Abstract
Targeting dysregulated signaling pathways in tumors has led to the development of a novel class of signal transduction inhibitors, including inhibitors of the epidermal growth factor (EGF) receptor (EGFR). To dissect oncogenic pathways, identify key pathway determinants, and evaluate the efficacy of targeted agents, it is vital to develop technologies that allow the detection of temporal signaling events under physiological conditions. Here we report the application of a label-free optical biosensor to reveal the rapid response of cancer cells to EGF, expressed as a dynamic mass redistribution (DMR) signal. In response to EGF, squamous cell carcinoma of the head and neck cells exhibited a rapid rise in DMR signal, whereas lung adenocarcinoma cells showed a biphasic DMR profile, suggesting a cell type-dependent DMR response. Pharmacological studies suggested the importance of EGFR and the phosphatidylinositol-3 kinase pathway in mediating the EGF-induced DMR response. The defined DMR signatures offer a simple yet sensitive tool for evaluating EGFR-targeted agents, as shown with gefitinib and erlotinib. The assay can also be used for cell-based high-throughput screening of EGF pathway inhibitors, as demonstrated by its robust performance in a 384-well plate format (Z′ > 0.5). This technology is applicable to other oncogenic pathways for the discovery of novel therapeutic agents for the treatment of various cancers.
Keywords: Label-free, epidermal growth factor (EGF) receptor, dynamic mass redistribution (DMR), oncogenic pathway, high-throughput screening
Introduction
Cancer development and progression have been correlated with dysregulated intracellular signaling pathways that control cell proliferation, immortalization, apoptosis, invasion, and angiogenesis (1). Better understanding of growth factor-mediated signaling pathways and their roles in tumorigenesis is vital for the discovery of new therapeutic strategies and for the effective use of existing anticancer therapies. Among these pathways, epidermal growth factor receptor (EGFR) signaling has been found to be dysregulated in a variety of human diseases, such as cancer, including head and neck cancer and lung cancer.
EGFR is a transmembrane tyrosine kinase receptor and belongs to the ErbB family of receptor tyrosine kinases (RTK) (2,3). This receptor family comprises four related proteins: EGFR (HER1/ErbB1), ErbB2 (HER2), ErbB3 (HER3), and ErbB4 (HER4) (2,4,5). Upon binding to its ligand, such as the epidermal growth factor (EGF), EGFR undergoes a conformational change that allows activation by homo- or hetero-dimerization with other ErbB family receptors. Activation of EGFR results in the recruitment and phosphorylation of a range of adaptor proteins (e.g., Shc or Grb2) and activates a network of intracellular signaling pathways. The major EGFR signal transduction pathways in various cell types involve the Ras/Raf mitogen-activated protein kinase (MAPK) cascade (6), phosphatidylinositol 3-kinase (PI3K) and its downstream protein-serine/threonine kinase AKT (7,8), signal transducers and activators of transcription (STATs), and downstream protein kinase C (PKC) (9,10). Because of their vital importance in regulating cell growth and maintaining cell integrity, EGFR and/or the downstream signaling regulators are subject to frequent oncogenic alterations that have been correlated with tumorigenesis and tumor resistance to therapies. Therefore, EGFR and its signaling pathways have become valuable targets for therapeutic interventions for the treatment of cancer.
Patients with head and neck cancer and non-small cell lung cancer (NSCLC) often have only very limited treatment options with a poor prognosis (11–13). Recent studies have identified frequently overexpressed and/or abnormally activated EGFR in these patients, which has led to the development of EGFR-targeted therapies (14–18). Although there are several strategies for targeting EGFR, such as monoclonal antibodies against EGFR and selective and reversible or irreversible small molecule tyrosine kinase inhibitors (TKIs), only a small subset of patients respond to drug treatment and drug resistance often occurs (11,17,18). Thus, alternative strategies are highly desired. Innovative technologies are demanded for effectively dissecting oncogenic signaling pathways and their targeting for drug discovery. In this study, we examined the use of a label-free optical biosensor system for monitoring EGFR signaling in cancer cells. This technology offers distinct advantages over many conventional methods for signaling pathway detection. To enhance detection sensitivity and selectivity, conventional methods often require artificially manipulated cell lines, such as fluorophore-labeled or overexpressed targets or a reporter system, and generally measure a single cellular event. Label-free technologies allow the detection of signaling events under physiological conditions without the need for cell line manipulations. Among a variety of label-free technologies, such as surface plasmon resonance (SPR), and impedance (19), the use of optical biosensors in cell-based assays is gaining increased attention because of recent advances in instrumentation and experimental design (20).
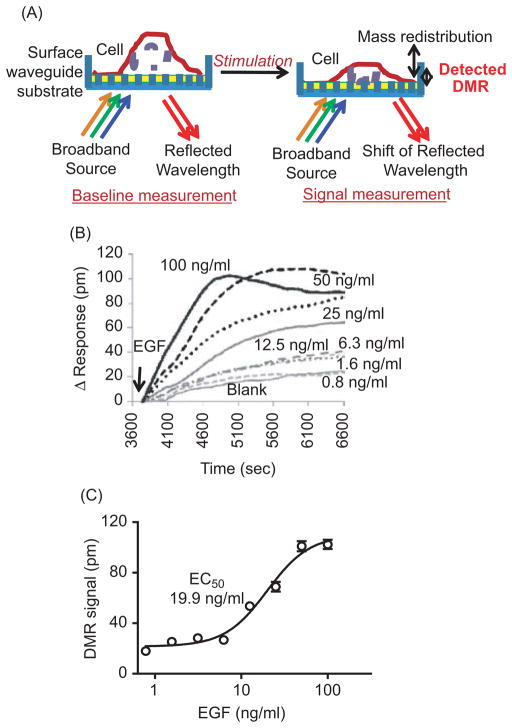
The Epic biosensor system is an optical biosensor that uses the resonant-waveguide grating (RWG) for monitoring bimolecular interactions (20–22). The principle of the Epic optical biosensor cell-based functional assay for monitoring a ligand-induced dynamic mass redistribution (DMR) signal is illustrated in Figure 1A (21,22). Cells can be placed onto the biosensor surface coated at the bottom of a 384- well microplate. When the biosensor with waveguide grating is illuminated with a broadband light source, only a “single” wavelength is resonant and reflected by the waveguide grating. The reflected wavelength is recorded by the Epic reader and reported in picometer (pm) units. Upon ligand stimulation, the molecular or cell mass near the bottom of the biosensor will be changed or redistributed, which leads to a change in the index of the refraction at the sensor surface. When the same well is illuminated with the broadband light source again, the reflected wavelength shifts compared to that without stimulation. The shift of the reflected wavelength after ligand stimulation can be detected and monitored in real time by the Epic reader. The reflected wavelength difference before (baseline measurement) and after ligand stimulation (signal measurement) is defined as the dynamic mass redistribution (DMR) signal (21,22). Because the energy of the broadband source decays over distance after penetrating the cells, only a change in mass within a 150-nm range from the biosensor surface can be detected. Therefore, only partial changes in cellular events at the bottom portion of the cells contribute to the measured DMR signal.
