Abstract
Melanoma is the most deadly form of skin cancer and DiGeorge syndrome (DGS) is the most frequent interstitial deletion syndrome. We characterized a novel balanced t(9;22)(p21;q11.2) translocation in a patient with melanoma, DNA repair deficiency, and features of DiGeorge syndrome (DGS) including deafness and malformed inner ears. Using chromosome sorting we located the 9p21 breakpoint in CDKN2A intron 1. This resulted in under-expression of the tumor suppressor p14ARF; the reduced DNA repair was corrected by transfection with p14ARF. UV-type p14ARF mutations in his melanoma implicated p14ARF in its pathogenesis. The 22q11.2 breakpoint was located in a palindromic AT-rich repeat (PATRR22). We identified a new gene, FAM230A that contains PATRR22 within an intron. The 22q11.2 breakpoint was located 800 kb centromeric to TBX1, which is required for inner ear development. TBX1 expression was greatly reduced. The translocation resulted in a chimeric transcript encoding portions of p14ARF and FAM230A. Inhibition of chimeric p14ARF-FAM230A expression increased p14ARF and TBX1 expression and improved DNA repair. Expression of the chimera in normal cells produced dominant negative inhibition of p14ARF. Similar chimeric mRNAs may mediate haploinsufficiency in DGS or dominant negative inhibition of other genes such as those involved in melanoma.
Keywords: melanoma, TBX1, deafness, p14ARF, PATRR22, DiGeorge syndrome
INTRODUCTION
We identified a patient (DD129BE) with melanoma, DNA repair deficiency, and features of DiGeorge syndrome (DGS; MIM# 188400) including deafness and malformed inner ears. The patient has a novel balanced t(9;22) translocation that served as a “Rosetta stone” to permit determination of the DNA sequence in a chromosome gap region. This resulted in identification of a new gene that as a chimera appears to play an important dominant negative regulatory role in melanoma induction and in development of the inner ear.
Melanoma, the most deadly form of skin cancer, has been linked to chromosome 9p21(Kamb et al., 1994; Randerson-Moor et al., 2001), which contains CDKN2A (Cyclin dependent kinase inhibitor 2A) [MIM# 600160] and CDKN2B (Cyclin dependent kinase inhibitor 2B) [MIM# 613149]. CDKN2A encodes the tumor suppressors p16INK4a and p14ARF (Ozenne et al., 2010). p14ARF (p14 alternate reading frame) has been associated with melanoma predisposition (Freedberg et al., 2008; Harland et al., 2005; Hewitt et al., 2002; Rizos et al., 2001a; Rizos et al., 2001b; Sherr, 2001; Sherr et al., 2005), but the molecular mechanisms are not understood.
DGS is a characteristic disorder of pharyngeal development with virtually all the derivatives of the pharyngeal arches and pouches being affected (Butts et al., 2005; Shprintzen, 2008). Most DGS cases result from a heterozygous deletion within the 3-Mb DGS critical region (DGCR) in 22q11.2 (Driscoll et al., 1992; Scambler et al., 1992). Deletion of chromosome 22q11 is the most frequent interstitial deletion in humans with an estimated incidence of 1 in 4000 live births (Oskarsdottir et al., 2004). Only two balanced translocations with partial DGS, involving t(2;22) and t(21;22), have been reported (Budarf et al., 1995; Holmes et al., 1997). The 22q11.2 region contains eight DNA low copy repeats (LCR22-A through LCR22-H) (Shaikh et al., 2007). LCR22-B contains a palindromic AT-rich repeat (PATRR22) that plays an essential role in mediating clinically relevant rearrangements and is located in a gap region whose sequence has not been conclusively determined (Kurahashi et al., 2007; Shaikh et al., 2000). Hearing impairment occurs in 44–77% of DGS cases (Digilio et al., 1999; Reyes et al., 1999; Swillen et al., 1999; Ohtani and Schuknecht, 1984) and some DGS patients have malformed inner ears. TBX1(T-box transcription factor 1) [MIM# 602054] is involved in inner ear sensory organ development and other features of DGS in mice (Jerome and Papaioannou, 2001; Lindsay et al., 2001; Vitelli et al., 2003; Arnold et al., 2006b; Arnold et al., 2006a; Braunstein et al., 2008; Braunstein et al., 2009) and has been implicated as a DGS candidate gene (Packham and Brook, 2003). We performed a comprehensive analysis of the clinical features, chromosomal structure, DNA sequence, gene expression and function of cells from patient DD129BE by investigating his novel t(9;22)(p21;q11.2) chromosome translocation. We characterized a unique fusion transcript encoding portions of p14ARF and a novel gene, FAM230A. We demonstrate that this chimeric transcript exerts a dominant negative regulatory function on both p14ARF and TBX1 thereby providing a mechanism for his symptoms of melanoma and deafness.
MATERIALS AND METHODS
This study was approved by the National Cancer Institute, Institutional Review Board. Cultured cells were obtained from the Human Genetic Mutant Cell Repository (Camden, NJ, USA). Fluorescent in-situ hybridization (FISH) analysis was performed with probes listed in Supp. Table S1. The derivative chromosomes 9 and 22 were flow sorted as described previously (Stanyon and Stone, 2008) and analyzed by array comparative genome hybridization (aCGH). Polymerase chain reaction (PCR) was used to identify the t(9;22) junction from both the der (9) and der(22) chromosomes using genomic DNA. The fusion transcript was isolated by use of primers designed to flank the probable breakpoints within CDKN2A and a hypothetical gene (LOC653203) (Supp. Table S2). 3′ rapid amplification of cDNA ends (RACE) assay was used to extend the sequence. Transcript levels were assessed by microarray and quantitative reverse transcription polymerase chain reaction (qRT-PCR). DNA of the melanoma was isolated by laser capture microdissection and sequenced for mutations in CDKN2A. DNA repair capability of DD129BE cells was assessed using post-UV host cell reactivation (HCR) with the pCMVLuc reporter gene plasmid as described previously (Khan et al., 2002). For examination of p14ARF protein level cells were treated with 10 nM MG-132 (Calbiochem, La Jolla, CA) or 0.1% dimethyl sufoxide (DMSO) for 6 h and proteins were extracted for western blot analysis (Magro et al., 2004). For UV irradiation cells were seeded in 10 cm dishes at a concentration of 8×105 cells/dish. After a 24 h pre-incubation period, cells were exposed to 10 J/m2 of UVC (254 nm) and allowed to recover for 6, and 24 h (Khan et al., 2002). Immunofluorescence was performed on cells in glass coverslips fixed in 2% paraformaldehyde as described previously (Dawe et al., 2007). Confocal images were obtained using a LSM 510 Confocal microscope (Carl Zeiss). The number of nuclei containing at least one localized area of immunofluorescence was determined by examination of the confocal images. Small interfering RNA’s (siRNAs) were purchased from Ambion. Statistical evaluation was performed using the Student’s unpaired t-test. Data are presented as mean ± s.e.m. and P ≤ 0.05 was considered statistically significant. For further details, please see the Supplementary Materials and Methods section.
