Abstract
A covalently linked protein–protein conjugate between ThiF and ThiS thiocarboxylate was found in a partially purified coexpressed ThiF/ThiS protein mixture by using Fourier transform mass spectrometry. The Cys-184 of ThiF and the C terminus of ThiS thiocarboxylate were identified to be involved in the formation of this complex by using both mutagenesis and chemical modification methods. A complementation study of Escherichia coli thiF− using thiF(C184S) suggests that this conjugate is an essential intermediate involved in the biosynthesis of the thiazole moiety of thiamin. This ThiF/ThiS conjugate is the first characterized example of a unique acyldisulfide intermediate in a biosynthetic system. This protein conjugate is also an example of an ubiquitin-E1 like protein–protein conjugate in prokaryotes and supports a strong evolutionary link between thiamin biosynthesis and the ubiquitin conjugating system.
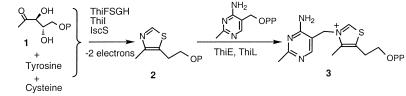
Thiamin pyrophosphate (3) is an essential cofactor in all living systems and plays a key role in the metabolism of carbohydrates and the biosynthesis of branched chain amino acids. The biosynthesis of the thiazole moiety (2) of thiamin involves the oxidative condensation of deoxy-d-xylulose-5-phosphate (1), tyrosine, and cysteine (Scheme S1) (1–4).
Scheme 1.
Substantial progress has been made on the elucidation of the details of sulfur transfer from cysteine to the thiazole moiety of thiamin. A protein ThiS (12), which has been posttranslationally modified by conversion of its carboxyl-terminal glycine to a thiocarboxylate (ThiS-COSH), has been identified as an intermediate sulfur carrier (5, 6), and the structure of this protein has been determined (7). Two enzymes involved in the formation of ThiS-COSH also have been identified and characterized: ThiF (5) catalyzes the formation of ThiS-COAMP, and IscS (8, 9) catalyzes the sulfur transfer from cysteine to the acyl adenylate of ThiS in a pyridoxal 5′-phosphate-dependent reaction. Using both of these enzymes, ThiS-COSH formation has been successfully reconstituted in vitro (3, 8). In addition, ThiI (9–12), a protein involved in thiouridine formation in tRNA, may participate in this sulfur transfer chemistry. However, ThiI is not essential and its role is not yet understood.
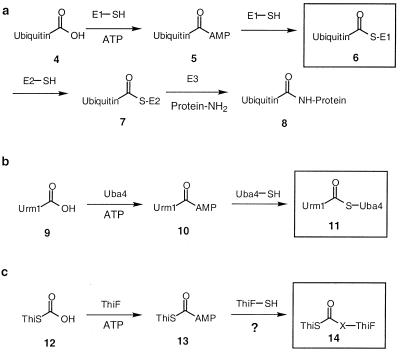
In eukaryotes, the ubiquitin conjugating system plays an essential role in several important cellular processes (13, 14) including the tagging of proteins for proteolytic degradation. In the first step of this tagging sequence, ubiquitin (4) is converted to its carboxyl-terminal acyl adenylate (5) by the ubiquitin-activating enzyme (E1). This acyladenylate then forms a thioester (6) with the active site cysteine of E1 (Scheme S2a). Ubiquitin then is transferred to one of the ubiquitin carrier proteins (E2) by a trans thioesterification reaction. Finally, E3 catalyzes the transfer of ubiquitin from 7 to lysine residues on the protein targeted for tagging. Besides the ubiquitin system, several other ubiquitin-like conjugating systems have been identified in yeast (15). One example of such a system is the Urm1/Uba4 system (16) (Scheme S2b).
Scheme 2.
The structure of ThiS is very similar to the structure of ubiquitin (7). In addition, ThiF shows striking sequence similarity to both E1 and Uba4. These observations suggest that the ThiS/ThiF sulfur transfer system may follow a similar pathway to the ubiquitin conjugating system, and a potential crosslinked intermediate, analogous to ubiquitin-COS-E1 (6) and the Urm1-COS-Uba4 (11), might exist between ThiS and its activating enzyme ThiF (14, Scheme S2c) (17).
In this paper, we describe the identification and characterization of an intermediate in which ThiS-COSH is crosslinked to ThiF through an acyldisulfide linkage and propose a biosynthetic role for this intermediate in the formation of the thiazole.
The discovery of this intermediate further strengthens the proposal that the ThiF/ThiS-catalyzed sulfur transfer chemistry in thiamin biosynthesis may be the bacterial ancestor of the ubiquitin and ubiquitin-like conjugation systems found in eukaryotes.