Figure 1.
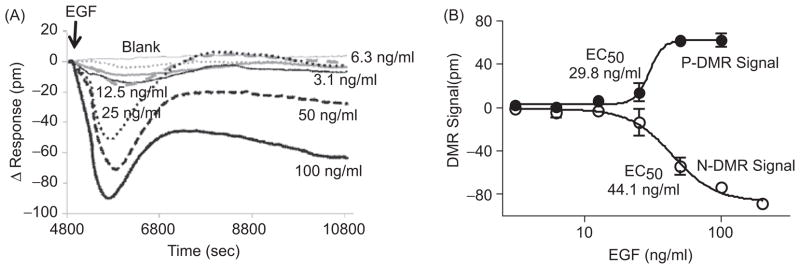
Characterization of the DMR response induced by EGF in UPCI-37B SCCHN cancer cells. (A) Principles of cell-based signature assay using the Epic optical biosensor system, which measures changes in the local index of refraction resulting from a ligand-induced dynamic mass redistribution (DMR) of the cell monolayer, as indicated by a shift in resonant wavelength. The shift in resonant wavelength is detected by the Epic reader. The reported unit for the DMR signal from the Epic reader is picometers (pm) and is used throughout this study. (B) Real-time DMR response of serum-starved (24 hr) UPCI-37B cells to increasing concentrations of EGF stimulation. The solid arrow indicates the time of EGF addition. (C) Δ Response after 20 min EGF addition calculated from the graph (B) as described and plotted against EGF concentrations. The EC50 value was analyzed by using Prism 4.0 software.
The Epic cell-based signature assay does not require label or manipulation of the cellular targets. The measured DMR signal is an integrated cellular response to a particular environmental stimulus. To develop the next generation of EGFR pathway targeted agents for the treatment of cancer, we established EGF-induced DMR response profiles for both squamous cell carcinoma of the head and neck (SCCHN) and lung adenocarcinoma cells with the use of the Epic biosensor system. We further revealed the signaling pathways that contribute to the ligand-induced DMR response and evaluated the potential applications of this technology for high-throughput screening of pathway-based anticancer agents.
Materials and methods
Reagents
EGF was purchased from Sigma Chemical Co. (St. Louis, MO). Gefitinib (Iressa), erlotinib (Tarceva), SU 1498, PD98059, Wortmannin, U0126, and LY 294002 were purchased from LC Laboratories (Woburn, MA). Corning Epic 384-well fibronectin-coated biosensor cell assay microplates (cat. 5042) were obtained from Corning Inc. (Corning, NY).
Cell lines and cell culture
The squamous cell carcinoma of the head and neck (SCCHN) cell line UPCI-37B was a gift from Dr. Theresa Whiteside (University of Pittsburgh Cancer Institute). The cells were grown in DMEM/F12 medium (1:1) supplemented with 10% fetal bovine serum (FBS). The lung adenocarcinoma cancer cell line A549 was obtained from the American Type Cell Culture and maintained in RPMI1640 medium supplemented with 10% FBS. All cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Cell plate setup for Epic assay
The cells were resuspended in the medium with 10% FBS and 40 μL of cells containing 10,000 cells were dispensed to each well of a 384-well biosensor plate by using a Multidrop Combi dispenser (Thermo-Fisher Scientific). The dispense speed was set at high to avoid the generation of bubbles. The plates were incubated at 37°C in a humidified atmosphere with 5% CO2 for 24 hr. The growth medium was aspired by using a BioTeck Washer. To minimize the stress conditions introduced by washing cells, the plate washer was carefully calibrated and set up so that 10 μL of the liquid was left at the plate bottom after aspiration. Then, 50 μL of medium without FBS was added to each well with use of the Multidrop Combi dispenser. The process was repeated four times to exchange the growth medium containing 10% FBS to medium without FBS. The plates were put back into the incubator and subject to 24 hr serum starvation before the Epic assay.
After serum starvation for 24 hr, the medium was aspired by using a BioTek washer as described above and 50 μL of assay buffer [Hanks’ Balanced Salt Solution (HBSS) with 20 mM HEPES] was added by using a Multidrop Combi. After two cycles of assay buffer exchange, the plates were sent to the Corning Epic microplate reader (Corning, NY).
Epic biosensor measurement of DMR signal
All Epic measurements were carried out kinetically for each well with 1-min interval between each reading. The Epic biosensors are thermally sensitive; any difference in temperature could complicate the signal response to stimuli. The Corning Epic microplate reader (Corning, NY) has a temperature-controlled environment with stackers inside that can hold 20 plates. After 24 hr of serum starvation and buffer exchange to assay buffer, the cell plates were thermo- equilibrated inside the instrument for 1 hr before the baseline read was initiated. After 10 min of baseline measurement, different concentrations of EGF or assay buffer were added to the wells by using a 384-channel low-volume head Sciclone liquid handler (Caliper LifeSciences) to ensure that the reagent for each well was added simultaneously at each time point. The plate was immediately sent back to the Epic reader for continuous measurement for 45 min, or as indicated.
Epic biosensor measurement of the effect of compound on DMR signal
The Epic cell assay plate was prepared as described above, sent to the Epic reader, and thermo-equilibrated for 1 hr. After 10-min baseline reading, the compound or vehicle was added by using a Sciclone liquid handler with 384-channel low-volume head. All the compounds were dissolved in DMSO as 10 mM stock and diluted by using assay buffer. The final DMSO concentrations were 1%. After compound addition, the plate was immediately sent back to the Epic reader for continuous measurement. After incubating/reading with compound or vehicle for 30 min, EGF or assay buffer as background control was added by using the Sciclone. The plate was then continuously monitored by the Epic reader for 45 min or as indicated.
Epic data collection and analysis
The response signal from each well of the biosensor plate was individually tracked and measured kinetically. The dynamic mass redistribution (DMR) signal of each time point was detected by the Epic reader and reported as a shift in pm (recorded pm) relative to an initial measurement. This value indicates the pm shift of the reflected wavelength after subtracting the measured value at time 0 (the first reading of the baseline measurement). The Epic data were collected and converted to a Microsoft Excel (Microsoft Corporation, Redmond, WA) format for further analysis.
To minimize well-well variation in baseline measurements, the EGF-induced DMR signal for each well is expressed as Δ Response and calculated for each well at each time point according to the following equation:
The response curves were plotted on the basis of the Δ Response (pm) vs. time [second (sec)] throughout this study.
The DMR signal collected kinetically by the Epic reader after ligand stimulation can be defined and described according to the shape of the response curves. An increase in the response curve is termed a positive DMR (P-DMR) event, and a decrease in the response curve is referred to as a negative DMR (N-DMR) event.