RESULTS
Clinical assessment
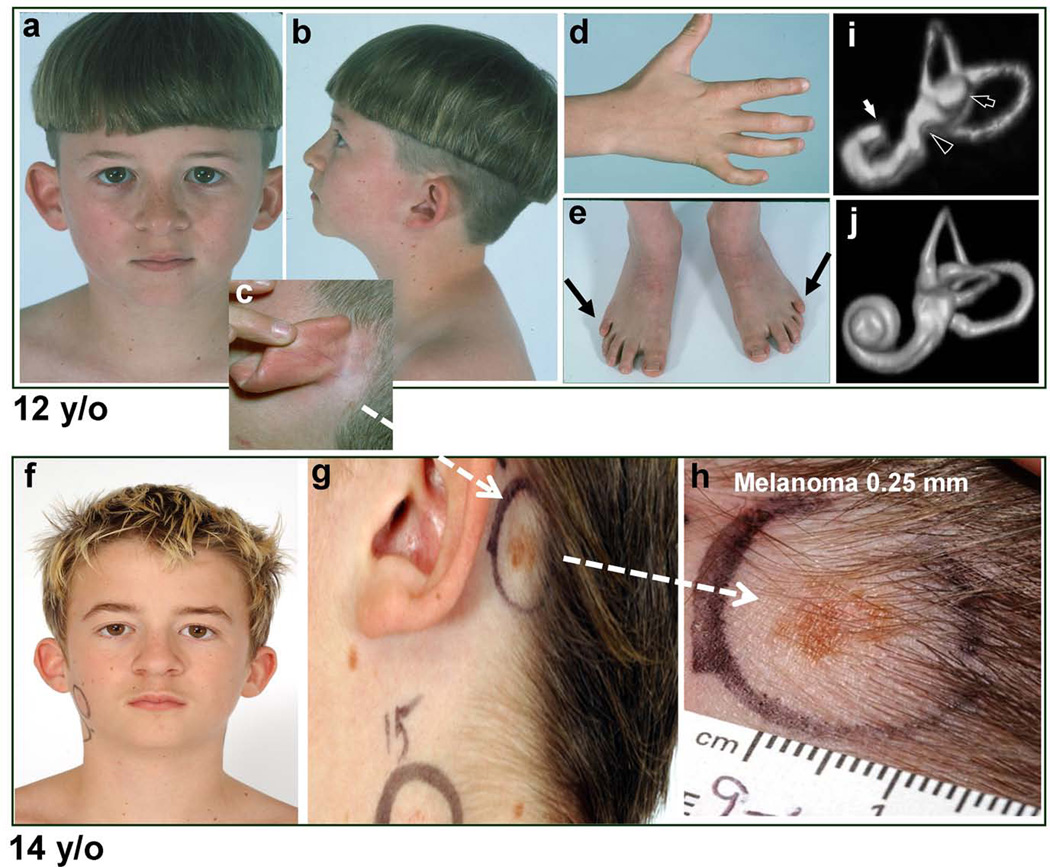
The proband (DD129BE) is a 20 year-old Caucasian male with an acquired melanoma, defective DNA repair, and profound congenital deafness with features suggestive of DGS (Figure 1a–h and Supp. Table S3). Computed tomography (CT) and magnetic resonance imaging (MRI) revealed bilateral incomplete partition of the cochleae, and hypoplasia of the lateral semicircular canals and vestibules. The ostea for the VIIIth cranial nerves were greatly diminished or absent bilaterally (Figure 1i–j). He had a dilated aortic root, bicuspid aortic valve, and short stature (<10%-ile) with normal intelligence, serum calcium, thyroid and parathyroid function. Cultured skin fibroblasts were hypersensitive (2x) to killing by ultraviolet radiation (UV) (data not shown).
Figure 1.
Patient DD129BE. (a–e) At 12 years of age (a) Facial dysmorphism with a broad-based nasal root. (b) Low-set, simply-formed, posteriorly rotated ear and multiple pigmented nevi; (c) Small, faint pigmented lesion behind left ear at hairline; (d and e) Bilateral clinodactyly and arthrogryposis of proximal and distal interphalangeal joints of the 5th finger and the 4th and 5th toes, and posterior 5th toes (arrows). (f–h) At 14 years of age, change in pigmented lesion behind left ear (compare c to g and h). (h) Close-up view of 5×10 mm pigmented lesion with irregular border and variation in color. Excision revealed a 0.25-mm deep melanoma. (i and j) Volume rendering of MRI of labyrinth of ear in the patient (i) and a normal control (j). (i) Labyrinth of patient: Cochlea is hypoplastic (white arrow) exhibiting less than one turn as compared to the normal ~ 2.5 turns. Lateral semicircular canal is only partially formed (black arrow) and vestibule is hypoplastic (black arrowhead). (j) Normal labyrinth for reference.
A constitutional translocation within AT-rich repeats in the CDKN2A gene on 9p21 and LCR22-B on 22q11.2
Chromosome analysis of cultured skin fibroblasts and whole chromosome painting identified a balanced translocation between chromosomes 9 and 22 [46, XY, t(9;22)(p22;q11.2)] (Supp. Figure S1 a–c). Fluorescent in situ hybridization (FISH) analysis localized the breakpoint between D9S1747 and D9S1752 on 9p21 and within LCR22-B on 22q11.2 (Supp. Figure S1d and e and Supp. Figures S2, S3c and Supp. Table S1). We isolated derivative chromosomes by bivariate flow sorting (Supp. Figure S4 a–d) and hybridization narrowed the breakpoint on chromosome 9 to a 2-kb region between CDKN2A exons 1β and 1α. The breakpoint on chromosome 22, however, could only be localized approximately to an 890-kb region within the 3-Mb DGCR due to inability to identify the sequences containing LCRs within the gap (Supp. Figure S4e and f).
The exact junction sequences of der(9) and der(22) were successfully amplified using a large series of primers (Figure 2, Supp. Figures S5 and S6 and Supp. Table S2). Both the chromosome 9 and 22 translocation breakpoints were located within short interspersed element (SINE) AT repeat regions. The translocation was almost perfectly balanced with a 4-bp region of overlap (GTAT/ATAC) and deletion of only 67 bp in ATRR flanking the der(9) breakpoint and 76 bp in PATRR22 flanking the der(22) (Figure 2).
Figure 2.
Sequence of the region flanking the t(9;22) breakpoint on 9p21 and 22q11.2 from patient DD129BE. Orientation of the sequence is centromere to telomere. PCR primers are indicated by italics. Forward and Reverse primer pairs are labeled with the same color. ATRR on chromosomes 9 and 22 are underlined. The nucleotide polymorphisms are boxed. (a) Normal chromosome 9. The portion of 9p21 proximal to the breakpoint and present in der(9) is depicted in bold type. The 127bp-SINE sequence on chromosome 9 is shown as a light blue box. The italicized bases in ATRR represent the deletion of 67 bp. (b) Derivative 9. The portions of 9p21 and 22q11.2 are depicted in bold and plain type respectively. Four base pairs [GTAT] which cannot be definitely assigned to chromosome 9 or 22 are outlined boxed in red. (c) Normal chromosome 22. The portion of 22 proximal to the breakpoint and present in der(22) is shown in bold. The 127bp-SINE sequence on chromosome 22 is presented in dark blue box. Two non-AT rich sequences in PATRR22 are shown in orange and yellow highlighting. (d) Derivative 22. The portions of 22q11.2 and 9p21 are depicted in bold and plain type respectively. Four base pairs [ATAC] which cannot be definitely assigned to chromosome 9 and 22 are boxed in red.
