, the initial hatching volume was diluted 75 times and unhatched eggs were removed. Experimental AeNAT5 dsRNA and negative control EGFP dsRNA, as well as DW-hatched larvae, were grown using an identical procedure to that described above. Larval mortality was recorded daily using visual observation under a stereo microscope. The counts were taken for intervals of 2, 4, and 8 days.
2.7. qPCR analysis
RNA isolation, cDNA synthesis and qPCR analysis were performed, essentially, as was previously described (Boudko et al., 2005a; Meleshkevitch et al., 2006), but following specific modifications in compliance with the MIQE guidelines (Bustin et al., 2009). 15–20 individual mosquitoes were used in each sample. RNA was isolated from 70% cold ethanol-fixed crushed tissues of Ae. aegypti larvae using an RNAqueous®-Micro kit (Ambion) and immediately processed. Purified DNase-treated RNA was converted to cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) with random hexamers. AeNAT5 expression levels were determined using SYBR Green dye technology on an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Columbia, MD, USA). Ae. aegypti 18S ribosomal RNA (NCBI Accession # AY433620) was selected as the most broadly employed and previously validated evenly expressed endogenous internal reference that is suitable to normalize the relative differences of total cDNA added to individual reactions, following MIQE guidelines. qPCR primers were designed using Primer Express v. 2.0 software (Applied Biosystems). Sequences of qPCR primers are summarized in Table 1. qPCR reactions were performed in triplicate in a total volume of 25 µl containing 12.5 µl SYBR Green PCR Master Mix (Applied Biosystems), 200 nmol l−1 AeNAT5 or 300 nmol l−1 18S rRNA of each primer, and approximately 1–10 ng cDNA template. Control runs included a subset of PCR components lacking the cDNA template. The PCR program was run as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s, 60°C for 1 min followed by a dissociation stage of 95°C for 15 s, 60°C for 20 s, and 95°C for 15 s. Dissociation curve ana lysis was performed to confirm the specificity of the reaction products. Relative expression values were calculated according to the comparative Ct method (Schmittgen and Livak, 2008). The amount of target normalized to an endogenous reference is given by the formula: 2-ΔCt. Ct refers to the cycle number at which the fluorescence rises above a set threshold. The efficiency (E) of each primer set was determined by plotting template dilutions against Ct values and is equal to 10[−1/slope]. Expression of AeNAT5 in each tissue was normalized to that found in whole larvae, which was set to a value of one. Data represent exactly three averaged replicates of two independent experiments with appropriate statistical validation of significant changes in AeNAT5 expression.
2.7. Data analysis
Excel software (Microsoft®) was used to analyze qPCR results and larval motility. Sigma Plot 11 (Systat Software, Inc.; CA) was used to generate the final graphs. Data were statistically tested using one parameter one-way ANOVA tests conducted for selected groups of samples using the QI Macros for Microsoft® Excel (qimacros.com). The significance was considered at p < 0.05.
3. Results
3.1. Molecular identity and phylogeny
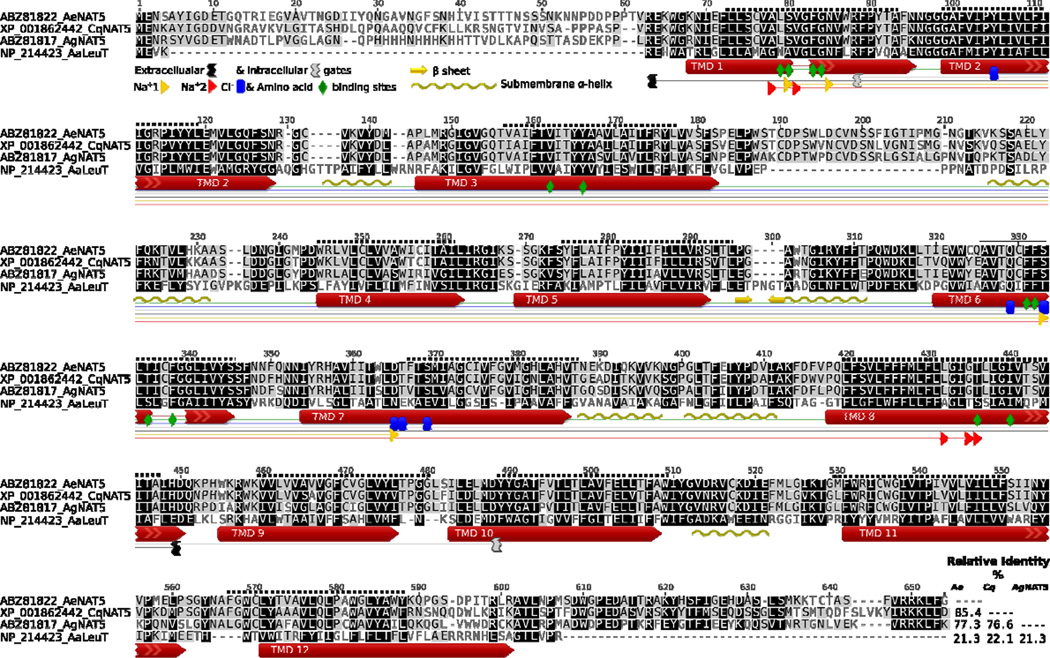
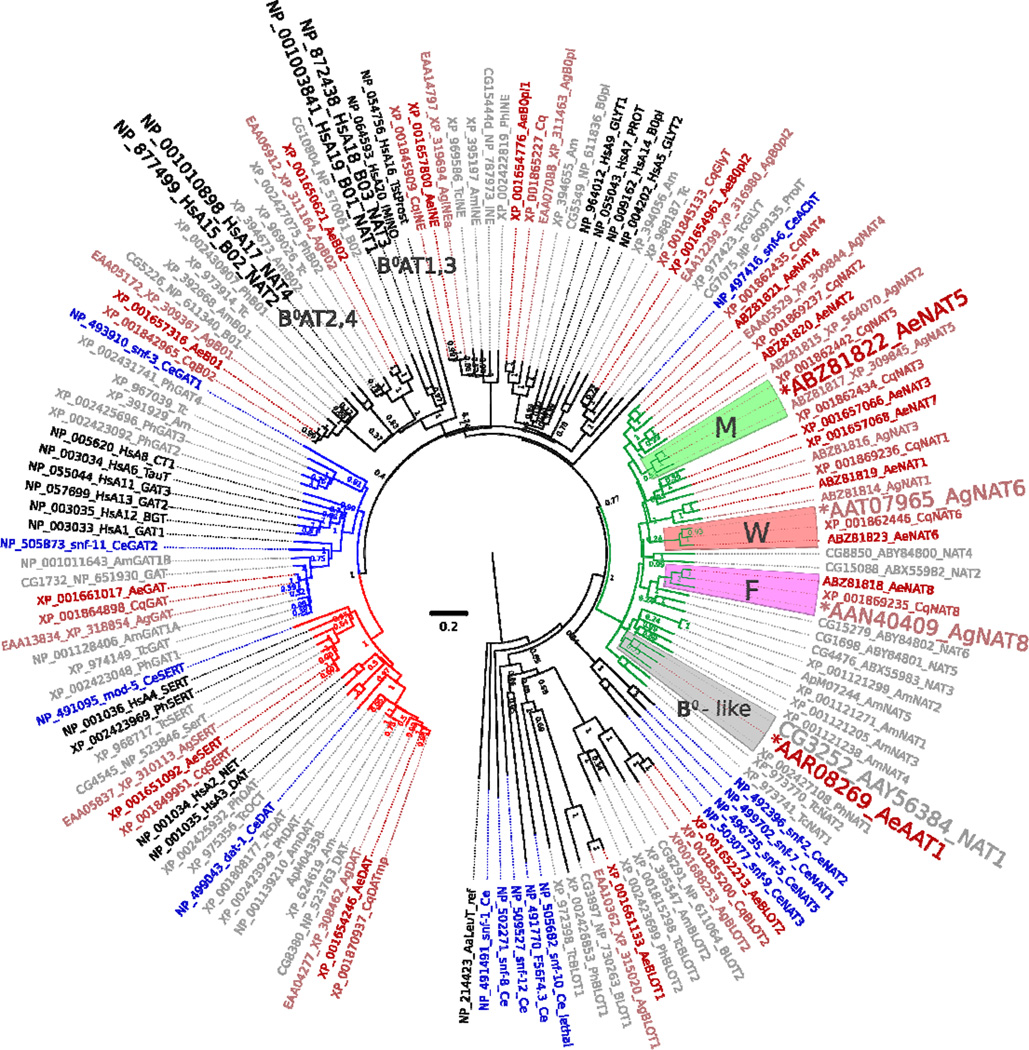
A transcript of a putative transport protein of the SLC6 family, named AeNAT5, was cloned from a larval cDNA collection of the yellow fever vector mosquito, Ae. aegypti, using the RACE procedure to confirm correct termini and splicing (AeNAT5, GB # EU267010 protein ABZ81822). It encodes a 652 amino acid protein with an estimated molecular mass of 70.678 kDA. A fragment of the transcript was earlier annotated in the Ae. aegypti genome (Peters et al., 2007). The predicted orientation and transmembrane domain topology (Fig. 1, dotted line above sequences), as well as sequence alignment of AeNAT5 with a crystalized bacterial homolog LeuT and its experimentally determined structural motifs (Fig. 1, graphical annotation below sequences), suggests a 12 transmembrane domain (TMD) protein with cytoplasmic N and C termini. The putative structural organization of AeNAT5 is consistent with the known organization of SLC6 transporters, allowing us to predict plausible positions of substrate binding motifs (Fig. 1, colored shapes and insert). Specifically, it reveals the identity of a putative substrate coordination framework in AeNAT5 and its closest relatives from other mosquitoes, An. gambiae and Culex quinquefasciatus (Fig. 1, colored shapes). The conservation of structural motifs and high percentages of sequence identity (Fig. 1, right bottom corner insert and darker alignment background) suggest homologous function and substrate specificity in the presented mosquito transporters. To reveal possible homology further, we constructed a comprehensive phylogenomic tree of SLC6 families selected from the best annotated genomes of nematode, insect, and mammalian species and include the most comprehensive sets of characterized SLC6 transporters (Fig. 2). The phylogenomic analysis showed that AeNAT5 forms an orthologous cluster with a single protein (gene) in other mosquito species. Hence, AeNAT5 represents a new member of the Nutrient Amino acid Transporter subfamily of the SoLute Carrier family 6 (NAT-SLC6) that arose and stabilized in the evolution of the SLC6 family of culicid insects. AeNAT5 forms a separate (paralogous) branch with all previously characterized mosquito transporters and has no orthologs in fruit fly, honeybee, beetle, or flea, nor in the nematode or mammalian genomes (Fig. 2). Taking into account that prediction of homologous function is not possible based on the described pattern of phylogenetic proximity, and considering that AeNAT5 is a representative of a phylogenetically separated group with unique substitutions in a frame of substrate binding residues that reflect evolutionary alteration of transport functions, we decided to functionally express and characterize the cloned transporter.