The DMR signal (P-DMR or N-DMR) in pm units is calculated on the basis of the sequence of the events induced by ligand stimulation:
For initial P-DMR or N-DMR event after EGF addition:
For the secondary DMR event:
The effect of compound treatment on the ligand-induced DMR response is expressed as % of Control and defined as the following equation:
DMR signalblank is defined as the DMR signal from the wells with vehicle (DMSO) only without EGF; DMR signal compound is the DMR signal from wells with EGF in the presence of compound; and DMR signalEGF is the DMR signal from wells with EGF in the presence of vehicle (DMSO) without compound.
All the experiments were performed at least in triplicates and repeated at least three times with similar results. The representative response curves are shown. All experimental data were analyzed by using Microsoft Excel (Microsoft Corporation, Redmond, WA) and Prism 4.0 (Graphpad Software, San Diego, CA).
Assay evaluation for high-throughput screening (HTS)
To evaluate the suitability of the Epic assay performance for HTS, the signal-to-background ratio (S/B) and the Z′ factor were calculated on the basis of the following equations (23):
The Z′ factor reflects the quality of the assay itself for HTS without the intervention of test compounds. The assay is considered robust and suitable for HTS when the Z′ value is between 0.5 and 1.
Results
EGF induces a unique optical DMR response signature in SCCHN cells
We chose the UPCI-37B SCCHN cell line as a model system to characterize and profile the EGF-induced DMR signal in living cells using the Epic biosensor. We first examined EGF dose responses. After serum starvation for 24 hr, increasing concentrations of EGF were introduced to the cells, and the DMR response was recorded in real time. As shown in Figure 1B, EGF stimulation triggered a DMR response in serum-starved UPCI-37B SCCHN cells. The addition of assay buffer alone as a treatment control generated only a slight increase in the DMR signal. Upon EGF addition, there was a rapid increase in the detected DMR response. The amplitude of the recorded pm values increased dose dependently with increasing concentrations of EGF. The increase in the measured DMR response is referred to as a positive DMR (P-DMR) event (24). The DMR response could be saturated, and the P-DMR signal, which is defined by the maximum difference in the recorded pm after EGF addition, was dose dependent on EGF concentration. The time to reach the maximum DMR signal was inversely correlated with the amount of EGF: The greater the EGF concentration, the shorter the time to reach a saturated DMR. After reaching the maximum, the DMR response curves showed only slight differences with increasing concentrations of EGF. Higher concentrations of EGF (50 and 100 ng/mL) resulted in a slight decrease in the DMR signal over time. The decay of the response was EGF dose-dependent. When the decay event occurred at higher concentrations of EGF (e.g., 100 ng/mL), the DMR response was approaching the maximum for the lower concentrations of EGF. These data suggest that EGF induces an Epic optical biosensor DMR profile in SCCHN cells that is distinct from that previously reported in A431 cells (24). This study defines a representative DMR signature of one SCCHN cell line in response to EGF.
The dose-response curve of the EGF-induced DMR signal was then evaluated. The DMR signal after EGF addition for 20 min was calculated and plotted against EGF concentration. To simplify the data analysis, the response after 20 min EGF addition was chosen because, at this time point, the maximum response was seen at the highest concentration of EGF (100 ng/mL). As shown in Figure 1C, the DMR signal dose dependently increased with increasing concentrations of EGF. The dose-response curve fitted well with a nonlinear regression with R2 of 0.97. The resulting EC50 was 19.9 ng/mL, which is similar to the reported EC50 in A431 cells (24). These results indicate that the EGF-induced DMR signal in SCCHN cells is dependent on EGFR activation.
The EGF-induced DMR optical signature in SCCHN cells requires EGFR
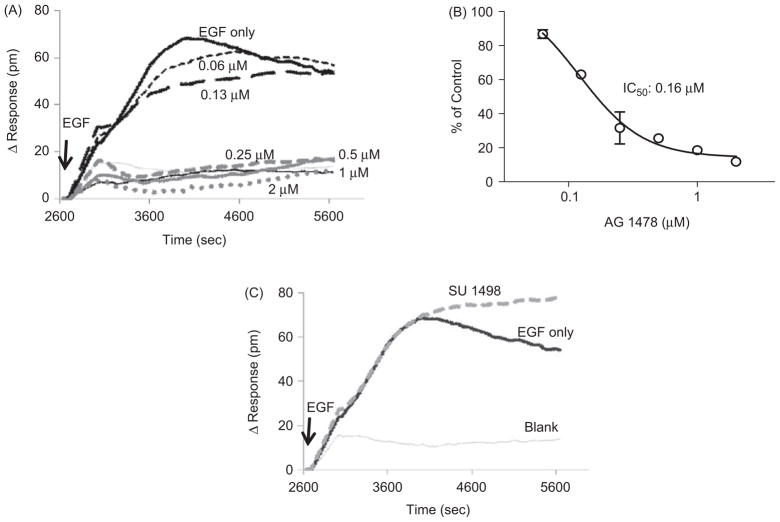
To address whether the DMR signal induced by EGF occurs via the EGFR pathway, we evaluated the effect of an EGFR tyrosine kinase inhibitor, AG 1478, on the EGF-induced DMR response. The vascular endothelial growth factor (VEGF) receptor tyrosine kinase inhibitor, SU 1498, was used as a specificity control. This compound is a known weak inhibitor of EGFR-kinase and Her-2 kinase (25). Serum-starved UPCI-37B cells were pretreated with increasing concentrations of AG 1478 for 30 min before the addition of EGF. The DMR signals were continuously monitored for another 45 min. As shown in Figure 2A, preincubation with AG 1478 resulted in a delayed and decreased DMR signal compared to the vehicle control. The DMR response was dose dependently attenuated with increasing concentrations of the inhibitor, leading to a complete blockage at 0.5 μM. The inhibitory effect of AG 1478 on EGF-induced P-DMR signal was normalized to the control wells with EGF only, which was plotted against inhibitor concentrations to give an estimated IC50 value of 0.16 μM (Fig. 2B). The obtained IC50 value in UPCI-37B cells is consistent with the previously reported value obtained in A431 cells (24), even though the DMR signal signature was different. In support of a specific EGFR-mediated event, the VEGFR inhibitor SU 1498 did not attenuate the EGF-induced DMR signal (Fig. 2C). These data show that the EGF-induced DMR signals in SCCHN cells are dependent on EGFR tyrosine kinase activity and are specific to EGFR activation.
Figure 2.
Evaluation of the specificity of the DMR signal induced by EGF in UPCI-37B cells. (A) Effect of the tyrosine kinase inhibitor AG 1478 on the EGF-induced DMR signal. The cells were preincubated with increasing concentrations of AG 1478 before introducing EGF (100 ng/mL). The real-time DMR response was continuously monitored for 45 min. (B) Dose-response curve of the AG 1478 effect on EGF-induced DMR response. The inhibitory effects of AG 1478 on the P-DMR signal induced by EGF were normalized to the vehicle control wells in the presence of EGF but without compound. The IC50 was calculated from the dose-response curve using Prism 4.0 software. (C) The effect of SU 1498 (10 μM), a VEGFR inhibitor as a negative control, on the DMR response induced by EGF (100 ng/mL).