The breakpoint occurred in intron 1 of CDKN2A on chromosome 9 and in PATRR22 in LCR-B on chromosome 22. We theorized that the t(9;22) breakpoint on chromosome 22 might lie in the same location as t(11;22)(Tapia-Paez et al., 2001) and t(17;22) (Kurahashi et al., 2003). A BLAST search revealed that the region surrounding the PATRR22 (Kurahashi et al., 2007) had 99% identity to a region including the first 5 exons of a predicted gene, LOC653203, located approximately 80 kb away from the gap (Supp. Figures S5b, S6a, S7 and S8b). Due to the duplicated-sequence character of LCR-B, we theorized that PATRR22 might lie in the gap within a new gene with high homology to LOC653203. This putative gene was named FAM230A by the Human Genome Organization (HUGO) (Supp. Figure S7). A 5-kb genomic DNA chimeric PCR product including intron 1 of CDKN2A on chromosome 9 and introns 1 to 3 of FAM230A was obtained from DD129BE, but not from the control cells, using long nested PCR (Supp. Figure S9). Our results indicate that the t(9;22) breakpoint on chromosome 22 lies within the gap in a similar PATRR22 location to recurrent translocations t(11;22) and t(17;22) (Supp. Table S4 and Supp. Figure S8) within FAM230A (GenBank accession number: JX456222). For further details, please see the Supplementary Results section.
The 9p21 breakpoint results in a unique chimeric transcript
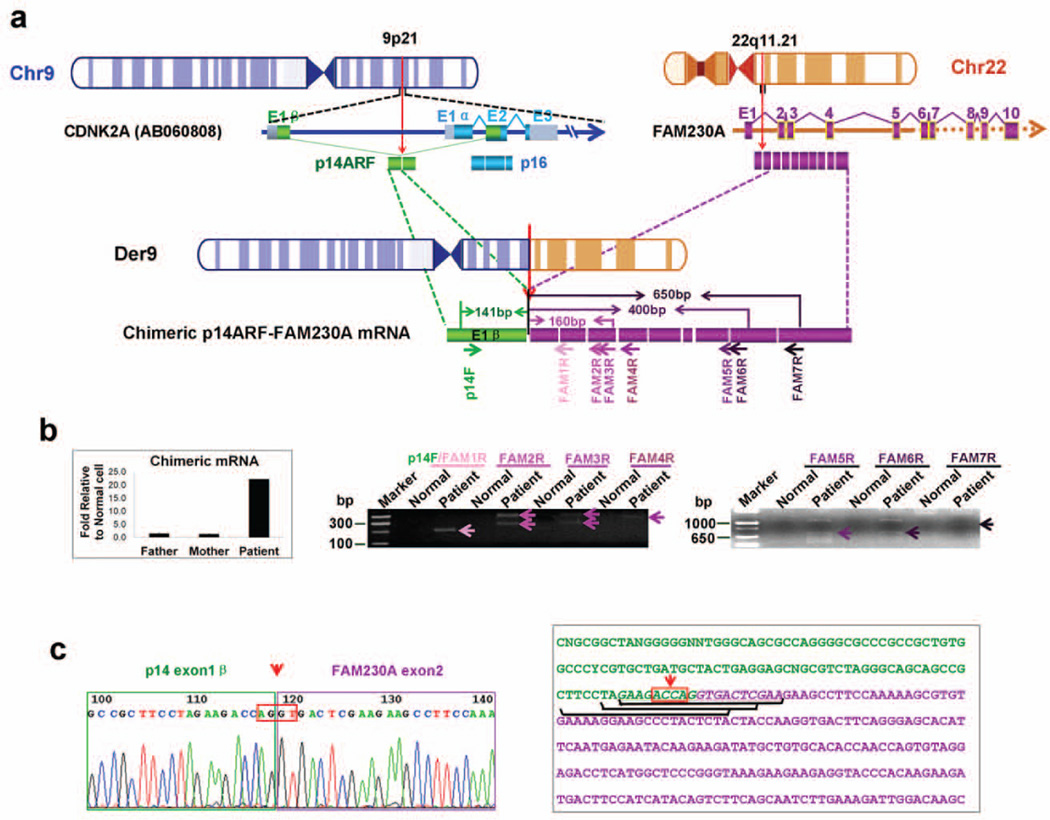
We characterized chimeric CDKN2A–FAM230A at the nucleotide sequence level. We hypothesized that the t(9;22) translocation might be involved in the generation of fusion transcript(s) (Figure 3a). Sequencing of RT-PCR products from DD129BE cells generated with forward primers in CDKN2A and reverse primers across exons 2 through 10 of FAM230A (Figure 3a and Supp. Table S2) revealed an in-frame fusion of exon 1β of p14ARF with exon 2 of FAM230A (Figure 3b and c) and containing the p14ARF initiation codon.
Figure 3.
Identification of chimeric p14ARF-FAM230A fusion transcripts. (a) A schematic depiction of chimeric p14ARF-FAM230A fusion transcripts. Small arrows depict PCR primers. CDKN2A exon1β is indicated by green boxes, DGCR15 by purple boxes and LOC653203 by yellow boxes. The identical exon sequences of DGCR15 and LOC653203 were drawn with purple boxes. (b) Left panel, expression of chimeric p14ARF- FAM230A fusion transcripts in DD129BE and parents by qRT-PCR as described in “Materials and methods”. The PCR analysis was performed by normalizing the chimeric p14ARF-FAM230A PCR products to the beta-actin gene products. Left panel, representative results of the PCR amplification of cDNA with the indicated primers. The PCR product was only detected in patient cDNA (arrow heads). The additional bands originating from heteroduplexes appear as PCR artifacts. (c) Sequences of p14ARF-FAM230A fusion products. This sequence was obtained by sequencing the junction fragments after PCR amplification using primers p14ARF and FAM230A 1–7R. The orientation of the sequence is centromere to telomere. The AGGT junction site is boxed. The three siRNA sequences specific for p14ARF-FAM230A fusion mRNA are underlined. In (a) and (c), the arrows with red dotted line indicate the location of breakpoints.
Using “mRNA walking” we were able to obtain an approximately 800-bp cDNA product from exon 1β of CDKN2A to exon10 of FAM230A3 (Figure 3c and Supp. Figure S10c). This chimeric p14ARF-FAM230A transcript variant 2 contained at least 10 exons (GenBank accession number: JX456220), that allowed us extend the 13.8-kb sequenced genomic region surrounding PATRR22 in LCR-B to 21 kb (Supp. Figure S8). A full-length ~1.3-kb p14ARF-FAM230A chimeric mRNA transcript variant 1 (GenBank accession number: JX456221) from DD129BE was characterized by 3’ RACE (Supp. Figure S10a, d and e). At the protein level, using bioinformatics and the in-frame fusion transcript, we predicted formation of a fusion protein (Supp. Figure S10b). Using anti-p14ARF antibody We detected extra bands of the predicted size in the DD129BE cells compared to normal cells using anti-p14ARF antibody (data not shown). Thus there was evidence of chimeric mRNA and protein in the DD129BE cells.