Figure 1. Protein sequence alignment of AeNAT5 and orthologous mosquito NATs versus the sequence and structural motifs of the LeuT.
The background intensity indicates sequence similarity that was rendered using a Blosum 45 matrix and 0 threshold optional settings. Colored shapes show relative positions of specific structural motifs and 12 TMDs in LeuT (long red arrows and specific #, colored shapes legend insert). The uncoiled fragments of TMDs 1 and 6 are shown as lines. The dotted line above the sequence is the relative position of theoretically predicted TMDs in AeNAT5. The graphical annotation of aligned positions and inserts demonstrates 12 amino acid binding sites, 4 and 5 sites coordinating the 1st and 2nd Na+ ions, 5 putative sites coordinating Cl−, and the charged residues of intracellular and extracellular gates of the transporters. The beta sheet hairpin and sub-membrane helices identified in the LeuT structure are indicated by yellow arrows and wave lines, respectively. The right corner insert displays a pairwise identity matrix (%) for aligned NATs without N and C termini.
Figure 2. Phylogenetic position of AeNAT5 relative to M, F, W and B0-like system NATs.
The consensus phylogenetic tree, inferred by the Neighbor-Joining method (Saitou and Nei, 1987), is taken to represent the evolutionary hierarchy of the SLC6 family members from genomes of selected organisms. The normalized bootstrap values from 2000 replicates (Felsenstein, 1985) are shown next to the branches. Branch lengths render relative evolutionary distances computed by the JTT matrix-based method (Jones et al., 1992). Scale bar reveals number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analysis involved 145 amino acid sequences representing all SLC6 transporters of 8 selected model organisms. Accession numbers and unique names of the transporters are shown as branch projections. The tree was built using MEGA 5 (Tamura et al., 2011), rendered with FigTree 1.4.0 software (tree.bio.ed.ac.uk), and visualized with Inkscape 0.48. Groups of representative transport systems are marked by background colors and capital letters. A larger font indicates characterized NATs. The green branch is the subfamily of insect-specific NAT-SLC6. Blue and red branches render the subfamilies of GABA and monoamine neurotransmitter transporters, respectively. Transporters from different organisms are depicted by different colors and unique two letters identifiers integrated in to the accession-name strings: blue, nematode Caenorhabditis elegans (Ce), black, Homo sapiens (Hs); red gradation, Aedes aegypti (Ae), Anopheles gambiae (Ag), and Culex quinquefasciatus (Cq) mosquitoes; gray, selected model insects including fly Drosophila melanogaster (CG prefix in the gene accession #), bee Apis mellifera (Am), beetle Tribolium castaneum (Tc), and louse Pediculus humanus corporis (Ph).
3.2. Transport function
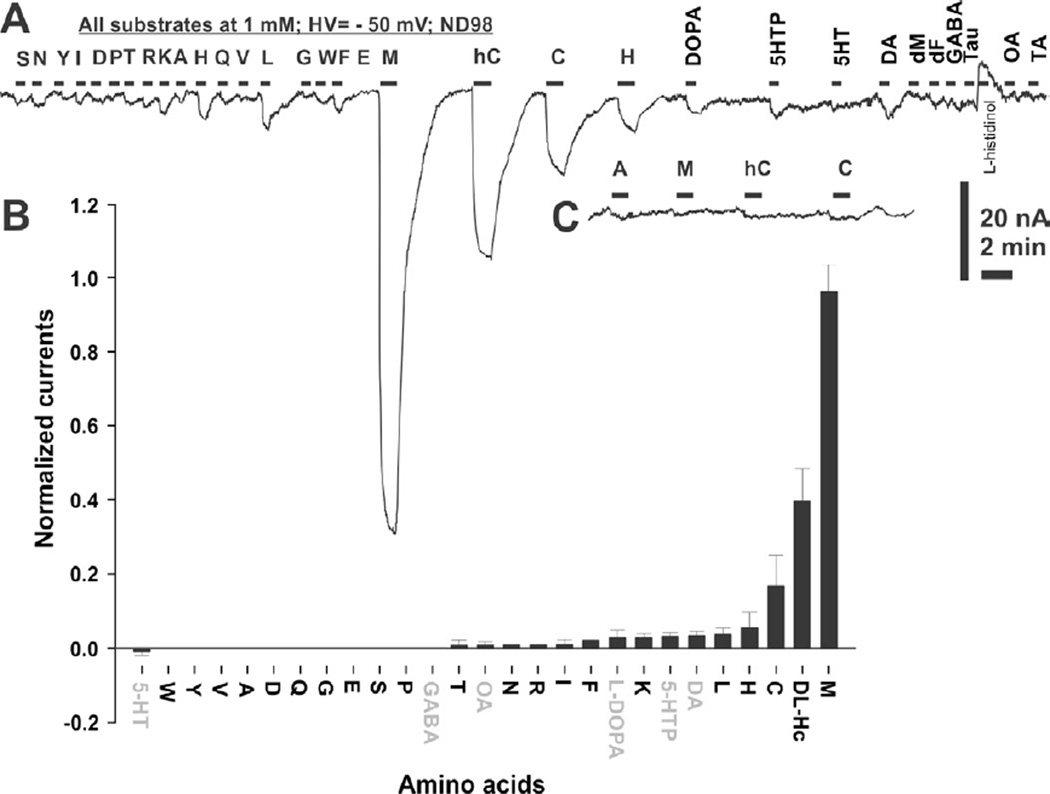
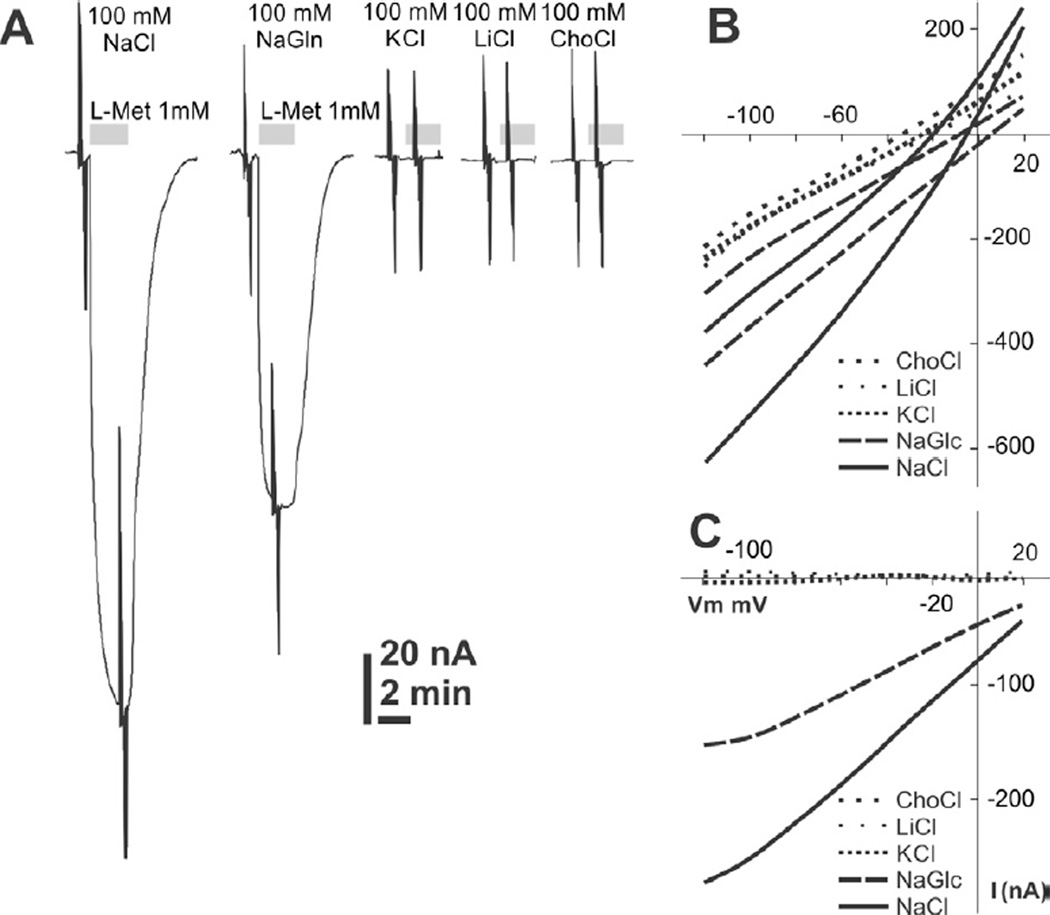
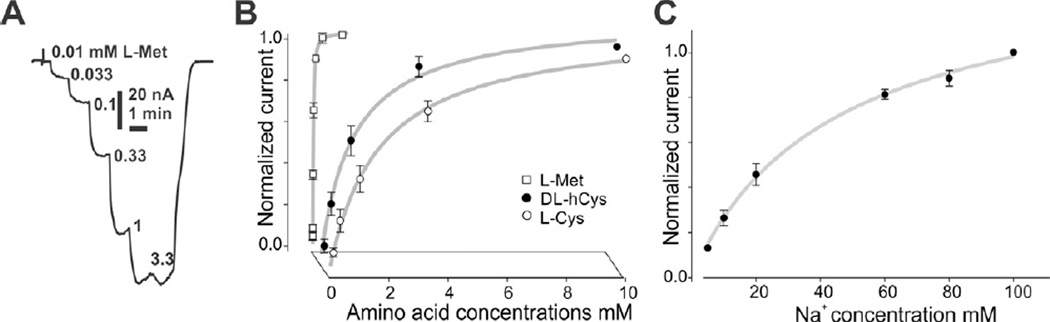
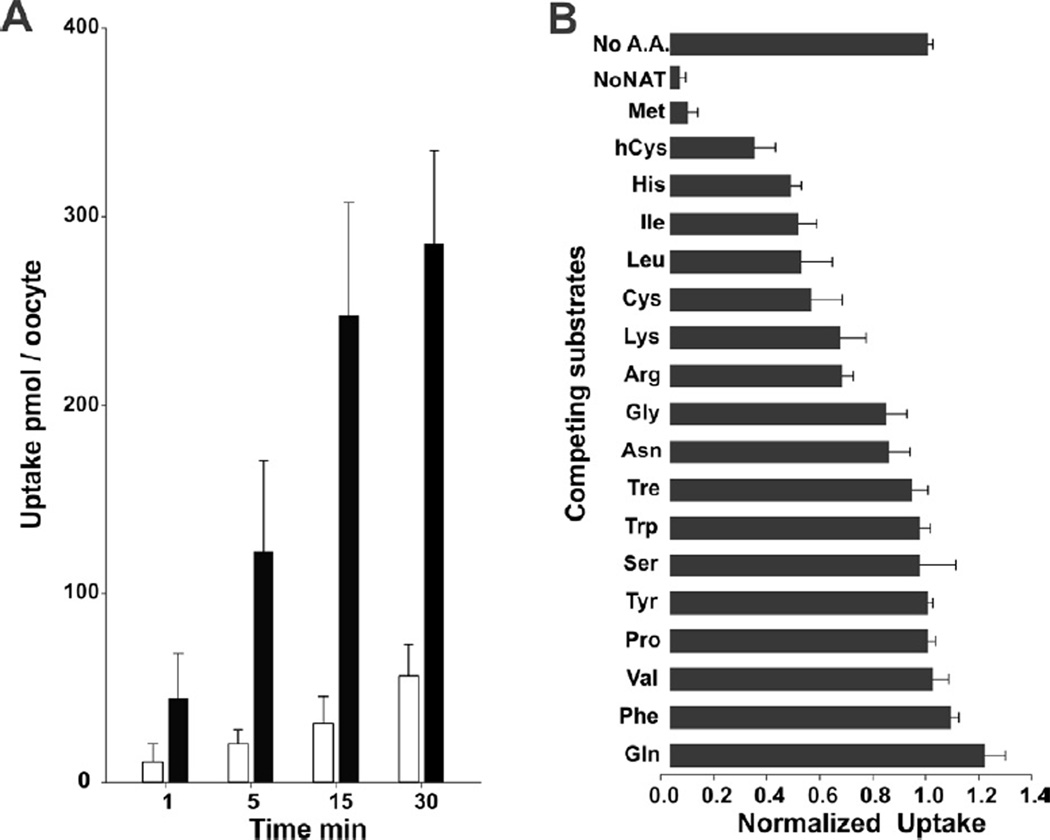