Materials and Methods
Wizard PCR preps was purchased from Promega. Escherichia coli BL21(DE3), the E. coli (DE3) lysogenization kit, and the pET overexpression system were purchased from Novagen. Protein purification was performed on a Waters 650 chromatography instrument. DEAE 8 HR prepacked column was from Waters. Gel filtration column (Superdex 75) was from Amersham Pharmacia. Protein concentration was assayed by using Coomassie Plus Protein Assay Reagent from Pierce. SDS/PAGE gels were prepared and run as described (18).
Standard methods were used for DNA restriction endonuclease digestion, ligation, and transformation (19, 20). Automated DNA fluorescence sequencing was performed at the Cornell BioResource Center. Plasmid DNA was purified with the Wizard Plus SV DNA miniprep kit (Promega). DNA fragments were separated by agarose gel electrophoresis, excised, and purified with the QIAEX II Gel Extraction Kit (Qiagen, Valencia, CA). E. coli strain DH5α was used as a recipient for transformations during plasmid construction and for plasmid propagation and storage. A Perkin–Elmer GeneAmp PCR System 2400 and platinum Pfx DNA polymerase (GIBCO/BRL) were used for PCR. T4 DNA ligase and all restriction endonucleases were purchased from New England Biolabs.
Construction of pCLK1404 (thiFSGH in pET-22b).
The plasmid pCAC111 (thiFS in pRSET) (21) was digested with NdeI/SalI, and the fragment containing thiFS and the 5′ end of thiG was purified. The plasmid pADB153 (thiSGH in pRSET) (22) was similarly digested, and the fragment containing the vector, thiH, and the 3′ end of thiG was purified. The fragments derived from pCAC111 and pADB153 were ligated to yield pCLK403 (thiFSGH in pRSET). The thiamin genes were excised from pCLK403 by digestion with NdeI and HindIII and ligated into similarly digested pET-22b (Novagen) to give pCLK1404.
Construction of pCLK1405 (thiFS in pET-22b).
The thiGH genes were deleted from pCLK1404 (see above) by digestion with SalI and XhoI. The fragment containing the vector and thiFS was purified, and the compatible cohesive ends of the two restricted sites were ligated to yield pCLK1405, thiFS.
Construction of pCLK433 (thiF in pET-22b).
The thiF gene was PCR-amplified from pVJS716 (23) by using the following primers: upstream primer 5′-GGA GTT GCA TAT GAA TGA CCG TGA CTT TAT GCG-3′ (inserts an NdeI site at the start codon of the gene); downstream primer 5′-GCA TCG CTG GAT CCT TAA ACA GGA TCT GCA TTG CTT CC-3′ (inserts a BamHI site at the end of thiF). The purified PCR product was digested with NdeI and BamHI and ligated into similarly digested pET-22b(+). Colonies were screened for the presence of the insert, and a representative plasmid was designated pCLK433.
Construction of pCLK411 (thiS in pET-22b).
The thiS gene was PCR-amplified from pVJS716 (23) by using the following primers: upstream primer 5′-GCG GAG GAA CAT ATG CAG ATC CTG TTT AAC GAT CAA GCG-3′ (inserts an NdeI site at the start codon of the gene); downstream primer 5′-CGG ATC GCC TCG AGC ATC AGT TGT GAA GAA GCG AAT TTC CC-3′ (inserts a BamHI site at the end of thiS). The purified PCR product was digested with NdeI and BamHI and ligated into similarly digested pET-22b(+). Colonies were screened for the presence of the insert, and a representative plasmid was designated pCLK411.
Construction of the thiF (C184S) Overexpression Plasmid.
The preparation was carried out by using Stratagene's QuickChange Site-Directed Mutagensis kit. The forward primer was GGA GCC AGA ACG CAA CA(T)G CCG T(C)AC GGC GGG CGT GGT TGG C, and the reverse primer was GCC AAC CAC GCC CGC CGT A(G)CG GCT(A) GTT GCG TTC TGG CTC. Two microliters (10 ng/μl) of pCLK433 (thiF overexpression plasmid), 1.5 μl of forward primer (125 ng), and 1.5 μl of reverse primer (125 ng) were used. Four colonies from the transformant plate were selected to prepare the plasmid.
Construction of the thiFS (thiF-C184S) Overexpression Plasmid.
The forward primer was GGA GCC AGA ACG CAA CA(T)G CCG T(C)AC GGC GGG CGT GGT TGG C, and the reverse primer was GCC AAC CAC GCC CGC CGT A(G)CG GCT(A) GTT GCG TTC TGG CTC. Two microliters (10 ng) of pCLK1405 (thiFS overexpression plasmid), 1.5 μl of forward primer (125 ng), and 1.5 μl of reverse primer (125 ng) were used. Four colonies from the transformant plate were selected to prepare the plasmid.
Construction of the thiFSGH (thiF-C184S) Overexpression Plasmid.