Reduced CDKN2A expression
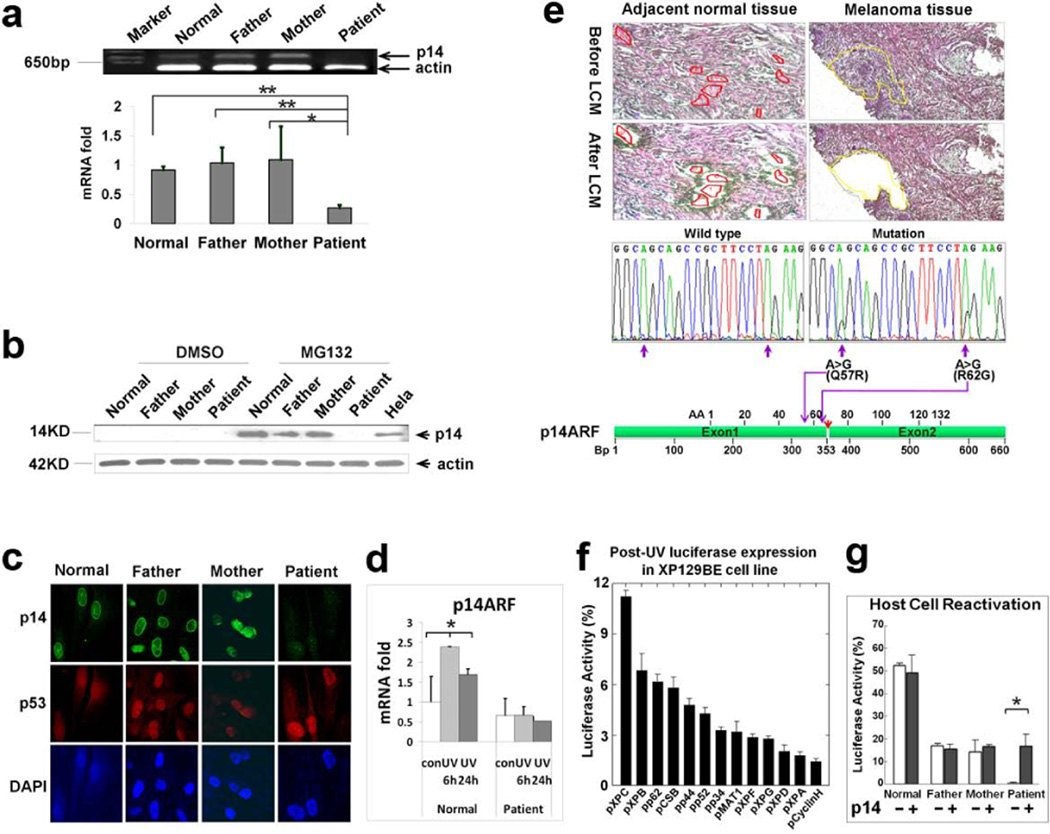
cDNA microarray analysis of RNA isolated from DD129BE fibroblasts indicated that expression levels of two genes, CDKN2A and TBX1, were substantially lower (Supp. Figure S11) in comparison to cells from his parents and two normal controls. Low mRNA and protein levels of p14ARF, but not of p16INK4a, was found in DD129BE by qRT-PCR, western blot and immunohistochemistry indicating that the abnormality was restricted to p14ARF within the CDKN2A locus (Figure 4a–c and Supp. Figure S12). In contrast to the response in normal cells, p14ARF mRNA levels did not increase in the patient cells at 6 hr or 24 hr after treatment with UV (Figure 4d), indicating impaired post-UV responsiveness associated with the abnormal p14ARF.
Figure 4.
Mutation, expression and functional analysis of p14ARF. (a) Reduced level of p14ARF mRNA was measured by semi-quantitative RT–PCR (top) and quantitative-real time PCR (bottom) in cells from patient DD129BE, compared to his parents and a normal control. (b) p14ARF protein was measured by western blot. The p14ARF protein level increased markedly in paternal, maternal and normal control fibroblasts in the presence of the proteasome inhibitor (MG132) but p14ARF was not detected in DD129BE cells. (c) Immunofluorescence showed low levels of p14ARF but normal levels of p53 in the patient cells. (d) Relative quantity of p14ARF mRNA in normal and patient fibroblast after UV exposure: cells were irradiated with 10 J/m2 UV and incubated for 6h and 24h. cDNA was synthesized from RNA collected and transcript expression was examined by qRT-PCR. p14ARF mRNA levels did not increase in the patient’s cells at 6 hr or 24 hr after treatment with UV. An asterisk denotes statistically significant difference between control and irradiated normal cells (P < 0.05) while there was no significant difference between control and irradiated patient cells. (e) Identification of p14ARF mutation in melanoma cells isolated by laser capture microdissection (LCM). Adjacent normal and melanoma tissues were microdissected and p14ARF was PCR amplified and sequenced. Two missense mutations were detected in melanoma cells (middle right) but not in adjacent normal tissue (middle left). Locations of two missense mutations are indicated by purple arrows, and the location of the breakpoint in p14ARF is indicated by a red arrow (bottom). (f and g) Post-UV host cell reactivation (HCR). UV-treated or untreated reporter gene plasmid (pCMVLuc) was co-transfected with plasmids expressing wild type cDNA of each of the known XP genes or p14ARF cDNA. (f) Reduced luciferase activity in DD129BE cells was not corrected by co-transfection with known XP genes except for partial correction with wild type XPC cDNA. (g) Co-transfection with wild type p14ARF cDNA-containing plasmid into triplicate cultures of normal (AG13354) (yellow bar), paternal (GM16735) (blue bar), maternal (GM16733) (green bar) and patient (DD129BE (pink bar) fibroblasts. The low luciferase activity in DD129BE cell was increased by co-transfection with wild type p14ARF cDNA. The data represent three independent experiments, and the error bars represent the standard deviation. (a), (d) and (g) mean±s.d., n≥3. *P≤0.05; **P≤0.01.
DNA repair capability of DD129BE cells was reduced in comparison to parental cells as measured by post-UV plasmid host cell reactivation (HCR). Impaired DNA repair is associated with increased risk of melanoma in patients with xeroderma pigmentosum (XP) (Bradford et al., 2011; Wei et al., 2003). DNA damage induced by UV is repaired by the nucleotide excision repair (NER) pathway which involves XP proteins (DiGiovanna and Kraemer, 2012). However, transfection of patient cells with XP NER genes did not increase DNA repair (Figure 4f) but transfection with p14ARF led to a markedly increased post-UV HCR (Figure 4g). This is evidence that p14ARF function plays a critical role in the DNA repair deficiency in the DD129BE cells.
No base substitution mutations were found in p14ARF or p16INK4a in blood cells or fibroblasts from DD129BE. In contrast, in laser-captured melanoma tissue, but not in adjacent normal tissue, we found two heterozygous missense mutations [(p.Q57R) and (p.R62G)] in exon 1β of CDKN2A (GenBank NG007485) (Figure 4e). These mutations occurred at sites of adjacent pyrimidines, consistent with UV mutagenesis (Wang et al., 2009). Thus the truncated p14ARF may contribute to development of the melanoma via a “two-hit” model (Knudson, 2001). The first hit would be the translocation and the second hit would be mutations induced by UV exposure, however, we were not able to determine conclusively that these melanoma-associated mutations are on the non-translocated allele.