On the 4th day post AeNAT5 mRNA injection, Xenopus laevis oocytes exhibited robust increase of amino acid induced transmembrane currents with a previously unknown ultimate selectivity for sulfur-containing amino acids. Specifically, L-Methionine induced large inward currents in AeNAT5 expressing oocytes (Fig. 3AB). Such currents were absent in naïve or water-injected control oocytes harvested concurrently (Fig. 3C and data not shown). Significant currents were also induced by application of Homocysteine and L-Cysteine, and to a much lesser extent L-Histidine (Fig. 3A). In contrast, other tested sulfur-containing amino acids: sulfonic acid, Taurine and dimeric Cystine elicited barely detectable current responses (Fig. 3A, and data not shown). Similarly, neurotransmitter substrates of the SLC6 family induced only minor currents (> 5 nA) that were comparable to those in control oocytes. No current changes were induced by applications of the D - enantiomer of Methionine (Fig. 3A). The ion dependency of AeNAT5, analyzed by measuring L-Met induced currents in media with defined ionic compositions, showed that substitution of Na+ with K+, Li+, or choline+ nearly abolished the response to application of 1 mM L-Met (Fig. 4A). In contrast, substitution of Cl− with the larger Gluconate− ion reduces current by only ~ 40% (Fig 4A, NaGln). AeNAT5 displays near linear current-voltage (IV) characteristics in the physiological range of transmembrane voltage (Fig. 4B; −100 – +20 mV). The comparative and subtracted IV plots further support the results of the ion substitution experiments and show that AeNAT5 has near-linear voltage dependency for its Na+ - and Cl− - dependent components. Apparently, it cannot use the reversed K+ gradient to drive transport as a few other characterized insect NATs. The amino acid elicited currents were saturable at high concentrations of organic substrates and Na+ ions (Fig. 5A). The estimated apparent affinity constants (K0.5), derived from the Hill Equation, were 0.02 ± 0.01, 0.89 ± 0.09, 2.16 ± 0.41 mM respectively for L-Met, DL-hCys, and L-Cys; and 46.3 ± 17.1 mM for Na+ (Fig. 5BC; statistical result for n > 3 samples ± s. e. and regression analysis). The estimated Hill constants for the preferred organic substrates and Na+ were close to 1, suggesting 1 amino acid:1 Na+ coupling stoichiometry under the specified conditions (Fig. 5; representative graph for L-Met, and data not shown). The substrate-induced current correlates with a robust uptake of 3H-labeled L-Met in AeNAT5 expressing oocytes vs. water-injected control oocytes (Fig. 6; p < 0.001, n = 3). The uptake was linear for the first 15 minutes (Fig. 6A) and dramatically reduced by addition of competing unlabeled L-Met and DL-Homocysteine (Fig. 6B). In contrast, other amino acids were significantly less effective as competing substrates (Fig. 6B), which supports the definition of AeNAT5 as a sulfur-containing amino acid selective transporter.
Figure 3. Substrate induced current inAeNAT5 expressing oocytes.