The forward primer was GGA GCC AGA ACG CAA CA(T)G CCG T(C)AC GGC GGG CGT GGT TGG C, and the reverse primer was GCC AAC CAC GCC CGC CGT A(G)CG GCT(A) GTT GCG TTC TGG CTC. Two microliters (10 ng) of pCLK1404 (thiFSGH overexpression plasmid), 1.5 μl of forward primer (125 ng), and 1.5 μl of reverse primer (125 ng) were used. Four colonies from the transformant plate were selected to prepare the plasmid.
Construction of the thiFS (thiS-ΔGG) Overexpression Plasmid.
The thiFS genes were PCR-amplified from pVJS716 (20) by using the following primers: upstream primer 5′-GGA GTT GCA TAT GAA TGA CCG TGA CTT TAT GCG-3′ (inserts an NdeI site at the start codon of the thiF ORF); downstream primer 5′-ACG TAA CAT GGA TCC TCA TGC AAT AAC CTG AAA AAG CAG-3′ (changes Gly-65 to a stop codon and inserts a BamHI site after the end of thiS). The purified PCR product was digested with NdeI and BamHI, purified, and ligated into similarly digested pET-22b(+). Colonies were screened for the presence of the insert, and a representative plasmid was designated pCLK1406.
Purification of ThiF and ThiF(C184S).
The plasmid pCLK433 (thiF) was overexpressed in BL21(DE3) and grown in LB/ampicillin (AMP) medium at 37°C to A595 of 0.6. The expression was induced with 0.3 mM of isopropyl β-d-thiogalactoside (IPTG). The growth was continued at 37°C for 3 h, and the cells were harvested and stored at −70°C. Cell paste from 2 liters of cell culture was thawed, resuspended in lysis buffer (50 mM Tris/200 mM NaCl/5% glycerol/1 mM DTT/1 mM PMSF, pH 7.5), lysed with lysozyme (2 mg) on ice for 1 h, and treated with DNase I (8 μg) on ice for 30 min. The cell lysate was centrifuged at 30,000 g for 30 min. The S-30 lysate was diluted in buffer B (50 mM Tris/10% glycerol/1 mM PMSF/14 mM 2-mercaptoethanol, pH 7.5) and was fractionated by FPLC using a DEAE column. The protein eluted under the following condition: 5% C (buffer B + 1 M KCl, pH 7.5) 95% B to 6% C in 35 min to 12% C in 35 min to 15% C in 30 min and kept at 15% C for an additional 10 min. The ThiF-containing fractions were pooled and concentrated, and the protein was further purified on a G-75 Sephadex column. ThiF was eluted with 20% C 80% B in about 3.5 h. The final protein was stored in a concentrated form in 50 mM Tris, 10% glycerol, 1 mM DTT, pH 7.5 at −70°C. ThiF(C184S) was purified in an identical manner.
Purification of ThiF/ThiS, ThiF(C184S)/ThiS, and ThiF/ThiS(ΔGG).
Buffer D was 25 mM Tris/2 mM DTT, pH 8, and buffer E was buffer D + 1 M NaCl, pH 8. The plasmid pCLK1405 (thiFS) was overexpressed in BL21(DE3) and grown in LB/AMP medium at 37°C to A595 of 0.6. Expression was induced with 0.3 mM of IPTG, and the zinc concentration in the culture medium was brought to 0.1 mM by the addition of ZnSO4. The growth continued at 37°C for 3 h, and the cells were harvested. Cell pellets derived from 1 liter of culture were thawed and resuspended in lysis buffer and lysed by using a French press. The cell lysate was centrifuged at 30,000 g for 30 min. The supernatant was brought to 50% ammonium sulfate, and the resulting precipitate was isolated by centrifugation, resuspended in buffer D, and dialyzed against the same buffer. The dialysate was filtered to remove the precipitated protein and applied to an FPLC equipped with a DEAE column. The protein was eluted under the following conditions: 0% E 100% D for 15 min, then to 25% E, 75% D in 80 min. The fractions were analyzed by gel electrophoresis, and the desired fractions were pooled and concentrated to give a mixture of protein with 50% purity. The final protein was stored in a concentrated form in 50 mM Tris, 10% glycerol, 1 mM DTT, pH 7.5 at −70°C. ThiF(C184S)/ThiS and ThiF/ThiS(ΔGG) were prepared in an identical manner.
Preparation of ThiS.