Reduced TBX1 expression
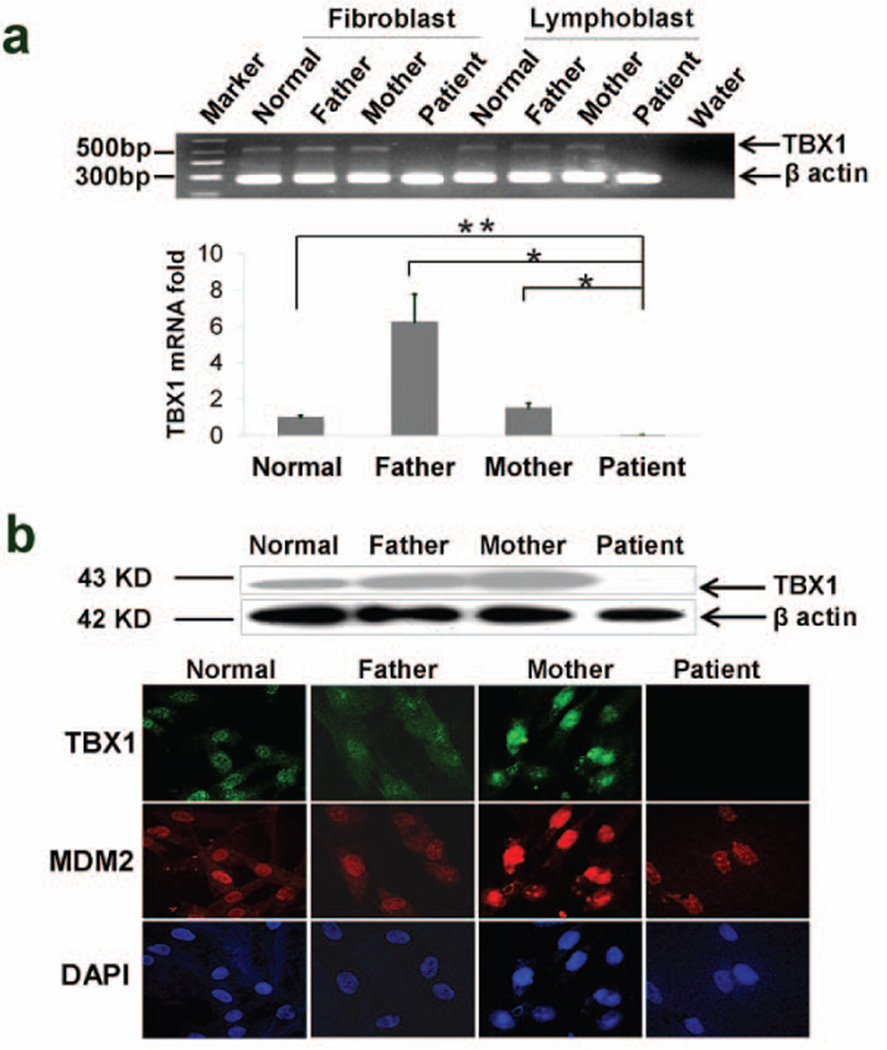
The markedly low expression of TBX1 in the microarray (Supp. Figure S11b) was confirmed by qRT-PCR, western blot and immunohistochemistry (Figure 5a and b). TBX1 is approximately 800 kb away from the breakpoint in PATRR22 (Supp. Figures S7 and S13). The high density aCGH of the unsorted and sorted chromosomes (Supp. Figures S3c and S4e) did not reveal deletions of TBX1. We sequenced TBX1 (including individual TBX1 exons belonging to each homologue from the sorted chromosomes) and found no mutations or deletions to account for the reduced expression (data not shown). Several other genes [DGCR6(DiGeorge syndrome critical region 6), SEPT5(septin 5) , COMT (catechol-o-methyltransferase), DGCR8 (DiGeorge syndrome critical region 8), ZNF74 (zinc finger protein 74) and MMP11(matrix metalloproteinase 11)] within the 3-Mb DGCR as well as putative TBX1-related genes [(SHH (sonic hedgehog) , GLI (glioma-associated oncogene homolog) , FGF8 (fibroblast growth factor 8), and FGF10(fibroblast growth factor 10)] did not have reduced expression (Supp. Figure S13). Interestingly, CTNNB1 (catenin, beta 1), a Wnt pathway negative regulator of TBX1 in DGS model mice (Huh and Ornitz, 2010), showed increased expression in DD129BE cells (Supp. Figure S13).
Figure 5.
Expression of TBX1. (a) Reduced TBX1 mRNA level was detected by semi-quantitative RT–PCR (gel-top), and QRT- PCR (bar graph – bottom) (b). Reduced TBX1 protein was detected in patient cells by western blotting (top) and by immunohistochemistry showing undetectable levels of staining of TBX1 and normal levels of MDM2 and DAPI (bottom). (a) mean±s.d., n≥3, *P≤0.05; **P≤0.01.
Dominant negative regulation by chimeric p14ARF-FAM230A
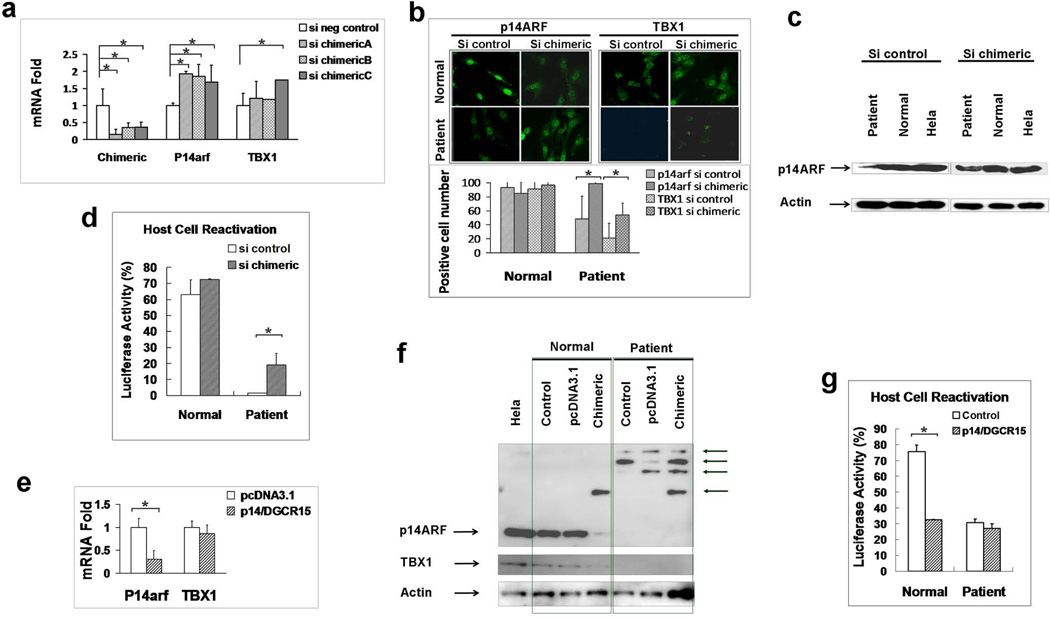
Functional analysis of p14ARF-FAM230A is shown in Figure 6 a–g. siRNA duplexes targeted to the chimeric p14ARF-FAM230A in the patient’s cells increased mRNA and protein expression of p14ARF and TBX1 (Figure 6a,b and c) and increased post-UV DNA repair (Figure 6d). Using anti-p14ARF antibody we were able to detect extra bands of the predicted size for chimeric p14ARF-FAM 230A protein in the DD129BE cells compared to normal cells by western blotting (Figure 6f – normal control vs patient control lanes ). Transfection of p14ARF-FAM230A cDNA into normal cells inhibited expression of p14ARF and TBX1 mRNA and protein as measured by qRT-PCR and western blotting (Figure 6e and f). More importantly, the post-UV DNA repair ability of the cells was significantly impaired after the transfection of the chimeric cDNA into normal cells (p<0.05) (Figure 6g). Thus p14ARF-FAM230A was a dominant negative regulator of p14ARF, TBX1 and post-UV DNA repair in normal cells.