A. Representative current trace recorded during perfusion of selected amino acids and amino acid derivatives. All substrates were added at 1 mM final concentration during the periods indicated by the black bars above the actual trace. 98 mM NaCl was used as the major salt. A voltage clamp mode with a holding transmembrane voltage of −50 mV was used during this recording. The single letter code corresponds to the canonical abbreviations for amino acids. Other abbreviations are: DOPA, L-3,4-dihydroxyphenylalanine; 5HTP, 5-Hydroxytryptophan; 5-HT. 5-Hydroxytryptamine; DA, dopamine; dM, D-Methionine; GABA, γ-Amino butyric acid; Tau, Taurine; OA, Octopamine; TA, Tyramine. B. Result of statistical evaluation of AeNAT5 induced currents (bars are the normalized average value for n > 3, different batches/oocytes). The neurotransmitters are indicated by gray font. C. Amino acid induced responses in control DW injected oocytes at the same amplitude and time scale as in panel A and insert.
Figure 4. Current voltage relationship and ion dependency of AeNAT5.
A. 1 mM L-Met induced currents in AeNAT5 expressing oocytes perfused with different ion composition solutions. Gray bars indicate L-Met application episode. The major salts used for the solutions are shown above the traces. Spikes are actual ramp stimulations that were used to build current-voltage relationship (IV) plots shown in panels B and C. B. Representative current/voltage profile of an AeNAT5 expressing oocyte. Different line patterns depict different ion compositions in the perfused media (see legend on the plot insert). The pairs of identical lines represent current in the presence and absence of 1 mM L-Met C. IV characteristics of AeNAT5 built by subtraction of substrate-independent current component from 1 mM L-Met induced currents.
Figure 5. Saturation profile ofAeNAT5.
A. Representative recording of saturable AeNAT5 - coupled current upon staircase increase of external L - Methionine concentration as indicated by numbers in mM. B. Amino acid - induced currents in ND98 (98 mM NaCl media).The half-maximum saturation constants were determined for normalized currents in X. laevis oocytes expressing AeNAT5. Shape positions correspond to normalized values of currents vs. maximum currents of the set ± s.d. for n > 2 oocytes. Curves are nonlinear regression of the data fitted with the Hill equation: f = axη / (K0.5η+xη); where K0.5 is the half maximum current saturation constant and η is a Hill constant. Estimated apparent constants are: L-Met K0.5 = 0.02 ± 0.01 mmol l−1 [s.e.], η = 1.19 ± 0.01; DL-hCys K0.5 = 0.89 ± 0.09 mmol l−1, η = 1.31 ± 0.23; K0.5 L-Cys = 2.16 ± 0.41 mmol l−1, η = 0.88 ± 0.07. C. Na+ - induced currents at 1 mM L-Met concentration, K0.5 Na+ = 46.3 ± 17.1 mmol l−1, η = 0.966 ± 0.12. The estimated stoichiometry of AeNAT5 at presented conditions are ~ 1:1.
Figure 6. L-Methionine uptake and selectivity of AeNAT5.
A. Time courses of 3H L-Met uptake in AeNAT5 and DW - injected oocytes are shown as black and empty bars, respectively. B. Cis-inhibition of 3H L-met uptake by competing L-amino acids and DL-homocysteine. Bars are normalized mean quantities of the radiolabeled substrate absorbed during 15 min by AeNAT5 - expressing oocytes that were bathed in ND98 with 1:100 diluted stock of radiolabeled L-Met and 5 mmol l−1 of specified amino acids. No A.A. bar represents positive control, 3H L-Met uptake in AeNAT5 expressing oocytes without competing amino acid added; and No NAT represents negative control, 3H L-Met uptake in DW - injected oocytes. Error bars are standard deviation values for n ≥ 3 oocytes.
3.3. Transcription pattern
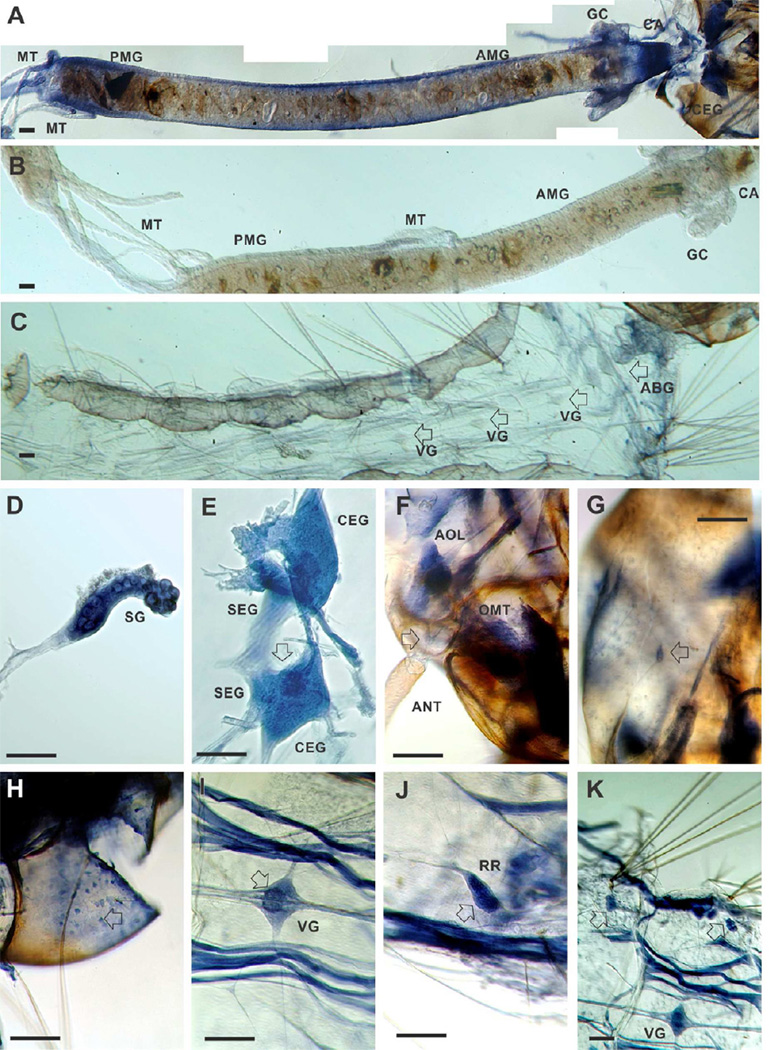
To determine the involvement of AeNAT5 in biological functions, we examined the accumulation of AeNAT5 transcript in larval tissues, considering that an elevated level of transcription likely correlates with elevated expression and contribution this transporter to a tissue specific function. We used whole mount preparations that are better for the assessment of relative levels and global patterns of transcript accumulation than other techniques. Antisense AeNAT5-DIG probes showed region specific in situ hybridization in the larval alimentary canal, central nervous system (CNS), and muscle systems (Fig. 7; AeNAT5 specific transcript blue stain (A) in the gut and head vs. nonspecific brown background pigmentation (B and C) in the peritrophic packet and head capsule). Control preparations processed with EGFP antisense DIG probes lack the specific labeling in the gut and integument tissues, including the nervous system (Fig. 7B and C, respectively). The patterns of alimentary and neuronal accumulation of AeNAT5 transcript were different in comparison to those previously identified for broad substrate transporter AeAAT1 from Ae. aegypti (Boudko et al., 2005a) and aromatic amino acid-specific transporters AgNAT6 (Meleshkevitch et al., 2009) and AgNAT8 (Meleshkevitch et al., 2006) from An. gambiae. Specifically, AeNAT5 hybridized in a much broader gut area, with particularly intense bands of hybridization in the middle portion of the larval gut and posterior midgut (Fig. 7A), while the transcripts of previously characterized transporters were mainly concentrated in the posterior and anterior midgut but virtually absent in the middle gut regions. AeNAT5 is also strongly expressed in the cardia, an esophageal invagination that secretes peritrophic membrane, the anterior imaginal disc of the gut, specific regions of the gastric caeca (Fig. 7A, CA and GC), and the salivary glands (Fig. 7D, SG). It was also detectable in the proximal parts of the Malphigian tubules (MT); however, labeling was weak in the rest of MT and in the rectum (not shown). Neuronal labeling was identified in the cerebral ganglia, with maximum intensities in the upper ocelli projection area (Fig. 7E, arrow). In contrast, the subesophageal ganglia produced relatively weak labeling. Profound AeNAT5 expression was detected in the peripheral nervous system, including in the larval ommatidia (compound eyes, OMT) (Fig. 7F), the antennal olfactory lobe (AOL), a cluster of bipolar cells projecting to the larval antenna, axonal branches and cell bodies that likely correlate to mechanosensory sensilli (Fig. 7F, arrow), and a plexus of multipolar cells lining the inner surface of the head capsule (Fig. 7H, arrow). Hybridization specific labeling was detected in ganglia of the ventral nerve cord, with particular concentration within a specific cell population of the central region occupying ~ 30 % of the total volume of the individual ganglia (Fig. 7I, arrow). AeNAT5 expression was also identified in the complex of striated muscles, reproductive rudiments (Fig. 7I and F), and a paired cluster of large cells proximal to the abdominal stellate hair tufts (Fig. 7G, arrows). The identified pattern of transcription suggests broad involvement of AeNAT5 in alimentary and neuronal functions. However, the significance of these contributions was enigmatic because the biological role of NATs has never been experimentally assessed in insects, nor has it been systematically addressed in other model organisms beyond reported links of several human NAT-SLC6 members contributing to an aminoacidurias complex (Broer and Palacin, 2011) and the recent demonstration of a role for C. elegans SNF-5 in nematode development (Metzler et al., 2013).