The plasmid pCLK411 (thiS overexpression plasmid) was overexpressed in BL21(DE3) and grown in LB/AMP medium at 37°C to A595 to 0.6. The expression was induced with 0.5 mM of IPTG. The growth was continued at 37°C for 3 h, and the cells were harvested and stored at −70°C. Cell paste from 2 liters of cell culture was thawed, resuspended in lysis buffer (50 mM Tris/200 mM NaCl/5% glycerol/1 mM DTT/1 mM PMSF, pH 7.5), lysed with lysozyme (2 mg) on ice for 1 h, and treated with DNase I (8 μg) on ice for 30 min. The cell lysate was centrifuged at 30,000 g for 30 min. The S-30 lysate was diluted in buffer B (50 mM Tris/10% glycerol/1 mM PMSF/14 mM 2-mercaptoethanol, pH 7.5) and was fractionated by FPLC using a DEAE column. The protein was eluted under the following condition: 5% C (buffer B + 1 M KCl, pH 7.5) 95% B. The ThiS-containing fractions were pooled and concentrated, and the protein was further purified on a G-75 Sephadex column. ThiS was eluted with 20% C 80% B in about 5 h with greater than 90% purity. The final protein was stored in a concentrated form in 50 mM Tris, 10% glycerol, 1 mM DTT, pH 7.5 at −70°C.
Purification of IscS.
IscS was prepared by following the protocol developed by Kambampati and Lauhon (9).
SDS/PAGE Assay for the Formation of ThiS Thiocarboxylate.
The reaction mixture containing ThiF (74.9 μM) or ThiF(C184S) (74.9 μM), ThiS (24.1 μM), [35S]Cys (75 μM), IscS (2 μM), pyridoxal 5′-phosphate (50 μM), ATP (5 mM), and MgSO4 (2 mM) in a final volume of 10 μl was incubated at 37°C for 30 min, diluted with 10 μl of reaction buffer (50 mM Tris/1 mM DTT, pH 7.5) and spin-filtered through a BioSpin 6 gel filtration column. The filtrate was analyzed by PAGE using the Tris-tricine buffer system (18). The resulting gel was fixed, stained, destained, and dried. [35S]ThiS-COSH was detected by using a PhosphoImager.
Alkylation of ThiF, ThiF(C184S), ThiF/ThiS, and ThiF(C184S)/ThiS.
ThiF (≈100 μg, 10–15 μl in 50 mM Tris, 1 mM DTT, 10% glycerol, 5 mM NaCl, pH 7.5) was thawed, and iodoacetic acid was added (10–15 μl of 10 mM solution in 50 mM Tris, pH 7.5). The reaction mixtures were incubated at room temperature for 5–60 min and quenched by the addition of 3–5 μl of acetic acid. All other proteins were alkylated in an identical manner.
Glu-C Proteolysis.
Iodoacetic acid-derivatized ThiF (50 μg) in 50 mM Tris, pH 7.5 was incubated with an equal volume of Glu-C protease (1 μg in lyophilized form, redissolved in H2O) for 3 h at 37°C. The reaction mixture then was quenched by the addition of 3–5 μl of acetic acid.
MS Analysis.
Intact protein and crude digests were desalted by loading onto a reverse-phase protein or peptide trap (Michrom Bioresources, Auburn, CA). The trap was washed with 2 ml of 2:96:2 CH3OH/H2O/CH3COOH and eluted with 150 μl 70:26:4 CH3OH/H2O/CH3COOH. This eluent was loaded into a nanospray electrospray ionization (ESI) emitter with a 2–4 μm i.d. tip; a voltage of 1.0–1.5 kV applied to the solution supported ESI and determined the flow rate (1–50 nl/min). The resulting ions were guided through a heated metal capillary, skimmer, and three radio frequency-only quadrupoles into the ion cell (10−9 torr) of a modified 6-T Finnigan Fourier transform mass spectrometry (FTMS) device (24). The outer trapping plates, conventionally used for electron containment, were used for ion trapping. Transients were stored as 256–512 K data sets and analyzed by using a Finnigan (San Jose, CA) FTMS Odyssey data system. Assignments of the fragment masses and compositions were made with the computer program thrash (25). The mass difference (in units of 1.00235 Da) between the most abundant isotopic peak and the monoisotopic peak is denoted in italics after each Mr value. Fragmentation was achieved by nozzle-skimmer collisionally activated dissociation (100–200 V) (26), “in-beam”-activated ion electron capture dissociation (27), and blackbody infrared radiative dissociation (28) for ions entering the instrument or by isolating specific ions by using stored-waveform inverse Fourier transform (29), followed by infrared multiphoton dissociation (30).
Complementation Analysis.