PI3K is a critical mediator of EGF-induced DMR in SCCHN cells
To investigate pathways that contribute to the EGF-induced DMR signal in SCCHN cells, we tested a panel of small molecule inhibitors of different targets known to be downstream of EGF ligand binding in various cells. The pharmacological properties of the inhibitors used are summarized in Table 1. The concentrations of each inhibitor were chosen to achieve the maximum specific inhibition based on the EC50 values reported in the literature.
Table 1.
The properties of pathway inhibitors and their effects on EGF-induced DMR signal in UPCI-37B SCCHN cells.
| Inhibitor | Properties | % of Control DMR signal |
|---|---|---|
| SU1498 | Potent VEGF receptor TKI, very weak EGFR TKI and HER-2 TKI (25) | 98.6 |
| U0126 | Selective inhibitor of the MAPK kinases, MEK-1 and MEK-2, with 100-fold greater potency than PD98059 (35–37). | 103.6 |
| PD98059 | Potent, selective and cell-permeable inhibitor of MAPK kinase (38–40). | 104.6 |
| Wortmannin | Potent, irreversible, and specific inhibitor of PI3K (41–43). | 16.2 |
| LY 294002 | Specific inhibitor of PI3K (44). | 30.4 |
VEGF, vascular endothelial growth factor.
TKI, tyrosine kinase inhibitor.
MAPK, mitogen-activated protein kinase.
PI3K, phosphatidylinositol 3-kinase.
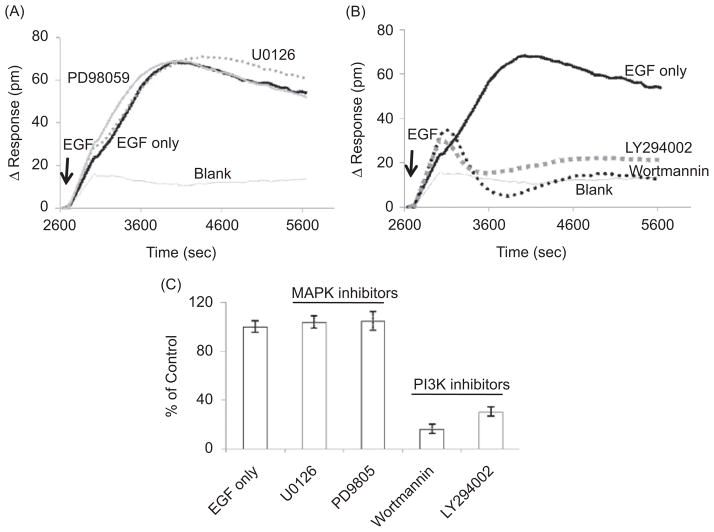
First, we examined the role of the Ras/MAPK pathway on EGF-induced DMR response. The Ras/MAPK pathway transmits EGF signaling in many cell types (10,26). For this purpose, the MEK1/MEK2 inhibitors U0126 and PD98059 were used. It was surprising that neither U0126 (1 μM) nor PD98059 (10 μM) showed any inhibition of the EGF-induced DMR signal in UPCI-37B cells (Fig. 3A). These results indicate that EGF-induced cellular response detected by the Epic biosensor in SCCHN cells may not be mediated by the activation of the Ras/MEK1/2/Erk1/2 MAPK pathway.
Figure 3.
Probing the pathways and mechanisms involved in the EGF-mediated DMR signal in UPCI-37B SCCHN cells. A panel of specific inhibitors targeting two major EGF downstream pathways, MAPK and PI3K, were evaluated. (A) The effect of MAPK pathway inhibitors on the EGF-induced P-DMR signal. The MAPK inhibitor U0126 (1 μM) and PD98059 (10 μM) did not attenuate the EGF-induced DMR signal. (B) The effect of PI3K pathway inhibitors on the EGF-induced DMR signal. Pretreatment with PI3K inhibitors LY 294002 (25 μM) and wortmannin (125 nM) completely blocked the DMR signal in response to EGF. (C) The inhibitory effects of the pathway inhibitors on the P-DMR signal were normalized to the EGF vehicle control wells without compound and summarized; 100 ng/mL EGF was used for all experiments.
We then evaluated the involvement of another major downstream EGF signaling pathway, the PI3K pathway, in EGF-induced DMR signaling. The PI3K kinase inhibitors LY 294002 and wortmannin were incubated with UPCI-37B cells for 30 min before EGF addition. It was interesting that both LY 294002 (25 μM) and wortmannin (0.13 μM) significantly blocked the EGF-induced DMR signal (Fig. 3B). These data indicate that the EGF-induced DMR signal in UPCI-37B SCCHN cells is primarily mediated by the PI3K pathway as detected by the Epic optical biosensor. In contrast, the EGF-induced DMR signal in A431 cells was determined by the MAPK pathway (24); thus, our data suggest that the downstream cellular events in response to growth factor stimulation might be cell line and cell type specific. The Epic biosensor may be used as a novel tool for cell line characterization and for profiling specific receptor signaling pathways.
The normalized data showing the effect of the tested panel of specific pathway inhibitors are summarized in Figure 3C and Table 1. Taken together, these data suggest that the EGF-induced DMR signal in UPCI-37B SCCHN cells was primarily due to the activation of PI3K pathway, illustrating a cell type-specific DMR response and supporting the use of Epic biosensor pathway-based drug discovery.
The EGF-induced DMR signal is cell line and cell type specific: the unique DMR signature of the adenocarcinoma NSCLC A549 cell line in response to EGF
To test whether different cancer cells have a distinct DMR response to EGF, we tested EGF-induced signaling in an additional cancer cell line, adenocarcinoma NSCLC A549 cells. It was interesting that the A549 lung cancer cells displayed a totally different DMR response to EGF than that in the SCCHN cancer cells (Fig. 4A). EGF stimulation led to an rapid initial decrease in the detected DMR signal, termed a negative DMR (N-DMR) event (24). After reaching the maximum N-DMR signal (the bottom of the curve), the DMR signal increased, leading to a positive DMR (P-DMR) event. The maximum N-DMR signal was about −70 pm, whereas the maximum calculated P-DMR signal was about 50 pm. The P-DMR event occurred more slowly than the N-DMR event and slightly decayed over the measurement time peroid (1 hr).
Figure 4.
Characterization of the optical signatures of the EGF-induced DMR response in the A549 human lung cancer cell line. (A) EGF induced a unique DMR response in A549 cells. It contained two DMR events: an initial decrease in signal (N-DMR) followed by an increase in signal (P-DMR). The dose-response of the EGF-induced DMR signal was evaluated. Both N-DMR and P-DMR signals increased dose dependently with increasing concentrations of EGF. (B) The DMR signals were calculated for N-DMR and P-DMR events and plotted against EGF concentrations. The EC50 values were calculated by using Prism 4.0 software.