Figure 7. In situ hybridization of AeNAT5 in whole mount preparations of Ae. aegypti larvae.
Labeling was performed in integrated preparations while photographed fragments were isolated for picture clarity. A. High resolution panoramic stitching reconstruction of the larval head and alimentary canal isolated from integument. B and C are representative panels of the isolated gut and integument that were treated equally to the experimental preparation except for using an EGFP antisense probe. D. Isolated salivary gland. E. Cerebral ganglia of the larval central nervous system; arrow indicates upper ocelli projections in the neuropile. F. A high magnification image of a head fragment prepared from a stack of different focal plane images using high dynamic range image fusion techniques, arrow indicates the cluster of olfactory neurons with dendrites projected in to the antenna. G. Labeling in a putative mechanosensory neuron in the larval head capsule (arrow). H. A plexus of multipolar neuronal cells lining the internal surface of the larval head capsule. I. Example labeling of the visceral ganglia; a visceral ganglion of 4th segment is shown; the arrow indicates intensified labeling of cells and neuronal branches in the central region. J. Reproductive rudiments (arrow). K. A low magnification image represented relative labeling intensity in the visceral ganglion, striated muscles, and paired clusters of large cells identified in abdominal segments (arrows; see also C for control labeling). Abbreviations indicate: ABG, Abdominal Ganglia; AMG, anterior midgut; ANT, antennae; AOL, Anterior Olfactory Lobe; CA, cardia; CEG, Cerebral Ganglia; GC, gastric caeca; MT, Malpighian tubes; OMT, Ommatidium; PMG, posterior midgut; SEG, Subesophageal Ganglia; SG, salivary gland; RR, Reproductive Rudiments; VG, visceral ganglia. Scale bars are 100 µM.
3.4. Hatching-induced RNAi silencing
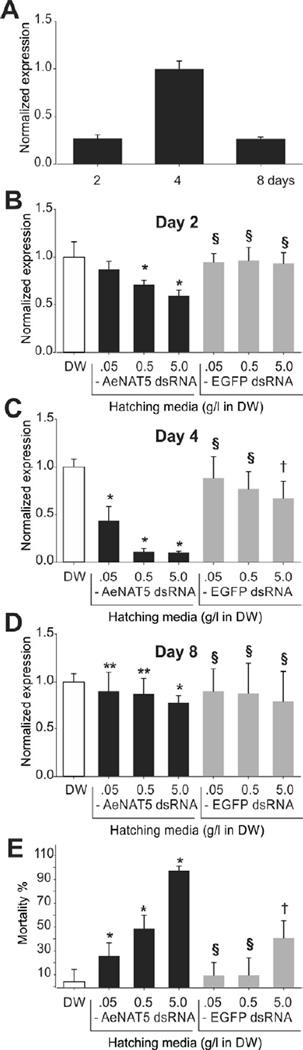
To test the contribution of AeNAT5 to mosquito biology, we developed a hatching-induced RNAi (hi-RNAi) technique that achieves reliable silencing of AeNAT5 gene product in the midgut of developing mosquito larvae (Fig. 8). The approach is based on the hatching of mosquito eggs in a drop of water with high concentrations of AeNAT5 specific dsRNA (see Methods for more details). Considering deviation of RNAi effects due to limited systemic and tissue-specific penetration of dsRNA, nonspecific probe toxicity, as well as possible compensation for AeNAT5 expression, we sampled and compared levels of AeNAT5 transcript in control groups that were hatched in deionized water (DW) and 3 gradual dilutions of EGFP dsRNA in DW relative to experimental groups that were hatched in 3 equimolar gradual dilutions of AeNAT5 dsRNA (Fig. 8). In DW controls, AeNAT5 displays maximum expression on day 4 that was approximately 3-fold higher vs. Day 2 and Day 8 samples (Fig. 8A). hi-RNAi with AeNAT5 dsRNA induces a reduction of the AeNAT5 transcript, especially significant on days 2 and 4 relative to EGFP controls (Fig. 8 B and C; one-way ANOVA, p < 0.01). The maximum RNAi silencing effect coincided with the maximum expression of AeNAT5 (Fig. 8 A Day 4 and Fig. 8 C). These hi-RNAi effects were concentration dependent (Fig. 8B and C). In contrast, samples treated with EGFP dsRNA showed no significant changes in AeNAT5 expression when compared to DW control samples (Fig. 8 B – D; p > 0.1). The AeNAT5 silencing effect decreases in late larval development, exhibiting no significant differences between surviving hi-RNAi and control larvae at day 8 after hatching (Fig. 8D). Addition of AeNAT5 dsRNA with the food suspension or bathing media at 0.5 g/l final concentrations does not alter the levels of hi-RNAi-induced silencing of AeNAT5, nor does it produce detectable silencing if applied to larvae that pass 2nd instar (data not shown). Therefore, we conclude that the targeted RNAi effect was primarily caused by hatching-induced or very early post-hatching mediated absorption of dsRNA by mosquito larvae. We did not distinguish between these two possibilities experimentally, but we anticipate that the initial rehydration of the hatching larvae may be key, as during this unique and short episode of life the larvae absorb a significant amount of external media. In parallel to determining the effect of RNAi on AeNAT5 transcript, we compared larval morphology and mortality in control and hi-RNAi groups. No apparent change of larval body sizes, morphology, and behavior were observed between control and hi-RNAi groups by visual inspection under a dissection microscope. In contrast, we observed substantially reduced success of early ecdysis, pupation, and adult emergence summarized as overall mortality in experimental groups when compared to control groups hatched in DW and EGFP dsRNA (Fig. 8E; one-way ANOVA, p < 0.01 for n = 8). In contrast, hatching eggs in EGFP dsRNA has no significant effect on mosquito survival when compared to DW hatched larvae (p > 0.1), except for the highest tested concentration, 5 g/l of EGFP dsRNA, which significantly increases mortality vs. DW sample (p < 0.01). Nonetheless, equal concentrations of AeNAT5 dsRNA still produce a significantly stronger effect (Fig. 8E; p < 0.01 for n = 8). The hi-RNAi experiment reveals the importance of the systemic contributions of AeNAT5 to mosquito development.
Figure 8. RNAi silencing of AeNAT5 in Ae. aegypti larvae.
A. qPCR assay of AeNAT5 transcript in developing mosquito larvae on days 2, 4 and 8, which in generally correspond to 2nd, 3rd, and 4th instar larval stages. Bars are mean relative quantity of AeNAT5 transcript ± Standard deviation for n = 3 samples. All samples were normalized relative to rRNA of ribosomal protein subunit Ae18S. B, C and D. Similar assays as those in panel A, except for larvae have been hatched in the indicated concentrations of AeNAT5 dsRNA (black bars), DW (empty bars), or EGFP dsRNA (grey bars) used as control to validate nonspecific toxicity following the hatching induced RNAi silencing protocol described in the Methods. E. hi-RNAi induced mortality in experimental (AeNAT5 dsRNA) and control (EGFP dsRNA) samples. The bars are mean values normalized to deionized water (DW, empty bars) ± Standard deviation for n = 3 (B, C, and D), or ± Standard error for n=8 (E). Markers indicate: * statistically significantly different vs. identical concentration of control EGFP dsRNA (effect); § statistically insignificantly different vs. DW control (no effect), and † statistically significantly different vs. DW (nonspecific dsRNA effects), respectively (one-way ANOVA test was used, considering significance level p < 0.05).