Figure 6.
Functional analysis of p14ARF-FAM230A (a) Knockdown of p14ARF-FAM230A by siRNA. Reduced expression of p14ARF-FAM230A RNA levels in DD129BE cells by 63.5–84.3% by use of 3 different siRNA’s targeted to the chimeric p14ARF-FAM230A (left graph) associated with increased levels of expression of p14ARF (middle graph) and TBX1 (right graph) in DD129BE cells. (b) p14ARF (left) and TBX1 (right) protein levels detected by immunofluorescence and numbers of positive cells (bottom) before and after p14ARF-FAM230A siRNA treatment. (c) Reduced p14ARF protein levels in patient lymphoblasts (left) was increased by knock down of p14ARF-FAM230A (right). (d) HCR assay showing increased post-UV DNA repair capability after treatment of patient cells with p14ARF-FAM230A siRNA. (e) Reduced mRNA expression of p14ARF in normal cells by transfection of the chimeric p14ARFFAM230A. (f) Reduced protein expression of p14ARF and TBX1 in normal cells by transfection of the chimeric p14ARF-FAM230A. The extra protein bands in patient cells were indicated by arrows. (g) Impaired post-UV DNA repair capability in normal cells by transfection of the chimeric p14ARF-FAM230A. (a, b, d, e and g) mean±s.d., n≥3. *P≤0.05.
DISCUSSION
This study has generated a comprehensive picture of the molecular mechanism of a unique, nearly balanced translocation from complete clinical evaluation, cytogenetic and molecular assessment to functional characterization. Although aCGH is a powerful tool for rapid detection of genomic imbalances a major weakness of this approach is that it might yield negative results when the translocated chromosomes are contaminated by the presence of normal chromosomes or when there is little loss of genomic DNA (Supp. Figure S3). Mapping of the breakpoint was accomplished by using bivariate flow sorting to separate the chromosomes of interest followed by aCGH (Stanyon and Stone, 2008) which showed an unambiguous breakpoint in the intron of CDKN2a on der(9) (Supp. Figure S4f). However, we were not able to precisely map the breakpoint on the sorted der(22) by aCGH because the highly repetitive AT sequence in this region did not permit designing a unique tiling array (Supp. Figure S4e). Determination of the precise breakpoint required use of several hundred primer pairs spanning the break.
p14ARF-FAM230A chimera
At the genome level, the breakpoints were located in intron 1 of CDKN2A and intron 1 of FAM230A on chromosomes 9 and 22, respectively. This was an almost completely balanced translocation with only a 4-bp overlap within AT repeat (SINE) regions and deletion of 71-bp (Figure 2). By using the sequences present in the chimeric transcript, we were able to identify a new gene, FAM230A in the unclonable gap on chromosome 22 associated with formation of a functional chimera: p14ARF-FAM230A (Figure 7). At the RNA level, the CDKN2A breakpoint resulted in the split of p14ARF between exon 1β and exon 2 and a chimeric transcript p14ARF-FAM230A on der(9). The entire p16INK4a was translocated to der(22) ( Supp. Figures S1b, S4, S5 and S6). However, we were not able to isolate this transcript. Interestingly, a mouse model with interruption of p19arf in this intron has high susceptibility to UV induction of melanoma (Ha et al., 2007; Merlino, 2008).
Figure 7.
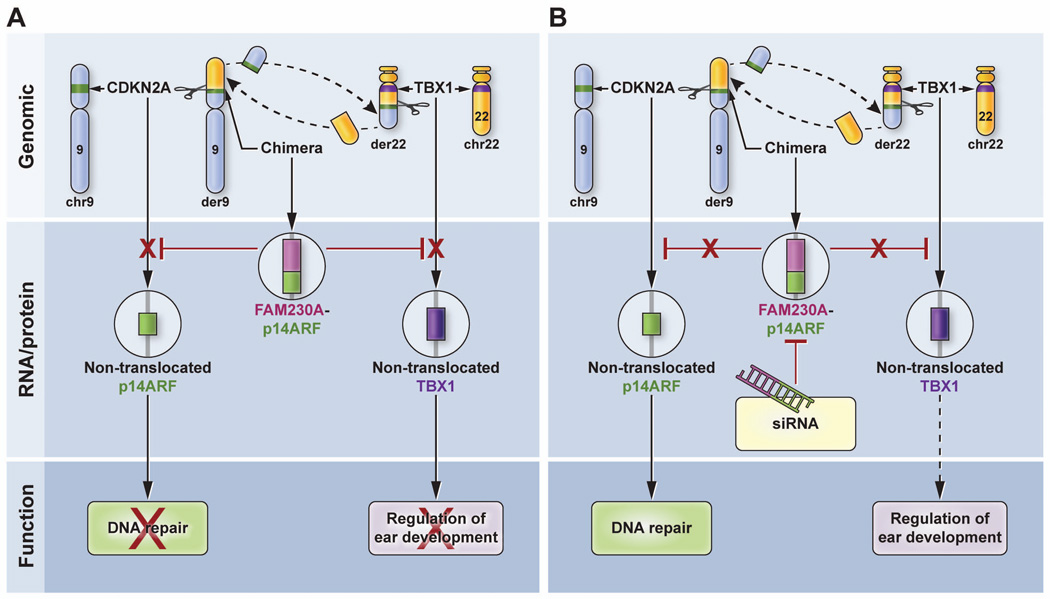
Model for the action of chimeric p14arf-FAM230A as a dominant negative regulator. A. (Upper panel) The chimeric der9 chromosome is formed by translocation of a portion of the long arm of chromosome 22 (yellow) to the short arm of chromosome 9 (blue) thereby splitting the CDKN2A gene (green band) within p14arf intron 1. The other chromosome 9 is intact with an unaffected CDKN2A gene (green band). The der22 chromosome is formed in a parallel manner. The TBX1 gene (purple band) is located 800kb away from the break and is not involved in the translocation. The other chromosome 22 is unaffected. (Middle panel) Chimeric DGCR mRNA is produced that results in decreased expression of the non-translocated normal p14ARF allele on the other chromosome 9 and decreased expression of both non-mutated TBX1 alleles on chromosome 22. (Lower panel) This dominant negative regulation results in reduced DNA repair and altered fetal development giving rise to DGS-like features. B. Treatment with siRNA (middle panel) results in inhibition of chimeric mRNA which would no longer inhibit expression of p14ARF and TBX1. This would result in increased expression of p14ARF and TBX1. (Lower panel) The increased expression of p14ARF would increase DNA repair and presumably, would facilitate normal ear development.