Plasmids pCLK1404 (thiFSGH), its thiF (C184S) mutant, and pET22b(+) were individually transformed into VJS2895 (thiF mutant cell strain) (23). Each transformant was allowed to grow on an LB/AMP/kanamycin (KAN) plate for 12 h. A single colony was picked to grow in 1 ml of LB/AMP/KAN medium for 14 h to saturation. The resulting cell cultures were spun down, and the cell pellets were washed with M9/AMP/KAN four times (500 ml M9/AMP/KAN medium: 3.39 g Na2HPO4, 1.5 g KH2PO4, 0.25 g NaCl, 0.5 g NH4Cl, 0.1 mM CaCl2⋅2H2O, 2 mM MgSO4⋅7H2O, 0.4% glucose, 100 mg AMP, 50 mg KAN, 0.1 mM IPTG, 0.0084% arginine). The resulting cell pellets were resuspended in 1 ml of M9/AMP/KAN medium, and 80 μl of this suspension was used to inoculate 40 ml of medium. The cells were allowed to grow at 37°C, and the cell densities were monitored over a period of 24 h.
Results and Discussion
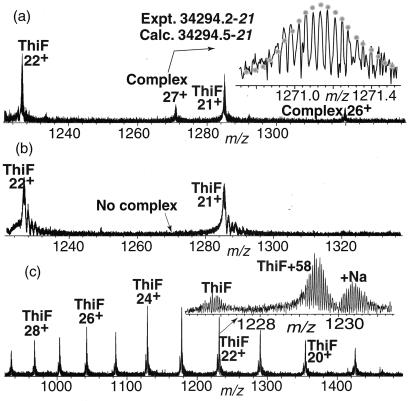
To look for the putative intermediate (14), a sample of ThiF/ThiS was rapidly purified to about 50% homogeneity from an overexpression strain (5). Analysis of this sample by using ESI/FTMS indicated the presence of a new component of mass 34294.2–21 Da (Fig. 1a). The stability of this species under relatively high (100–200 V) nozzle-skimmer voltage (26, 31) was consistent with a covalent complex. When the sample was treated with 100 mM DTT for 30 min at 37°C, the spectrum no longer showed this component (Fig. 1b). The mass and reactivity of this complex were consistent with a species in which ThiS-COSH (7326.6) was crosslinked to ThiF (26969.9) via a disulfide bond.
Figure 1.
(a) ESI/FTMS spectrum of ThiF/ThiS overexpressed and purified from E. coli. The best match of the theoretical isotopic distribution (dotted line) yielded a Mr value of 34294.2–21 where the terminal value in italics represents the mass difference (in units of 1.00235 Da) between the most abundant isotopic peak and the monoisotopic peak. (b) ESI/MS analysis of ThiF/ThiS after incubation with DTT demonstrated the absence of the ThiS-ThiF complex. (c) Direct ESI/MS analysis of the ThiF alkylation reaction mixture after incubation of ThiF with iodoacetic acid for 15 min showing about 80% conversion of ThiF to monoalkylated ThiF. No dialkylation was detected.
We used an indirect alkylation method to identify the cysteine residue on ThiF that was involved in the crosslinking because of the low abundance of the crosslinked complex in the ThiF/ThiS samples. This approach was based on the assumption that this cysteine is likely to be activated by base catalysis and therefore to be more reactive toward alkylation with iodoacetic acid. When ThiF was incubated with iodoacetic acid (10 mM, 1,200 equivalents) at 25°C for 15 min, ThiF with an additional mass of 58 Da was the only new species detected (≈80% alkylation, Fig. 1c). No +116-Da species, corresponding to dialkylation, was observed until the reaction time was increased to 1 h (data not shown).
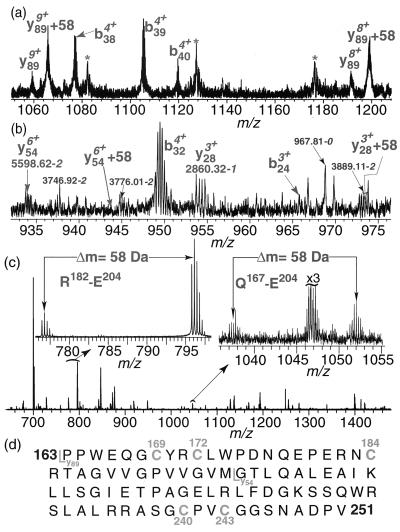
Infrared multiphoton dissociation (30) of the ThiF + 58 ion (Fig. 2a) gave the y89 and (y89 + 58) ions. These ions corresponded to an 89-aa fragment from the carboxyl terminus and localized the site of alkylation between residue 163 and 251. Nozzle-skimmer collisionally activated dissociation (26) of the y89 + 58 ion gave the C-terminal fragments y28 and y54, but not the corresponding y28 + 58 and y54 + 58 (Fig. 2b). The absence of alkylation of the y54 and the y28 ions also was confirmed by blackbody infrared radiative dissociation (28) and activated ion electron capture dissociation (27). Thus the modification is localized between residues 162 and 197, a region containing three cysteine residues: Cys-169, Cys-172, and Cys-184 (Fig. 2d).
Figure 2.