4. Discussion
With the identification of AeNAT5, the insect-specific NAT-SLC6 subfamily, to date, includes 5 characterized dipteran NATs: 4 from mosquitoes and one from Drosophila (depicted in Fig. 2 by large size font); and 2 lepidopteran NATs from caterpillar, Manduca sexta (not included in the tree in Fig. 2). All earlier characterized NATs form independent phylogenetic clusters and strictly differ by substrate specificity. Specifically, Aedes aegypti’s AeAAT1, representing a B0 - like amino acid transport system in insects (Boudko et al., 2005a), shares a cluster with another dipteran B0 - system transporter from fruit fly, DmNAT1 (Miller et al., 2008) (Fig. 2). Lepidopteran MsKAAT1 (Castagna et al., 1998) and MsCAATCH1(Feldman et al., 2000) also share B0-like spectra, but are distinguished by their order of substrate preference, and form an independent phylogenetic cluster from that of dipteran B0-like transporters, as shown in the previously published tree (Meleshkevitch et al., 2006). Anopheles gambiae AgNAT8 and AgNAT6 are aromatic amino acid selective transporters with unique preferences for phenylalanine/phenol-branched substrates (Meleshkevitch et al., 2006) and tryptophan/indole-branched substrates (Meleshkevitch et al., 2009), respectively. The characterized aromatic NATs anchoring two phylogenetically independent groups with exactly one ortholog per genome in other mosquito species were identified as representatives of F (phenylalanine) and W (tryptophan) systems based on their ultimate substrate specificity and phylogenetic clustering (Meleshkevitch et al., 2009). The present study reveals a new transporter that is clearly paralogous, and differs by substrate profile to all previously characterized SLC6 members, including the aforementioned F and W systems. The ultimate conservation of substrate binding motifs (Fig. 1) reflects functional homology of its orthologs. Together, our results suggest that AeNAT5 is the first molecular representative of a previously unknown transport system of sodium dependent transporters with ultimate selectivity for sulfur-containing amino acids. Following the canonical nomenclature of amino acid transport systems (Christensen et al., 1994), it is defined here as system M (Fig. 2, M cluster; M for Methionine and capital for sodium dependent transporters). The proposed systematic of the currently known insect NATs is summarized in Table 2 along with NCBI protein accession numbers and ordered by preferred substrate of characterized representatives.
Table 2.
A summary of amino acid selectivity and expression profile for currently characterized insect NATs.
| System | Spec | SLC member | NCBI | Substrate specificity profile* | SCS | Expre ss |
References |
|---|---|---|---|---|---|---|---|
| B0-like | Ms | KAAT1 | AAC24190 | (K+) FL123M>AI>HS> (Na+) MFYL120VAI>CHQSTG> | 2:1 | A** | 1 2 |
| B0-like | Ms | CAATCH1 | AAF18560 | (K+) T270>S>G>P1900>FQM>HA>>L (Na+) P>TG>>A | 2:1 | − | 3 |
| B0-like | Dm | SLC6-NAT1 | AAY56384 | (Na+) M40F60YVSL> (~ for D and L form) | 1:1 | A, N | 4 |
| B0-like | Ae | SLC6-AAT1 | AAR08269 | (Na+) F45CHASMIYTGNP> | 2:1 | A, N | 5 |
| F-selective | Ag | SLC6-NAT8 | AAN40409 | (Na+) YF>W>(DOPA)>> | 2:1 | A, N | 6 |
| W-selective | Ag | SLC6-NAT6 | AAT07965 | (Na+) W13>>5-HTP26Y40F>DOPA700>> | 2/1:1 | A, N | 7 |
| M-selective | Ae | SLC6-NAT5 | ABZ81822 | (Na+) M20 >>hC890 >C2200>LH>> | 1:1 | A, N | This work |
Species abbreviation: Ms, Manduca sexta; Dm, Drosophila melanogaster; Ae, Aedes aegypti; Ag, Anopheles gambiae. SCS, apparent Substrate Coupling Stoichiometry in numbers of Na+ ions per amino acid molecules. Abbreviation for the Identified sites of expression are: A, alimentary; N, neuronal
Character strings represent decaying orders of apparent substrate preferences that are summarized from several published records (see References). The properties of specified transporters have been acquired under different experimental and should be compared between transporters with caution. Superscript numbers show reported apparent affinities in µM. The adjacent substrates have similar substrate induced currents or uptake rate while characters > and >> indicate significant or strongly significant differences between proximal substrates
Expression in the alimentary epithelia has been reported only for MsKAAT1, while neuronal expression was not explicitly analyzed for both caterpillar transporters. References: 1, induced current (Castagna et al., 1998); 2, uptake competition (Vincenti et al., 2000); 3 (Feldman et al., 2000); 4, induced current (Miller et al., 2008); 5, induced current (Boudko et al., 2005a); 6, induced current (Meleshkevitch et al., 2006); 7, induced current (Meleshkevitch et al., 2009).
The identification of a new NAT with narrow selectivity for essential L-Met further supports our earlier hypothesis that NATs evolved and act as an integrated mechanism for active absorption of essential amino acids (Boudko et al., 2005a; Meleshkevitch et al., 2006; Meleshkevitch et al., 2009). Specifically, it shows that adaptations inside the NAT subfamily expand beyond those of B0-like and aromatic amino acid selective F and W system transporters. The conservation of narrow specificity F, W, and M system transporters in mosquitoes, and the apparent absence of known orthologs and homologs of these transporters in other metazoan organisms, is a particular intriguing aspect of the molecular evolution and adaptation of NAT mechanisms. We anticipate that the identified narrow specificity NATs can improve acquisition of the most underrepresented essential amino acids by reducing the competition for transport mechanisms and energy with abundant amino acids. Moreover, the NAT gene duplication and specialization process yields more flexible regulation of integrated amino acid absorption through alteration of gene expression of individual NATs. The identified narrow selectivity NATs are particularly well fit for the nutrient ecology of mosquito larvae that tend to escape predatory pressure in aquatic niches with a limited flux of nutrients. Nutrient adaptations may also explain extinction or absence of narrow specificity NATs in omnivorous, predatory, or parasitic metazoa because broad spectra NATs could satisfy the essential amino acid demand in metazoan organisms that have access to a stable and balanced pool of nutrient amino acids.
AeNAT5 mediated transport of L-Met is strictly Na+ dependent. Such ultimate cationic selectivity is unusual among characterized insect NATs, most of which can opportunistically use an inverted K+ gradient in parallel or in substitute to the Na+ gradient, and therefore can transport amino acids under limited availability of Na+ ion (Castagna et al., 1998). For example, mosquito broad spectra AeAAT1 (Boudko et al., 2005a), as well as aromatic amino acid selective AgNAT6 (Meleshkevitch et al., 2009) and AgNAT8 (Meleshkevitch et al., 2006), all generate amino-acid dependent K+ currents below the K+ reversal voltage. Hence, Na+ selectivity is not a universal characteristic of either insect, mosquito, or Aedes NATs. Additional comparative analysis of NATs is necessary to justify if this is a randomly acquired trait, or a result that is parallel to the evolution of AeNAT5’s ultimate selectivity. The preferred stoichiometry of AeNAT5, Na+1:1AA, is common for NATs in different species (Boudko, 2012). However, the present analysis (Fig. 5B) emphasizes Na+ binding at a fixed, nearly saturated, amino acid concentration. NATs preferably using 2:1 and 3:1 coupling stoichiometry (Metzler et al., 2013), and even triggering between 1 and 2 Na+ ratios (Meleshkevitch et al., 2009), have been reported previously. The crystalized bacterial LeuT possesses two Na+ binding sites (Yamashita et al., 2005) with distinct ion binding mechanisms (Noskov and Roux, 2008) that are additionally modulated through the binding of the transporter by organic substrates and Cl−. The summary of the available data is consistent with a more generalized paradigm: that only one Na+ is obligatory and may be sufficient for transport event induction, while additional Na+ ions act as facultative providers of additional motive force that aids to concentrative accumulation of amino acids against an increasing transmembrane gradient.
Five putative Cl− binding sites, inferred from the identified Cl− binding residues of SLC6 transporters (Forrest et al., 2007), are strictly conserved in insect NATs (Fig. 1), which may suggest some similarities in the interaction of insect NATs with Cl−. Indeed, the moderate Cl− dependency of AeNAT5 is similar to other characterized insect NATs (Boudko, 2012). The putative Cl−binding motif of AeNAT5 is distinct from the motif in the strictly Cl− independent LeuT. Perhaps the most important difference is that AeNAT5 includes only one acidic amino acid with a negative charge in TMD7 (350D−pKa3.22TFTS motif; Fig. 1), while LeuT has two (N287E−pKa4.09K+A290E−pKa4.09). The differences in the total amount and distribution of negative charges in those motifs are apparent. It has been suggested that the presence of the negative charges compensates for the requirement of and reduces Cl− binding potential in Cl− - independent LeuT (Forrest et al., 2007). Similarly a fraction of this charge, as well as its minor structural relocation, may be responsible for the transient Cl− dependency of insect NATs. Obviously, Cl− binding would decrease the integrated motive forces of amino acid transport and should not be not treated as an adaptation that aids in the energetic performance of an SLC6 mechanism. Indeed, why it would be necessary to compensate for the Na+ charge in a transporter that uses it for its main function? The currently available experimental evidence consistently shows that SLC6 members use Cl- as a constitutive donor of a negative charge that aids in the recycling of the outside open conformation. Bidirectional (electroneutral) translocation would explain the reduction of transport current upon reduction of external Cl− concentration (Fig. 4C and previously published NATs), and is consistent with the structural constitutive compensation for the negative charge in that locus of LeuT. Nonetheless, the significance of Cl− interaction remains an incompletely understood and interesting aspect of SLC6 function that requires additional attention.