Dominant negative regulation by the chimera
The level of expression of p14ARF and of TBX1 was reduced in DD129BE cells despite the presence of normal chromosomes 9 and 22 which contain non-translocated p14ARF and TBX1 respectively. This appears to be a dominant negative regulation by the presence of the t(9;22) (Figure 7a). We hypothesized that the chimeric p14ARF-FAM230A protein would mediate negative regulation of p14ARF and TBX1. Consistent with this hypothesis, siRNA treatment of the DD129BE cells produced a reduction in the level of p14ARF-FAM230A mRNA and protein that resulted in a significant increase in expression of the p14ARF and TBX1 genes and an increase in DNA repair capacity (Figure 4f, g and 7b). We then synthesized an expression vector containing nearly full length p14ARF-FAM230A and transfected it into normal cells. This vector produced some of the same changes in the normal cells as in the DD129BE cells, including reduction of expression of p14ARF and reduced DNA repair.
The mechanism of this dominant negative p14ARF-FAM230A regulation of p14ARF and TBX1 is not known. The p14ARF portion of the chimeric cDNA has a high homology with p19ARF super family while the FAM230A portion has a high homology with Ret finger protein-like 3 antisense (RFPL3)(Bonnefont et al., 2008; Seroussi et al., 1999) [MIM# 605970]. Since the RFPL3 gene may have a role in the antisense regulation of the RFPL genes which are thought to mediate protein-protein interactions, we proposed that FAM230A might have similar antisense regulation of the protein-protein interactions as the RFPL genes. Future studies will be needed to explore the role of p14ARF-FAM230A and the complex relationship between the regulation of p14ARF, TBX1 and beta-catenin (Damalas et al., 2011; Huh and Ornitz, 2010) [MIM# 116806]. Interestingly, the t(8,21) fusion protein, AML1-ETO associated with acute myeloid leukemia represses transcription of p14ARF by binding to its promoter (Linggi et al., 2002).
Dominant negative inhibition of p14ARF by the chimera resulting in reduced DNA repair and melanoma
The role of CDKN2A, which codes for p16ink4a and p14ARF, in melanoma is well documented (Merlino, 2008; Udayakumar et al., 2010). DD129BE cells have one truncated p14ARF allele as a result of the translocation. While the other p14ARF allele is intact (Supp. Figure S1a and b and Figure 7a), the cells nevertheless have low levels of p14ARF. The reduced post-UV DNA repair capability could be corrected by transfection of p14ARF but not by any of the known XP complementation group genes, suggesting an essential role of the low levels of p14ARF in the DNA repair defect (Figure 4f and g). Consistent with our results, mouse cells with deficient ARF have reduced DNA repair (Sarkar-Agrawal et al., 2004) and mice lacking p19ARF are highly susceptible to UV-induced melanoma (Merlino, 2008). This is similar to the situation in patients with xeroderma pigmentosum who have markedly increased frequency of post-UV mutations and melanomas in association with defective DNA repair (DiGiovanna and Kraemer, 2012). Prior reports that ARF is involved in regulation of XPC (Dominguez-Brauer et al., 2009) are supported by this study in that transfection of DD129BE cells with XPC cDNA resulted in partial improvement in the DNA repair defect (Figure 4).
The melanoma from patient DD129BE had an intact p16INK4a but a truncated and mutated p14ARF demonstrating a crucial role for p14ARF in his melanoma. As with patients with xeroderma pigmentosum, the reduced DNA repair resulted in UV-type mutations and an increased frequency of melanomas (Wang et al., 2009). Laser capture microdissection followed by DNA sequencing revealed two UV-type missense p14ARF point mutations in the melanoma cells but not in the adjacent normal cells. Our data support a two-hit model for the melanoma in this patient in which one copy of p14ARF is disrupted by the translocation followed by mutations that affect the other allele. Genetic alteration of the p14ARF gene by t(9;22) alters DNA repair capability and suggests a possible therapeutic approach to melanoma control in patient DD129BE by use of siRNA directed toward p14ARF-FAM230A.
Dominant negative regulation of TXB1 and the DGS phenotype
The great majority of DGS cases results from a heterozygous micro-deletion in the 3-Mb DGS critical region (DGCR) in chromosome 22q11.2, making it difficult to characterize the DGS candidate gene(s)(Burn et al., 1993; Driscoll et al., 1992; Eymin et al., 2006; Greenberg et al., 1984; Scambler et al., 1992). Balanced translocations of t(2;22)(q14;q11.21)(Budarf et al., 1995) and of t(21;22) (Holmes et al., 1997) had features of partial DGS. The t(9;22) in this study is the third case of translocation involving the 22q11.2 region (Supp. Figure S8c) with features of DGS (Supp. Table S3).
Transcription factor TBX1, located in the 22q11.2 region has been implicated as a cause of DGS (Yagi et al., 2003; Braunstein et al., 2009; Jerome and Papaioannou, 2001; Scambler, 2010). TBX1 has a DNA binding T-box region and is involved in fetal development. Interestingly, mouse models with TBX1 over- or under-expression recapitulate the DGS/velocardiofacial syndrome (VCFS) phenotype (Liao et al., 2004). TBX1 has been shown to be involved in development of the cochlea (Arnold et al., 2006b; Arnold et al., 2006a; Braunstein et al., 2008; Braunstein et al., 2009; Jerome and Papaioannou, 2001). About 95% of the DGS patients have deletions in the 22q11.2 region and some of those patients without deletions have been reported to have mutations in the TBX1 gene (Yagi et al., 2003). DD129BE cells did not have a mutation or deletion in TBX1 but did have an extremely decreased level of TBX1 expression while other genes in the 3-Mb DGCR had normal expression. This reduced TBX1 expression, along with the profound hearing loss, suggests that TBX1 is a critical player in the formation of the cochleovestibula and the inner ear sensory organ in humans. While others have proposed using TBX1 FISH probe for diagnosis of DGS (Beaujard et al., 2009), consideration can also be given to analysis of TBX1 mRNA or protein expression (Figure 5a and b) in the absence of large deletions or mutations.
PATRR22 lies within intron 1 of the new gene FAM230A in the unclonable gap of 22q11.2
Chromosome 22q11.2 contains extremely repetitive regions comprised of 8 chromosome-specific low-copy DNA repeats (LCRs) A–H. The translocation-associated palindromic AT-rich repeat region on chromosome 22 (PATRR22)(Kato et al., 2012) is located in a 50-kb gap in LCR22-B that has been difficult to sequence. PATRR 22 plays an essential role in mediating clinically relevant rearrangements (Supp. Figure S8). A 595-bp PATRR22 was cloned (Kurahashi et al., 2007) and an approximately 10-kb region surrounding PATRR22 was reconstructed. However, little is known about the function of PATRR22.
Using information from the genomic and transcript sequences of der(9) and der(22), we detected a new gene, FAM230A, with at least 10 exons with high homology to the predicted gene LOC653203 (Supp. Figure S7). PATRR22 resides in intron 1 of FAM230A (Supp. Figure S6 and S7). By combining the sequences of der(9) and der(22) we were able to expand the 13-kb PATRR22 surrounding sequence to about 21 kb (Supp. Figure S6).