Localization of the alkylated cysteine on ThiF by using MS/MS analysis and proteolysis. (a) Infrared multiphoton dissociation of alkylated ThiF; about 80% of y89 was alkylated. (b) Nozzle-skimmer collisionally activated dissociation of alkylated ThiF; no alkylation of y28 and y54 was detected. (*, noise peak). (c) ESI/MS analysis of the alkylated ThiF from Glu-C proteolysis reaction mixture; all of the peptides containing Cys-184 show the +58 modification. (d) The sequence fragment of ThiF between residues 163 and 251. The cysteines in this fragment are numbered; both y89 and y54 fragments also are indicated.
To further localize the site of modification, the peptides resulting from proteolysis of 60% monoalkylated ThiF with Glu-C were analyzed by ESI/FTMS (Fig. 2c). Two of these peptides (R182-E204, Q167-E204) were particularly informative. R182-E204 is accompanied by an abundant peptide with an additional mass of 58 Da. Because Cys-184 is the only cysteine residue in this peptide, this observation demonstrates that this cysteine is the site of modification. The peptide Q167-E204, which contains Cys-169, Cys-172, and Cys-184, also showed the +58-Da modification and is consistent with this assignment.
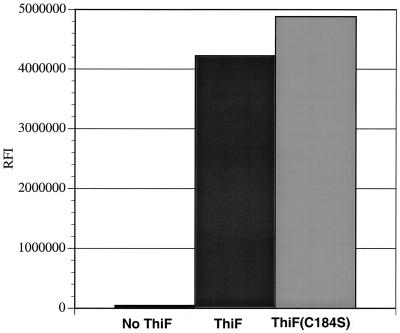
To directly probe the role of Cys-184 in the crosslink, a mutated enzyme, predicted to be unable to form the disulfide crosslink (ThiF(C184S)/ThiS), was prepared. ESI/FTMS analysis of this demonstrated the absence of the crosslinked complex (see Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org). In addition, treatment of ThiF(C184S)/ThiS with iodoacetic acid (10 mM, 1200 equivalents at 25°C for 15 min) did not give any alkylated ThiF (data not shown). ESI FTMS analysis of ThiF(C184S)/ThiS also demonstrated that this complex contained ThiSCOSH (see Fig. 6, which is published as supplemental data). In addition, biochemical assays demonstrated that ThiF and ThiF(C184S) were equally effective in catalyzing the formation of ThiSCOSH (Fig. 3). We therefore conclude that Cys-184 is directly involved in the crosslinking of ThiF to ThiS but is not essential for the formation of ThiS-COSH.
Figure 3.
In vitro formation of ThiS thiocarboxylate using ThiF and ThiF(C184S). ThiF(C184S) or ThiF (74.9 μM) and ThiS-COOH (24.1 μM) were incubated together with ATP (5 mM), MgSO4 (2 mM), pyridoxal 5′-phosphate (50 μM), IscS (2 μM), and 35S-Cys (75 μM) and analyzed by Tris-tricine SDS/PAGE (39) followed by autoradiography using a PhosphoImager. RFI is a measurement of the relative fluorescence intensity.
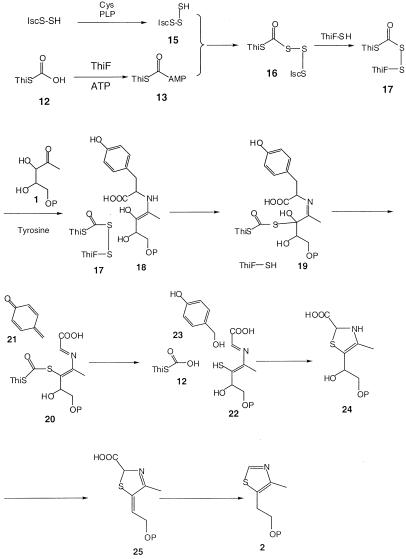
To identify the sulfur from ThiS involved in the crosslink, ThiF/ThiS (ΔGG), which lacked the carboxyl-terminal glycine pair of ThiS, was prepared. ESI/FTMS analysis of this demonstrated the absence of the crosslinked complex (see Fig. 7, which is published as supplemental data). This result demonstrated that the thiocarboxylate sulfur and not the sulfur of Cys-12 is required for the ThiF/ThiS crosslink and suggested that ThiF is crosslinked to ThiS by an acyldisulfide linkage (17, Scheme S3).
Scheme 3.