Because the DMR response of A549 lung cancer cells to EGF is completely different from that of UPCI-37B SCCHN cells, we examined whether such a DMR profile is EGF-dose dependent in A549 cells. As shown in Figure 4A, stimulation with increasing concentrations of EGF led to increases in both the N-DMR and P-DMR signals. The EC50 values were 44.1 ng/mL and 29.8 ng/mL for the N-DMR signal and P-DMR signal, respectively (Fig. 4B).
The unique DMR signature of A549 cells in response to EGF stimulation is consistent with the fact that the pathways and mechanisms involved in the regulation of cell signaling are cell type specific. These studies suggest that the DMR response of cells to EGF monitored by the Epic biosensor may serve as a promising approach for cell line characterization and for monitoring oncogenic signaling pathways.
Using the established DMR signature to evaluate the efficacy of known EGFR targeting drugs
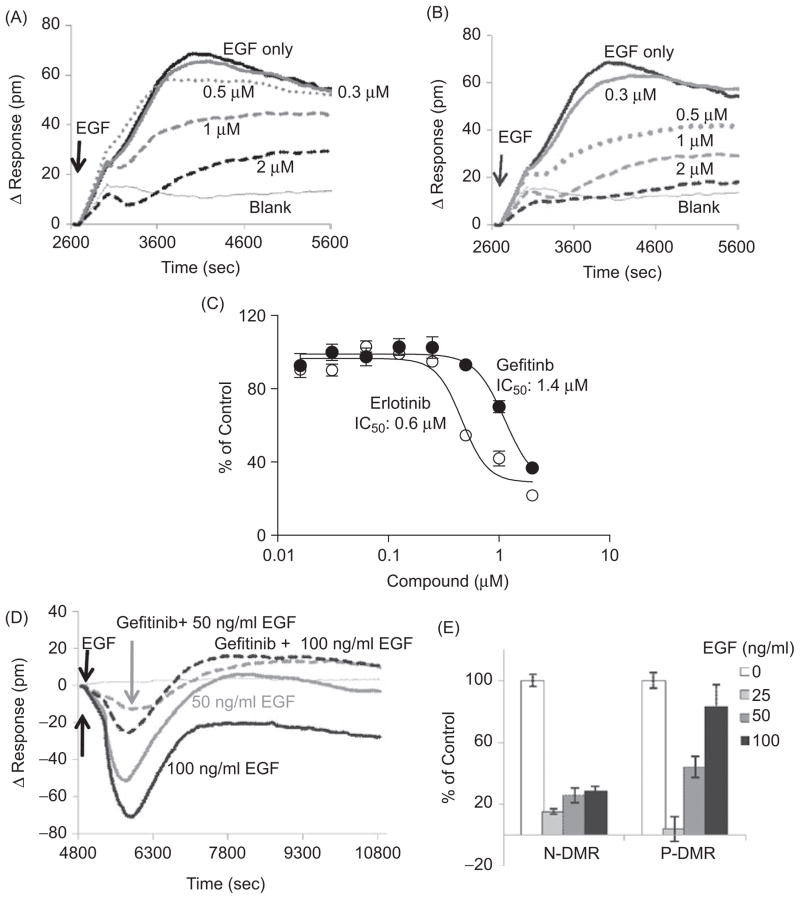
EGFR is frequently activated in SCCHN, lung, and other cancer types and is a validated cancer target. To further validate the above results and to explore the potential use of the Epic biosensor for the discovery of new EGFR signaling-targeted agents, we tested the effect of two small molecule EGFR kinase inhibitor drugs, gefitinib (Iressa) and erlotinib (Tarceva), on the EGF-induced DMR signal. Gefitinib and erlotinib have been approved by the FDA for the treatment of locally advanced or metastatic non-small cell lung cancer (NSCLC) since May 2003 and November 2004, respectively. UPCI-37B SCCHN cells were preincubated with gefitinib, erlotinib, or vehicle before EGF (100 ng/mL) stimulation. The EGF-induced DMR signal was then monitored continuously for 45 min. Treatment with gefitinib showed significant attenuation of the DMR response compared with that in vehicle-treated control wells (Fig. 5A). The inhibitory effect of gefitinib on EGF-induced DMR was dose dependent, with an estimated IC50 value of 1.42 μM (Fig. 5C). Erlotinib showed a similar inhibitory effect as gefitinib on the EGF-induced DMR signal, with an IC50 of 0.57 μM (Fig. 5B and 5C).
Figure 5.
Validation of the EGF-induced DMR signature assay using gefitinib and erlotinib, two FDA approved drugs targeting EGFR. (A) The effect of gefitinib on EGF-induced DMR response in UPCI-37B SCCHN cancer cells. Increasing concentrations of gefitinib were incubated with the serum-starved cells for 30 min before EGF (100 ng/mL) addition. The DMR signals were then monitored kinetically for 45 min. The real-time DMR response was attenuated dose dependently in the presence of gefitinib. (B) The effect of erlotinib on EGF-induced DMR response in UPCI- 37B SCCHN cancer cells was similar to that of gefitinib. (C) The dose-response curves of gefitinib and erlotinib effects on the P-DMR signal in UPCI-37B cells. The inhibitory effects of gefitinib and erlotinib were normalized to the EGF vehicle control wells without compound, and the IC50 values were calculated by using Prism 4.0 software. (D) The effect of 1 μM gefitinib on DMR response induced by increasing concentrations of EGF in A549 lung cancer cells. (E) The normalized inhibition effect of gefitinib (1 μM) on N-DMR and P-DMR signal in A549 lung cancer cells.
We then examined the effect of these EGFR-targeted drugs in A549 cells that have a distinct DMR response profile. Pretreatment of A549 cells with gefitinib (1 μM) attenuated both the EGF-induced N-DMR and P-DMR signals (Fig. 5D). However, in contrast to the results in UPCI-37B SCCHN cells, gefitinib treatment did not change the shape of the DMR response curve to EGF in A549 cells. To test the efficiency of gefitinib in blocking the defined EGF-induced DMR signature in A549 cells, we carried out drug dose-response experiments against a series of EGF concentrations. Treatment with gefitinib (1 μM) was able to attenuate both N-DMR and P-DMR signals induced by different concentrations of EGF (Fig. 5D). The normalized inhibitory effects of gefitinib at 1 μM on N-DMR signals induced by different concentrations of EGF (25–100 ng/mL) were similar (Fig. 5E). However, we noticed that the inhibitory effects on P-DMR signals were decreased with increasing concentrations of EGF (Fig. 5E). Despite their differences in DMR response signature profiles, both UPCI-37B and A549 cells exhibit a similar dependence on EGFR activation in response to EGF stimulation, as gefitinib could attenuate the EGF-induced DMR response in both cell lines (Fig. 5A and 5D).
These results show that the Epic biosensor can be used as an effective tool to monitor the efficacy of EGF-targeted therapy, as well as a novel approach to discover new therapeutic agents targeting abnormal pathways involved in human diseases.