By the identified substrate selectivity and morphological pattern of its expression, AeNAT5 fits perfectly in to an earlier proposed essential amino acid transport network (Boudko, 2010). Specifically, AeNAT5, similar to other mosquito NATs, is strongly expressed in the absorptive region of the larval alimentary canal. Although the presented transcript localization is unable to demonstrate membrane localization of AeNAT5 in the epithelial tissue, the previous immunolocalization of aromatic NATs in the apical membrane of larval posterior midgut and basal membrane in salivary glands and cardia (Okech et al., 2008) allows us to anticipate a similar docking pattern for AeNAT5. Likely, narrow and broad specificity NATs together form a transport mechanism for active absorption of free digested essential amino acids across the apical membrane of the posterior midgut. The narrow specificity NATs would be particularly important for the absorption of essential aromatic and sulfur-containing amino acids that are limited in nutrient proteins, as well as for supplying extensive secretion processes in the salivary glands and cardia.
Despite the evident fitting NATs in to the essential amino acid transport (Boudko, 2012) NAT essentially may be arguable because alternative mechanisms for acquisition of essential amino acids may exist. Considering that metazoan amino acid absorption is predominately mediated by secondary transporters of the Major Facilitator Superfamily, which are comprehensively identified in completed insect genomes with references to characterized transporters (Boudko, 2010; Boudko et al., 2005b), we can now address the theoretical contributions of different families of amino acid transporters (AAT) to the essential amino acid transport network. In addition to 9 SLC6-NATs, the Ae. aegypti genome includes 72 genes of 9 SLC families affiliated with amino acid transport in insects or other organisms. However, a large fraction of these transporters cannot participate in the essential amino acid transport network based on their substrate specificity and localization. In the Ae. aegypti genome these transporters are 4 SLC1 (acidic and small neutral AATs); 16 SLC17 (diverse organic/inorganic substrates and vesicular glutamate AATs); 1 SLC32 (vesicular GABA transporter); and 3 SLC38 (systems A and N AATs that specialized in sodium dependent transport of small dispensable amino acids) transporters. SLC15 (proton dependent peptide transporters) may contribute some alimentary accumulation of essential amino acids as small peptides. The Ae. aegypti genome includes 2 uncharacterized SLC15 transporters. However, SLC15 transporters need an inward proton gradient, while the proton gradient likely is outward in the alkaline gut of mosquito larvae (Boudko et al., 2001a; Boudko et al., 2001b). For the same reason, we may dismiss 12 AeSLC36 (proton coupled AATs). In contrast, 9 NAT-SLC6 and 12 SLC7 (Hansen et al., 2011) transporters represent theoretically plausible components of the essential amino acid transport network in Ae. aegypti based on known substrate specificity (Boudko, 2012). The SLC6-NATs form an active (cation gradient driven) apical component, while SLC7 contribute to facilitative basolateral component of alimentary absorption. Moreover, transporters of both these families participate in systemic distribution of essential amino acids (Boudko, 2012). In particular, stronger in situ hybridization signal (Fig. 7) suggests AeNAT5 contribution in a function of the salivary glands, secretion of peritrophic membrane by the cardia, as well as central and peripheral neuronal functions. The neuronal role of aromatic NATs as substrate providers for the synthesis of dopamine and serotonin neurotransmitters was suggested based on their substrate specificity and neuronal in situ hybridization (Meleshkevitch et al., 2006; Meleshkevitch et al., 2009). AeNAT5 substrates are not immediate precursors of known neurotransmitters and its in situ hybridization reveals more even expression in the central nervous system, with slightly elevated expression in the neuropile region (Fig. 7C, G). In contrast, AeNAT5 expression was cell-specific in the peripheral neurons associated with visual, mechanosensory, and chemosensory transduction (Fig. 7D, E, F). Recent studies of SNF-5, a broad spectra NAT with particularly high affinity to L-Met in a model of basal Ecdysozoa, C. elegans, has shown specific expression of this transporter in identifiable Amphid Sensory neurons: ASI, ASK, and ADF (Metzler et al., 2013). These cells detect amino acids and control worm development and metabolism. snf-5 deletion resulted in a similar phenotype as the ablation of SNF-5 expressing sensory neurons, and it was suggested that this transporter may act as a receptor monitoring the concentration of nutrient amino acids. It is possible that AeNAT5 aids in similar neuronal functions in mosquito larvae. Dietary L-Met has been identified as a key modulator of TOR signaling that controls development, metabolism, reproduction, and longevity in eukaryotic organisms ranging from yeast (Powers et al., 2006) to mammals (Orentreich et al., 1993). For example, L-Met alone increases fecundity without reducing lifespan in the dietary limited fruit fly (Grandison et al., 2009). Similarly, addition of L-Met to natural diet significantly increases fecundity, fertility, adult emergence, and longevity while decreasing mortality in B. mori (Laz, 2009). In contrast, L-Met deprivation typically results in metabolic disorders and death. Rats develop fatty liver diseases and anemia, and lose over 60% of their body weight during 35-day restriction of L-Met (Oz et al., 2008).
To address the biological significance of NATs directly, we developed a technique for the RNAi suppression of NATs in early stage mosquito larvae. Despite uniform extinction of several dsRNA transporting SID genes (Jose and Hunter, 2007) in dipteran insect genomes (personal unpublished observation), transient RNAi has been achieved in several mosquito species after abdominal injection of dsRNA in adult mosquitoes. Injection induced RNAi is much more challenging in larvae, especially in the early stage that is more adequate for testing development and nutrition related genes. Two approaches have been reported previously that limit artifacts of mechanical handling and simplify the experimental procedure of RNAi in mosquito larvae: feeding Ae. aegypti larvae with chitosan-coupled dsRNA nano particles (Zhang et al., 2010) and osmotic stress assisted delivery of dsRNA in Culex pipiens larvae (Lopez-Martinez et al., 2012). As both techniques induce considerable nonspecific mortality in early Ae. aegypti larvae, we developed a hatching induced RNAi (hi-RNAi) technique that reduces this undesired effect of chitosan and osmotic stress. As we placed partially desiccated mosquito eggs in a drop of concentrated dsRNA, the initial liquid drive and/or immediate exposure of hatched larvae to dsRNA was sufficient to induce transient silencing as recorded through a quantitative assay of AeNAT5 transcript: ~ 50% on day 2; ~85% on day 4; and ~ 20% on day 8. In contrast to previous reports, we did not achieve sustained silencing nor detectable morphological defects, which may reflect differences in the targeted genes, RNAi induction procedure, the loss of silencing due to the developmental sequence in our model organism and/or transporter, or compensatory up regulation of the AeNAT5 gene. Moreover, we should mention that our initial morphological assay lacks the resolution to define fine morphological and ultrastructural changes in mosquito organs and tissues. Nonetheless, partial AeNAT5 suppression was sufficient to increase larval mortality and nearly eliminate adult emergence, which for the first time demonstrates the indispensable contribution of a NAT-SLC6 transporter to the development of a metazoan organism. In conclusion, AeNAT5 function in the nutrient acquisition and distribution of essential L-Met and related sulfur-containing amino acids is critical for mosquito development and survival.
► Metazoan transporters for selective absorption of essential L-Met were unknown. ► The first representative of a sulfur-containing amino acids selective transport system M was cloned from mosquito larvae. ► The characterized transporter mediates alimentary absorption and distribution of essential methionine, cysteine, and homocysteine and its function is essential for development in vector mosquito. ► Novel indispensable metazoan transporter and transport system were identified and characterized.