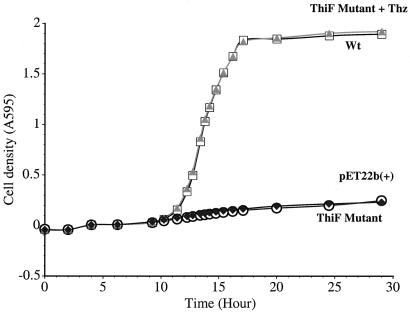
To probe the physiological significance of Cys-184, thiFSGH and thiF(C184S)SGH were separately inserted into pET22b(+) and expressed in E. coli (thiF−). The thiSGH genes were needed because the thiF mutant used was polar (27, 28). E. coli (thiF−) showed little growth at 37°C in minimal medium after transformation with thiF(C184S)SGH or pET22b(+) compared with the cells transformed with thiFSGH. When 50 μM of thiazole was added, they showed wild-type growth (Fig. 4). This finding demonstrates that Cys-184 is essential for the thiazole biosynthesis and suggests that the ThiS/ThiF acyldisulfide is a new intermediate on the thiazole biosynthesis pathway.
Figure 4.
Relative growth of E. coli thiF− strains (VJS2895) at 37°C in M9/glucose/AMP/KAN medium (500 ml) containing 0.4% glucose, 100 mg AMP, 50 mg KAN, 0.1 mM IPTG, and 0.0084% arginine. ThiF Mutant + Thz (▴): VJS2895 was transformed with thiF(C184S)SGH in pET22b(+) + additive (50 μM thiazole); wild type (Wt) (□): VJS2895 was transformed with thiFSGH in pET22b(+); pET22b(+) (⧫): VJS2895 was transformed with pET22b(+); ThiF mutant (○): VJS2895 was transformed with thiF(C184S)SGH in pET22b(+).
Although acyldisulfides are well characterized intermediates in organic chemistry (32, 33), the ThiF/ThiS acyldisulfide is a novel example of such an intermediate in biosynthesis. Its identification is particularly significant because of our previous failure to reconstitute thiazole biosynthesis by using ThiF/ThiS-COSH/ThiG/ThiH and demonstrates the value of sequence and structure analysis in deducing new chemistry on a complex biosynthetic pathway. In addition, this intermediate is an example of a prokaryotic protein–protein conjugating system, and its discovery further strengthens the proposal that the ThiF/ThiS-catalyzed sulfur transfer chemistry in thiamin biosynthesis may be the bacterial ancestor of the ubiquitin and ubiquitin-like conjugating systems found in eukaryotes (17, 34). Recently, Schindelin and coworkers (35) found a covalent link between MoaD and MoaE (ThiS/ThiG orthologs) in molybdopterin synthase. However, this crosslink was formed only after prolonged incubation and is unlikely to represent a biosynthetic intermediate.
A mechanistic proposal for the formation of the acyldisulfide intermediate and its role in thiazole formation is outlined in Scheme S3. Addition of the persulfide of IscS to the carboxyl-terminal acyl adenylate of ThiS would give 16, which could be converted to 17 by a disulfide interchange with Cys-184 of ThiF. An alternative mechanism involving initial sulfur transfer from 15 to Cys-184 can be excluded because ThiF(C184S) and ThiF catalyze the formation of ThiSCOSH at equal rates (Fig. 3). Eneamine (18), formed by condensation of tyrosine and deoxy-d-xylulose-5-phosphate (1), then could attack the acyldisulfide (17) to give 19. Analogous reactions are found in pyruvate dehydrogenase (36, 37) and in the glycine cleavage system (38–40). Quinone methide (21) extrusion followed by loss of water and thioester hydrolysis would give 22. Addition of water to 21 would give 23, a previously detected byproduct of thiazole biosynthesis (41). Ring closure followed by loss of water and decarboxylation would give thiazole phosphate (2).
Supplementary Material
Acknowledgments
This research was funded by grants from the National Institutes of Health (to T.P.B., DK44083; to F.W.M., GM16609) and by a gift from Hoffmann-La Roche.