Evaluation of the use of EGF-responsive DMR signal profiles for high-throughput screening (HTS) of EGFR pathway inhibitors
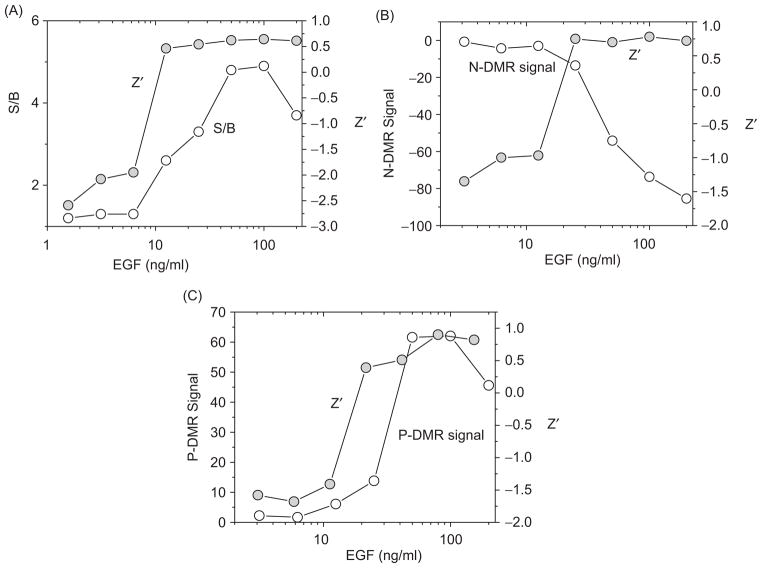
To evaluate the performance and suitability of Epic biosensor assays for HTS of new agents targeting EGF-induced signal transduction pathways, we examined the robustness of the DMR profile assays in a 384-well format. Z′ factors and signal-to-background (S/B) ratios were calculated for the detected P-DMR signal in both UPCI- 37B SCCHN and A549 lung cancer cells. As described above, EGF stimulation led to a single event, P-DMR, detected by the Epic biosensor in UPCI-37B SCCHN cancer cells. Z′ factors and S/B were calculated on the basis of the DMR signal from the wells with increasing concentrations of EGF compared with the buffer only wells. The Z′ factor and S/B ratio both increased with increasing concentrations of EGF (Fig. 6A). At EGF concentrations > 25 ng/mL, the Z′ value rose above 0.5 and the S/B ratio was higher than 3, indicating a robust assay for HTS.
Figure 6.
Evaluation of cell-based optical signature assays for high-throughput screening (HTS). (A) To evaluate the suitability and optimal conditions of the biosensor EGFR signaling assay for HTS, the Z′ factors and signal-to-background ratio (S/B) were calculated and plotted against increasing concentrations of EGF in UPCI-37B SCCHN cells. (B) Z′ factors were calculated for EGF-induced N-DMR and P-DMR signal in A549 cells and plotted along with N-DMR signal against EGF concentrations. The Z′ factor incorporates both the signal window and the well-to-well variations and is a criterion for evaluating the suitability and conditions of the assay for HTS. A Z′ factor between 0.5 and 1 indicates that the assay is robust for screening chemical modulators of the targets.
Unlike UPCI-37B SCCHN cells, A549 lung cancer cells gave rise to an initial N-DMR signal followed by a P-DMR event. Z′ factors were evaluated for both N-DMR and P-DMR signals. Because there was almost no change in DMR signal from background/blank control wells (Δ Response was close to 0 pm), the S/B ratios were not calculated for A549 cells. When stimulated with higher concentrations of EGF (≥50 ng/mL), the Z′ factors were above 0.5 for both N-DMR (Fig. 6B) and P-DMR events (Fig. 6C). The Z′ was above 0.8 when 100 ng/mL of EGF was tested for N-DMR and P-DMR signals. Therefore, the Epic assay offers a new approach for screening compounds targeting abnormal EGFR pathways in both SCCHN and lung cancer cells.
Discussion
Growth factor receptors and their downstream pathways play a crucial role in normal cell growth and differentiation as well as in tumor proliferation and survival. Traditional cell-based assays for motioning growth factor-regulated signaling pathways rely on the measurement of a single cellular event, quite often involving the use of fluorescence-tagged molecules or engineered/overexpressed cellular systems. These assays are time consuming in terms of assay development and are difficult to adapt to a high-throughput format for the discovery of new compounds under physiological conditions. However, cellular function is regulated by an integrated signaling network that involves the interactions of multiple pathways and cellular events. In this report, we have developed and validated a cell-based functional assay for characterizing and monitoring integrated signaling pathways using a novel label-free detection technology—a resonant-waveguide grating-based Epic optical biosensor, which allows the measurement of ligand-induced dynamic mass redistribution (DMR) in living cells under physiological conditions. It does not require fluorescence-tagged molecules or an overexpression system. The resulting DMR signal could be due to many cellular events, such as receptor internalization, recruitment of cellular components to the activated receptors, cell movement, spreading and detachment (24,27); therefore, it measures an integrated cell response to a specific ligand-binding event.
EGFR is a key to the pathogenesis of a variety of cancers, including SCCHN and adenocarcinoma of the lung, and has been extensively studied as a therapeutic target with strong clinical validation. Approximately 90% of cancers of the head and neck are squamous cell carcinomas (28). The EGFR is expressed at much higher levels in SCCHN than in normal epithelial tissue, and EGFR expression correlates with poor prognosis. Non-small cell lung cancer (NSCLC) comprises 75%–85% of newly diagnosed lung cancers (29). EGFR is also frequently overexpressed in NSCLC and has been implicated in the pathogenesis of this disease (14–16). Therefore, much effort is currently directed toward targeting aberrant EGFR signaling for effective drug discovery.
The resonant waveguide grating (RWG) technology, as used in the Corning Epic optical biosensor system, measures the change in cellular dynamic mass redistribution (DMR) near the biosensor surface of the live cells in response to ligand stimulation. The system contains two components: the biosensor-coated microplate for cell growth and the Epic reader for recording the DMR signal. Here, using the EGFR signaling pathway as a model system, we have first characterized the EGF-induced DMR optical signature in SCCHN cancer cells using the Epic biosensor system. The EGF-induced DMR signal has been well established in epidermoid carcinoma A431 cells (24). However, there were no reports characterizing integrated cell signaling using this label-free technology in other types of cancer cells. Therefore, it was unclear whether different cells may have distinct DMR optical signatures in response to a specific ligand.
Many cellular events contribute to the detected DMR signal after ligand stimulation. The resulting DMR events can be classified into two classes: P-DMR and N-DMR (18,22,24). We found that the optical signature in response to EGF is different and unique in UPCI-37B SCCHN cells compared with that in A431 cells (24). Unlike A431 cells, which underwent both P-DMR and N-DMR events, EGF stimulation in serum-starved UPCI- 37B cells led to only a single P-DMR event (Fig. 1B). The detected P-DMR signal was dose and time dependent on EGF. The use of the specific tyrosine kinase inhibitor AG 1478 further validated the EGFR-mediated DMR signal and supports the importance of EGFR tyrosine kinase activity in transmitting EGF-induced cellular signaling. These data suggest that distinct cellular events in different cell types may contribute to EGF-induced DMR signals.