Acknowledgment
This work was supported by a R56AI030464 NIH-NIAID research grant (D.B.). M.P. was supported by a NSF summer research fellowship, Research Experience for Undergraduates (REU). We appreciate Drs. L. Ebihara and H. Sackin and their lab personnel for providing X. laevis eggs, and Dr. M. Nanazashvili for the helpful suggestions and editing of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Blitzer EJ, Vyazunova I, Lan Q. Functional analysis of AeSCP-2 using gene expression knockdown in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2005;14:301–307. doi: 10.1111/j.1365-2583.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- Boudko DY. Molecular ontology of amino acid transport. In: Gerencser G, editor. Epithelial Transport Physiology. Springer, NY: Humana-Springer Verlag; 2010. pp. 379–472. [Google Scholar]
- Boudko DY. Molecular basis of essential amino acid transport from studies of insect nutrient amino acid transporters of the SLC6 family (NAT-SLC6) J Insect Physiol. 2012;58:433–449. doi: 10.1016/j.jinsphys.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko DY, Kohn AB, Meleshkevitch EA, Dasher MK, Seron TJ, Stevens BR, Harvey WR. Ancestry and progeny of nutrient amino acid transporters. Proc Natl Acad Sci U S A. 2005a;102:1360–1365. doi: 10.1073/pnas.0405183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko DY, Moroz LL, Harvey WR, Linser PJ. Alkalinization by chloride/bicarbonate pathway in larval mosquito midgut. Proc Natl Acad Sci U S A. 2001a;98:15354–15359. doi: 10.1073/pnas.261253998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko DY, Moroz LL, Linser PJ, Trimarchi JR, Smith PJ, Harvey WR. In situ analysis of pH gradients in mosquito larvae using non-invasive, self-referencing, pH-sensitive microelectrodes. J Exp Biol. 2001b;204:691–699. doi: 10.1242/jeb.204.4.691. [DOI] [PubMed] [Google Scholar]
- Boudko DY, Stevens BR, Donly BC, Harvey WR. Nutrient Amino acid and Neurotransmitter transporters. In: Lawrence I, Gilbert KIaSSG, editors. Comprehensive Molecular Insect Science. First edition. Amsterdam: Elsevier; 2005b. pp. 255–309. [Google Scholar]
- Bright SW, Lea PJ, Miflin BJ. The regulation of methionine biosynthesis and metabolism in plants and bacteria. Ciba Found Symp. 1979:101–117. doi: 10.1002/9780470720554.ch7. [DOI] [PubMed] [Google Scholar]
- Broer S, Palacin M. The role of amino acid transporters in inherited and acquired diseases. Biochem J. 2011;436:193–211. doi: 10.1042/BJ20101912. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Castagna M, Shayakul C, Trotti D, Sacchi VF, Harvey WR, Hediger MA. Cloning and characterization of a potassium-coupled amino acid transporter. Proc Natl Acad Sci U S A. 1998;95:5395–5400. doi: 10.1073/pnas.95.9.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NH, Reith ME, Quick MW. Synaptic uptake and beyond: the sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Arch. 2004;447:519–531. doi: 10.1007/s00424-003-1064-5. [DOI] [PubMed] [Google Scholar]
- Christensen HN, Albritton LM, Kakuda DK, MacLeod CL. Gene-product designations for amino acid transporters. J Exp Biol. 1994;196:51–57. doi: 10.1242/jeb.196.1.51. [DOI] [PubMed] [Google Scholar]
- Cooper AJ. Biochemistry of sulfur-containing amino acids. Annu Rev Biochem. 1983;52:187–222. doi: 10.1146/annurev.bi.52.070183.001155. [DOI] [PubMed] [Google Scholar]
- Feldman DH, Harvey WR, Stevens BR. A novel electrogenic amino acid transporter is activated by K+ or Na+, is alkaline pH-dependent, and is Cl--independent. J Biol Chem. 2000;275:24518–24526. doi: 10.1074/jbc.M907582199. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Forrest LR, Tavoulari S, Zhang YW, Rudnick G, Honig B. Identification of a chloride ion binding site in Na+/Cl -dependent transporters. Proc Natl Acad Sci U S A. 2007;104:12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gache C, Vacquier VD. Transport of methionine in sea-urchin sperm by a neutral amino-acid carrier. Eur J Biochem. 1983;133:341–347. doi: 10.1111/j.1432-1033.1983.tb07468.x. [DOI] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen IA, Boudko DY, Shiao SH, Voronov DA, Meleshkevitch EA, Drake LL, Aguirre SE, Fox J, Attardo GM, Raikhel AS. AaCAT1 Of The Yellow Fever Mosquito, Aedes Aegypti: A Novel Histidine-Specific Amino Acid Transporter From The Slc7 Family. J Biol Chem. 2011:10803–10813. doi: 10.1074/jbc.M110.179739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen T, Grunnet M, Angelo K, Klaerke DA, Olesen SP. Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques. 2002;32:536–538. doi: 10.2144/02323st05. 540. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jose AM, Hunter CP. Transport of sequence-specific RNA interference information between cells. Annual review of genetics. 2007;41:305–330. doi: 10.1146/annurev.genet.41.110306.130216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laz R. Effects of methionine and tryptophan on some quantitative traits of silkworm, Bombyx mori L.(Lepidoptera: Bombycidae) University Journal of Zoology, Rajshahi University. 2009;28:15–19. [Google Scholar]
- Lopez-Martinez G, Meuti M, Denlinger DL. Rehydration driven RNAi: a novel approach for effectively delivering dsRNA to mosquito larvae. J Med Entomol. 2012;49:215–218. doi: 10.1603/me11122. [DOI] [PubMed] [Google Scholar]
- Matz MV. Amplification of representative cDNA samples from microscopic amounts of invertebrate tissue to search for new genes. Methods Mol Biol. 2002;183:3–18. doi: 10.1385/1-59259-280-5:003. [DOI] [PubMed] [Google Scholar]
- Meleshkevitch EA, Assis-Nascimento P, Popova LB, Miller MM, Kohn AB, Phung EN, Mandal A, Harvey WR, Boudko DY. Molecular characterization of the first aromatic nutrient transporter from the sodium neurotransmitter symporter family. J Exp Biol. 2006;209:3183–3198. doi: 10.1242/jeb.02374. [DOI] [PubMed] [Google Scholar]
- Meleshkevitch EA, Robinson M, Popova LB, Miller MM, Harvey WR, Boudko DY. Cloning and functional expression of the first eukaryotic Na+-tryptophan symporter, AgNAT6. J Exp Biol. 2009;212:1559–1567. doi: 10.1242/jeb.027383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler R, Meleshkevitch EA, Fox J, Kim H, Boudko DY. A SLC6 transporter of the novel B0,- system aids in absorption and detection of nutrient amino acids in Caenorhabditis elegans. J. Exp. Biol. in press. 2013 doi: 10.1242/jeb.081497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MM, Popova LB, Meleshkevitch EA, Tran PV, Boudko DY. The invertebrate B(0) system transporter, D. melanogaster NAT1, has unique d-amino acid affinity and mediates gut and brain functions. Insect Biochem Mol Biol. 2008;38:923–931. doi: 10.1016/j.ibmb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Neal JJ, Wu D, Hong YS, Reuveni M. High affinity transport of histidine and methionine across Leptinotarsa decemlineata midgut brush border membrane. Journal of Insect Physiology. 1996;42:329–335. [Google Scholar]
- Noskov SY, Roux B. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J Mol Biol. 2008;377:804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okech BA, Meleshkevitch EA, Miller MM, Popova LB, Harvey WR, Boudko DY. Synergy and specificity of two Na+-aromatic amino acid symporters in the model alimentary canal of mosquito larvae. J Exp Biol. 2008;211:1594–1602. doi: 10.1242/jeb.017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- Oz HS, Chen TS, Neuman M. Methionine deficiency and hepatic injury in a dietary steatohepatitis model. Dig Dis Sci. 2008;53:767–776. doi: 10.1007/s10620-007-9900-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SH, Loomis WF. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryotic Cell. 2006;5:272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:W30–W34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Levy JI, Muilenberg ML, Coull BA, Spengler JD. Efficacy of integrated pest management in reducing cockroach allergen concentrations in urban public housing. J Asthma. 2007;44:455–460. doi: 10.1080/02770900701421971. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KH. Optimization and troubleshooting in PCR. Cold Spring Harb Protoc 2009. 2009 doi: 10.1101/pdb.ip66. pdb ip66. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Soriano-Garcia JF, Torras-Llort M, Ferrer R, Moreto M. Multiple pathways for L-methionine transport in brush-border membrane vesicles from chicken jejunum. J Physiol. 1998;509(Pt 2):527–539. doi: 10.1111/j.1469-7793.1998.527bn.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotschel C, Follmann M, Nettekoven JA, Mohrbach T, Forrest LR, Burkovski A, Marin K, Kramer R. Methionine Uptake in Corynebacterium glutamicum by MetQNI and by MetPS, a Novel Methionine and Alanine Importer of the NSS Neurotransmitter Transporter Family. sBiochemistry. 2008 doi: 10.1021/bi801206t. [DOI] [PubMed] [Google Scholar]
- Verrey F, Closs EI, Wagner CA, Palacin M, Endou H, Kanai Y. CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. 2004;447:532–542. doi: 10.1007/s00424-003-1086-z. [DOI] [PubMed] [Google Scholar]
- Vincenti S, Castagna M, Peres A, Sacchi VF. Substrate selectivity and pH dependence of KAAT1 expressed in Xenopus laevis oocytes. J Membr Biol. 2000;174:213–224. doi: 10.1007/s002320001046. [DOI] [PubMed] [Google Scholar]
- Wolfersberger MG. Amino acid transport in insects. Annual Review of Entomology. 2000;45:111–120. doi: 10.1146/annurev.ento.45.1.111. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae) Insect Mol Biol. 2010;19:683–693. doi: 10.1111/j.1365-2583.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- Zhao H, Kim G, Levine RL. Methionine sulfoxide reductase contributes to meeting dietary methionine requirements. Arch Biochem Biophys. 2012;522:37–43. doi: 10.1016/j.abb.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]