Abbreviations
- IPTG
isopropyl β-d-thiogalactoside
- ESI
electrospray ionization
- FTMS
Fourier transform mass spectrometry
- KAN
kanamycin
- AMP
ampicillin
References
- 1.Begley T P, Kinsland C, Taylor S V, Tandon M, Nicewonger R, Wu M, Chiu H, Kelleher N, Campobasso N, Zhang Y. Top Curr Chem. 1998;195:93–142. [Google Scholar]
- 2.Begley T P, Xi J, Kinsland C, Taylor S V, McLafferty F W. Curr Opin Chem Biol. 1999;3:623–629. doi: 10.1016/s1367-5931(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 3.Begley T P, Downs D M, Ealick S E, McLafferty F W, Van Loon A P G M, Taylor S V, Campobasso N, Chiu H, Kinsland C, et al. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 4.Spenser I D, White R L. Angew Chem Int Ed Engl. 1997;36:1032–1046. [Google Scholar]
- 5.Taylor S V, Kelleher N L, Kinsland C, Chiu H, Costello C A, Backstrom A D, McLafferty F W, Begley T P. J Biol Chem. 1998;273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 6.Kinsland C, Taylor S V, Kelleher N L, McLafferty F W, Begley T P. Protein Sci. 1998;7:1839–1842. doi: 10.1002/pro.5560070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Xi J, Begley T P, Nicholson L K. Nat Struct Biol. 2001;8:47–51. doi: 10.1038/83041. [DOI] [PubMed] [Google Scholar]
- 8.Lauhon C T, Kambampati R. J Biol Chem. 2000;275:20096–20103. doi: 10.1074/jbc.M002680200. [DOI] [PubMed] [Google Scholar]
- 9.Kambampati R, Lauhon C T. Biochemistry. 1999;38:16561–16568. doi: 10.1021/bi991119r. [DOI] [PubMed] [Google Scholar]
- 10.Kambampati R, Lauhon C T. J Biol Chem. 2000;275:10727–10730. doi: 10.1074/jbc.275.15.10727. [DOI] [PubMed] [Google Scholar]
- 11.Mueller E G, Palenchar P M. Protein Sci. 1999;8:2424–2427. doi: 10.1110/ps.8.11.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palenchar P M, Buck C J, Cheng H, Larson T J, Mueller E G. J Biol Chem. 2000;275:8283–8286. doi: 10.1074/jbc.275.12.8283. [DOI] [PubMed] [Google Scholar]
- 13.Vierstra R D, Callis J. Plant Mol Biol. 1999;41:435–442. doi: 10.1023/a:1006323317890. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Genes Dev. 1998;149:677–692. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 15.Jentsch S, Pyrowolakis G. Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa K, Mizushima N, Noda T, Ohsumi Y. J Biol Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 17.Hochstrasser M. Nat Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 18.Schaegger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Costello C A. Dissertation. Ithaca, NY: Cornell University; 1996. [Google Scholar]
- 22.Backstrom A D. Dissertation. Ithaca, NY: Cornell University; 1996. [Google Scholar]
- 23.Vander Horn P B, Backstrom A D, Stewart V, Begley T P. J Bacteriol. 1993;175:982–992. doi: 10.1128/jb.175.4.982-992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beu S C, Senko M W, Quinn J P, Wampler F M, III, McLafferty F W. J Am Soc Mass Spectrom. 1993;4:557–565. doi: 10.1016/1044-0305(93)85017-R. [DOI] [PubMed] [Google Scholar]
- 25.Horn D M, Zubarev R A, McLafferty F W. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 26.Loo J A, Udseth H R, Smith R D. Rapid Commun Mass Spectrom. 1988;2:207–210. [Google Scholar]
- 27.Horn D M, Ge Y, McLafferty F W. Anal Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 28.Ge, Y., Horn, D. M. & McLafferty, F. W. (2001) Int. J. Mass Spectrom., in press.
- 29.Marshall A G, Wang T, Chin L, Ricca T L. J Am Chem Soc. 1985;107:7893–7897. [Google Scholar]
- 30.Little D P, Speir J P, Senko M W, O'Connor P B, McLafferty F W. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 31.Speir J P, Senko M W, Little D P, Loo J A, McLafferty F W. J Mass Spectrom. 1995;30:39–42. [Google Scholar]
- 32.Robert J, Anouti M, Paris J. J Chem Soc Perkin Trans. 1997;2 3:473–478. [Google Scholar]
- 33.Liu C-F, Rao C, Tam J P. Tetrahedron Lett. 1996;37:933–936. [Google Scholar]
- 34.Hochstrasser M. Science. 2000;289:563–564. doi: 10.1126/science.289.5479.563. [DOI] [PubMed] [Google Scholar]
- 35.Rudolph M J, Wuebbens M M, Rajagopalan K V, Schindelin H. Nat Struct Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 36.Tazaki M. Phosphorus Sulfur Silicon Relat Elem. 1999;153–154:419–420. [Google Scholar]
- 37.de Kok A, Hengeveld A F, Martin A, Westphal A H. Biochim Biophys Acta. 1998;1385:353–366. doi: 10.1016/s0167-4838(98)00079-x. [DOI] [PubMed] [Google Scholar]
- 38.Motokawa Y, Fujiwara K, Okamura-Ikeda K. Antioxid Health Dis. 1995;2:389–407. [Google Scholar]
- 39.Patel M S, Smith R L. In: The Evolution of Antioxidants in Modern Medicine. Schmidt K, Diplock A T, Ulrich H, editors. Stuttgart, Germany: Hippokrates; 1994. pp. 65–77. [Google Scholar]
- 40.Stauffer G V. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 506–513. [Google Scholar]
- 41.White R H. Biochim Biophys Acta. 1979;583:55–62. doi: 10.1016/0304-4165(79)90309-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.