There are two major well-characterized signaling pathways downstream of EGFR: the Ras/Raf/MAPK pathway and the PI3K/Akt pathway. To elucidate the mechanism underlying the unique optical signature observed in UPCI-37B SCCHN cells in response to EGF and to investigate the molecular events involved in the DMR event, we evaluated a panel of specific inhibitors targeting these two major EGFR downstream pathways. First, the Ras/Raf/MAPK pathway was evaluated by using the MEK inhibitors PD98059 and U0126. Our results showed that neither PD98059 nor U0126 could attenuate the DMR signal. These results are different from those obtained in A431 SCCHN cells, in which the EGF-induced DMR is primarily mediated by the Raf/MEK pathway (24). In UPCI-37B SCCHN cells, on the other hand, inhibition of the Ras/Raf/MEK/Erk pathway had little effect on the EGF-induced DMR, supporting the notion that pathways other than the MAPK mediate the EGF-induced signaling network. Indeed, LY 294002 and wortmannin, two PI3K inhibitors, completely blocked the EGF-induced DMR signal (Fig. 3B), indicating that the PI3K/Akt pathway plays an important role in EGF-induced DMR response in these cells. P13K inhibitors did not attenuate the initial DMR signal, indicating that the P13K activation is the downstream even of EGFR activation induced by EGD binding. Therefore, the DMR response to a specific ligand appears to be cell line and cell type dependent. The different mechanisms involved in the DMR response may explain the difference of the DMR response profiles among different cell lines. The unique DMR response to a specific ligand can be used for monitoring oncogenic pathways and for cell line characterization.
We also noticed that different cells have distinct DMR responses to a specific ligand. UPCI-37B SCCHN cells displayed only a P-DMR event, whereas A549 cells contained two DMR events: an initial rapid decrease in signal (N-DMR) followed by a P-DMR (Fig. 4A). Although both A549 and A431 cells give rise to two DMR events, the sequence of their appearance was opposite: A431 cells showed an initial P-DMR followed by an N-DMR event (24). These results suggest that the molecular events in A549 cells in response to EGF are different from those in UPCI-37B or A431 cells. Further investigations are needed to elucidate the mechanisms involved in DMR signal response.
Given the prominent importance of EGFR signaling in cancer development, targeting EGFR represents a new paradigm in cancer therapy and has shown therapeutic promise in a wide range of tumor types, including SCCHN and NSCLC. Two highly selective and reversible tyrosine kinase inhibitors (TKIs), gefitinib and erlotinib, have been approved by the FDA for the treatment of NSCLC and are under phase II clinical evaluation for use in SCCHN. However, the clinical benefit of these TKIs is limited as only a subset of patients respond to drug treatment and drug resistance frequently develops in the responsive patients. Furthermore, gefitinib, erlotinib, and most EGFR TKIs currently under development target the EGFR ATP binding site. Thus, to screen for new and novel EGFR and EGFR pathway inhibitors, alternative strategies for monitoring the integrated signal pathways of EGFR are highly desirable. To evaluate whether the Epic biosensor for monitoring integrated cellular events under physiological conditions can be used as such an alternative assay, we evaluated the effects of gefitinib and erlotinib on the EGF-induced DMR signal in SCCHN cells. Both gefitinib and erlotinib blocked the DMR signal in a dose-dependent manner (Fig. 5). The resulting IC50 values for gefitinib (1.4 μM) and erlotinib (0.6 μM) are in a similar range as those obtained from cell-based assays, such as a growth inhibition assay (30), which are higher than the IC50 values obtained in direct kinase assays for EGFR phosphorylation (low nM range) (31,32). Because the DMR signal measures an integrated cellular response rather than a single cellular event, such as EGFR phosphorylation, such an assay allows the evaluation of pathway modulators. For the discovery of novel EGF pathway-targeting agents, the suitability of the assay for HTS was explored. Conditions for a robust assay performance with Z′ above 0.5 were determined. Therefore, EGFR pathway assays using the Epic biosensor provide a novel approach for clinical efficacy evaluation and for screening new classes of inhibitors targeting EGFR, or EGF-responsive signaling pathways, in a particular cancer cell environment.
The Epic optical sensor system offers a highly sensitive and versatile detection system for oncogenic signaling pathways. Because EGFR-targeted therapeutics only show activity in a selected population of patients (18,33,34), the Epic optical signature assay may be used to correlate with therapeutic efficacy to predict in vivo therapeutic response. Our current study has clearly shown cell line- and cell type-dependent DMR profiles in response to the same EGF signal. Understanding of the molecular basis of these distinct DMR profiles and their correlation with therapeutic response will aid in their applications as an in vitro predictive tool for in vivo efficacy. Continued probing of EGFR- and other growth factor receptor-induced signaling pathways with the use of the Epic biosensor is expected to reveal distinct DMR signal signatures for various oncogenic pathways in a variety of cancer types. Characterization of these optical signatures for a variety of cell types will help to establish a cell optical signature “library.” Enriched information will provide a powerful tool for cell line characterization, pathway studies, and future therapeutic development.
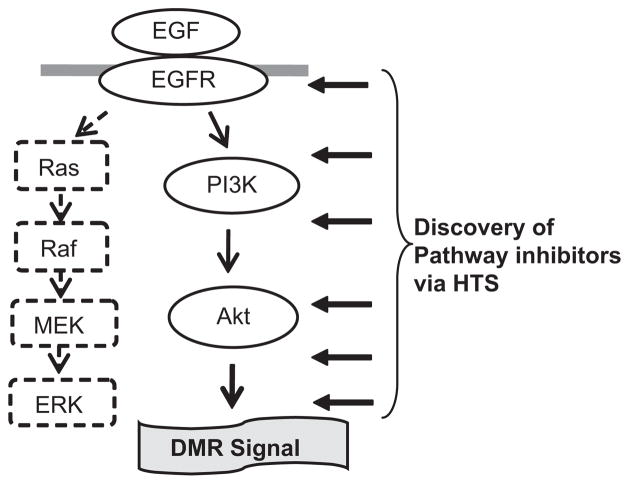
In summary, we have identified and characterized distinct optical signatures of EGF-induced signaling events in SCCHN and lung cancer cells using the label-free Epic optical biosensor. Our data show that the DMR signals are cell line and cell type specific in response to a specific ligand. The PI3K/Akt pathway, but not the Ras/Raf/MEK/ERK pathway, mediates the EGF-induced DMR signal in UPCI-37B SCCHN cancer cells as summarized in Figure 7. This technology offers not only a novel tool to study ligand-receptor signaling pathways under physiological conditions but also provides a powerful approach for targeting oncogenic signaling pathways for therapeutic agent discovery through HTS. The established DMR signal signatures in SCCHN and lung cancer cells may have value in examining the action of EGFR-targeting agents for clinical use. This technology is applicable to other oncogenic pathways for the discovery of novel therapeutic agents targeting various cancers.
Figure 7.
Model of the mechanism by which the EGF-induced DMR signal is mediated in UPCI-37B SCCHN cells and its application for HTS of EGFR pathway inhibitors. The PI3K/Akt (solid arrow), but not Ras/Raf/MEK/ERK (dotted arrow) pathway, mediates the EGF-induced DMR signal in UPCI-37B SCCHN cells. Our established cell-based optical signature assay is a novel approach for the discovery of inhibitors targeting the EGFR signaling pathway through HTS.
Acknowledgments
This research was supported by the National Institutes of Health grant 5 P50 CA128613 SPORE in Head and Neck Cancer (YD, ZGC, SYS, DMS, FRK, and HF) P01 CA116676 Lung Cancer Program (FRK, HF), and 5U54 HG003918 Emory Chemical Biology Discovery Center in MLSCN (HF). ZJL is an Emory Global Health Institute-Peking University Health Science Center exchange scholar. YD is a recipient of Emory University’s SPORE in Head and Neck Cancer Career Development award (P50 CA128613). HF is a Georgia Research Alliance Distinguished Investigator. DMS, FRK, HF and ZGC are Georgia Cancer Coalition Distinguished Cancer Scholars.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Seger R, Yarden Y, Kashles O, Goldblatt D, Schlessinger J, Shaltiel S. The epidermal growth factor receptor as a substrate for a kinase-splitting membranal proteinase. J Biol Chem. 1988;263(7):3496–500. [PubMed] [Google Scholar]
- 3.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev. 2005;5(5):341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 4.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. Embo J. 2000;19(13):3159–67. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 6.Lewis TS, Shapiro PS, Ahn NG. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 7.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 8.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 9.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 10.Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366(1):2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Saba NF, Khuri FR, Shin DM. Targeting the epidermal growth factor receptor. Trials in head and neck and lung cancer. Oncology (Williston Park) 2006;20(2):153–61. discussion 62, 66, 69 passim. [PubMed] [Google Scholar]
- 12.Khuri FR, Cohen V. Molecularly targeted approaches to the chemoprevention of lung cancer. Clin Cancer Res. 2004;10(12 Pt 2):4249s–53s. doi: 10.1158/1078-0432.CCR-040019. [DOI] [PubMed] [Google Scholar]
- 13.Harichand-Herdt S, Ramalingam SS. Targeted therapy for the treatment of non-small cell lung cancer: focus on inhibition of epidermal growth factor receptor. Semin Thorac Cardiovasc Surg. 2008;20(3):217–23. doi: 10.1053/j.semtcvs.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Rubin Grandis J, Melhem MF, Gooding WE, et al. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90(11):824–32. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 15.Ang KK, Berkey BA, Tu X, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62(24):7350–6. [PubMed] [Google Scholar]
- 16.Byers LA, Heymach JV. Dual targeting of the vascular endothelial growth factor and epidermal growth factor receptor pathways: rationale and clinical applications for non-small-cell lung cancer. Clin Lung Cancer. 2007;8 (Suppl 2):S79–85. doi: 10.3816/clc.2007.s.006. [DOI] [PubMed] [Google Scholar]
- 17.Vokes EE, Chu E. Anti-EGFR therapies: clinical experience in colorectal, lung, and head and neck cancers. Oncology (Williston Park) 2006;20(5 Suppl 2):15–25. [PubMed] [Google Scholar]
- 18.Sattler M, Abidoye O, Salgia R. EGFR-targeted therapeutics: focus on SCCHN and NSCLC. Scientific World Journal. 2008;8:909–19. doi: 10.1100/tsw.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xi B, Yu N, Wang X, Xu X, Abassi YA. The application of cell-based label-free technology in drug discovery. Biotechnol J. 2008;3(4):484–95. doi: 10.1002/biot.200800020. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y. Label-free cell-based assays with optical biosensors in drug discovery. Assay Drug Dev Technol. 2006;4(5):583–95. doi: 10.1089/adt.2006.4.583. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y, Ferrie A, Fontaine N, Ki Yuen P. Optical biosensors for monitoring dynamic mass redistribution in living cells mediated by epidermal growth factor receptor activation. Conf Proc IEEE Eng Med Biol Soc. 2005;1:666–9. doi: 10.1109/IEMBS.2005.1616501. [DOI] [PubMed] [Google Scholar]
- 22.Fang Y, Ferrie AM, Fontaine NH, Mauro J, Balakrishnan J. Resonant waveguide grating biosensor for living cell sensing. Biophys J. 2006;91(5):1925–40. doi: 10.1529/biophysj.105.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Fang Y, Ferrie AM, Fontaine NH, Yuen PK. Characteristics of dynamic mass redistribution of epidermal growth factor receptor signaling in living cells measured with label-free optical biosensors. Anal Chem. 2005;77(17):5720–5. doi: 10.1021/ac050887n. [DOI] [PubMed] [Google Scholar]
- 25.Strawn LM, McMahon G, App H, et al. Flk-1 as a target for tumor growth inhibition. Cancer Res. 1996;56(15):3540–5. [PubMed] [Google Scholar]
- 26.Kamath S, Buolamwini JK. Targeting EGFR and HER-2 receptor tyrosine kinases for cancer drug discovery and development. Med Res Rev. 2006;26(5):569–94. doi: 10.1002/med.20070. [DOI] [PubMed] [Google Scholar]
- 27.Fang Y, Ferrie AM, Li G. Probing cytoskeleton modulation by optical biosensors. FEBS Lett. 2005;579(19):4175–80. doi: 10.1016/j.febslet.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 28.Bernier J, Schneider D. Cetuximab combined with radiotherapy: an alternative to chemoradiotherapy for patients with locally advanced squamous cell carcinomas of the head and neck? Eur J Cancer. 2007;43(1):35–45. doi: 10.1016/j.ejca.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 29.Langer CJ. Emerging role of epidermal growth factor receptor inhibition in therapy for advanced malignancy: focus on NSCLC. Int J Radiat Oncol Biol Phys. 2004;58(3):991–1002. doi: 10.1016/j.ijrobp.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 30.Muller S, Su L, Tighiouart M, et al. Distinctive E-cadherin and epidermal growth factor receptor expression in metastatic and nonmetastatic head and neck squamous cell carcinoma: predictive and prognostic correlation. Cancer. 2008;113(1):97–107. doi: 10.1002/cncr.23557. [DOI] [PubMed] [Google Scholar]
- 31.Wakeling AE, Guy SP, Woodburn JR, et al. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62(20):5749–54. [PubMed] [Google Scholar]
- 32.Moyer JD, Barbacci EG, Iwata KK, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res. 1997;57(21):4838–48. [PubMed] [Google Scholar]
- 33.Rocha-Lima CM, Soares HP, Raez LE, Singal R. EGFR targeting of solid tumors. Cancer Control. 2007;14(3):295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 34.Taguchi T, Tsukuda M, Imagawa-Ishiguro Y, Kato Y, Sano D. Involvement of EGFR in the response of squamous cell carcinoma of the head and neck cell lines to gefitinib. Oncol Rep. 2008;19(1):65–71. [PubMed] [Google Scholar]