To date, five constitutional translocations involving PATRR22 have been reported (Gimelli et al., 2009; Gotter et al., 2004; Gotter et al., 2007; Kehrer-Sawatzki et al., 1997; Kurahashi et al., 2000; Kurahashi et al., 2003; Kurahashi et al., 2007; Ledbetter et al., 1989; Nimmakayalu et al., 2003; Sheridan et al., 2010; Kurahashi and Emanuel, 2001b) (Supp. Figure S8f and Supp. Table S4). The clinical phenotype of the PATRR22-associated translocations is widely variable (Supp. Table S4). Translocations were found in somatic cells and in neoplasms. For example a high rate of recurrent translocation t(11;22) was detected in sperm from normal males (Kurahashi and Emanuel, 2001b). Interestingly, the types of partner sequences of PATRR22 are different between recurrent and non-recurrent translocations. The partner sequences on recurrent translocation are PATRR and on non- recurrent translocations are non-PATRR suggesting that the PATRR structures are more susceptible to translocation. A possible explanation for the distinct phenotypes of disruption of PATRR22 might be that the individual positions of the translocation in PATRR22 result in different fusion genes or regulatory sequences which could be responsible for influencing gene expression. Our data suggests that the novel fusion gene in DD129BE regulates the expression of both p14ARF and TBX1. On the other hand, it is possible that the small deletions of less than 100-bp in or surrounding the PATRR22 (Figure 2) might be contributing to the different phenotypes. Since the PATRR22 sequence in FAM230A is a site of frequent translocations and of micro-deletions in DGS patients, these genetic alterations might create new chimeric mRNAs (Guo et al., 2011; Kato et al., 2012; Kurahashi et al., 2007). Chimeric mRNAs derived from FAM230A may mediate haploinsufficiency in DGS or dominant negative inhibition of other genes such as those involved in melanoma.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Cancer Institute, the National Institute of Deafness and Communication Disorders, and the Clinical Center, National Institutes of Health, Bethesda, MD. We thank Drs. Paul Bergstresser and Clay Cockerell for assistance in care of the patient, Drs. Vladimir Larionov, Robert Walker, and Alexander Kovalchuk for assistance in molecular analysis, Drs. Medha Bhagwhat and Paul Khavari for bioinformatics advice, Dr. Doris Wu for expertise in mouse ear development and Drs. Kurt Kohn and Allan Balmian for metabolic pathway analysis.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
DISCLOSURE DECLARATION: The authors have no conflicts of interest.
REFERENCES
- Anzick SL, Chen WD, Park Y, Meltzer P, Bell D, El-Naggar AK, Kaye FJ. Unfavorable prognosis of CRTC1-MAML2 positive mucoepidermoid tumors with CDKN2A deletions. Genes Chromosomes Cancer. 2010;49:59–69. doi: 10.1002/gcc.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JS, Braunstein EM, Ohyama T, Groves AK, Adams JC, Brown MC, Morrow BE. Tissue-specific roles of Tbx1 in the development of the outer, middle and inner ear, defective in 22q11DS patients. Hum Mol Genet. 2006a;15:1629–1639. doi: 10.1093/hmg/ddl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JS, Werling U, Braunstein EM, Liao J, Nowotschin S, Edelmann W, Hebert JM, Morrow BE. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development. 2006b;133:977–987. doi: 10.1242/dev.02264. [DOI] [PubMed] [Google Scholar]
- Beaujard MP, Chantot S, Dubois M, Keren B, Carpentier W, Mabboux P, Whalen S, Vodovar M, Siffroi JP, Portnoi MF. Atypical deletion of 22q11.2: detection using the FISH TBX1 probe and molecular characterization with high-density SNP arrays. Eur J Med Genet. 2009;52:321–327. doi: 10.1016/j.ejmg.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Bonnefont J, Nikolaev SI, Perrier AL, Guo S, Cartier L, Sorce S, Laforge T, Aubry L, Khaitovich P, Peschanski M, Antonarakis SE, Krause KH. Evolutionary forces shape the human RFPL1,2,3 genes toward a role in neocortex development. Am J Hum Genet. 2008;83:208–218. doi: 10.1016/j.ajhg.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford PT, Goldstein AM, Tamura D, Khan SG, Ueda T, Boyle J, Oh KS, Imoto K, Inui H, Moriwaki S, Emmert S, Pike KM, Raziuddin A, Plona TM, DiGiovanna JJ, Tucker MA, Kraemer KH. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J Med Genet. 2011;48:168–176. doi: 10.1136/jmg.2010.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein EM, Crenshaw EB, III, Morrow BE, Adams JC. Cooperative function of Tbx1 and Brn4 in the periotic mesenchyme is necessary for cochlea formation. J Assoc Res Otolaryngol. 2008;9:33–43. doi: 10.1007/s10162-008-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein EM, Monks DC, Aggarwal VS, Arnold JS, Morrow BE. Tbx1 and Brn4 regulate retinoic acid metabolic genes during cochlear morphogenesis. BMC Dev Biol. 2009;9:31. doi: 10.1186/1471-213X-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budarf ML, Collins J, Gong W, Roe B, Wang Z, Bailey LC, Sellinger B, Michaud D, Driscoll DA, Emanuel BS. Cloning a balanced translocation associated with DiGeorge syndrome and identification of a disrupted candidate gene. Nat Genet. 1995;10:269–278. doi: 10.1038/ng0795-269. [DOI] [PubMed] [Google Scholar]
- Burn J, Takao A, Wilson D, Cross I, Momma K, Wadey R, Scambler P, Goodship J. Conotruncal anomaly face syndrome is associated with a deletion within chromosome 22q11. J Med Genet. 1993;30:822–824. doi: 10.1136/jmg.30.10.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts SC, Tatum SA, III, Mortelliti AJ, Shprintzen RJ. Velo-cardio-facial syndrome: the pediatric otolaryngologist's perspective. Curr Opin Otolaryngol Head Neck Surg. 2005;13:371–375. doi: 10.1097/01.moo.0000186203.53214.ac. [DOI] [PubMed] [Google Scholar]
- Damalas A, Velimezi G, Kalaitzakis A, Liontos M, Papavassiliou AG, Gorgoulis V, Angelidis C. Loss of p14(ARF) confers resistance to heat shock- and oxidative stress-mediated cell death by upregulating beta-catenin. Int J Cancer. 2011;128:1989–1995. doi: 10.1002/ijc.25510. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Smith UM, Cullinane AR, Gerrelli D, Cox P, Badano JL, Blair-Reid S, Sriram N, Katsanis N, Attie-Bitach T, Afford SC, Copp AJ, Kelly DA, Gull K, Johnson CA. The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum Mol Genet. 2007;16:173–186. doi: 10.1093/hmg/ddl459. [DOI] [PubMed] [Google Scholar]
- Debeer P, Mols R, Huysmans C, Devriendt K, Van de Ven WJ, Fryns JP. Involvement of a palindromic chromosome 22-specific low-copy repeat in a constitutional t(X; 22)(q27;q11) Clin Genet. 2002;62:410–414. doi: 10.1034/j.1399-0004.2002.620510.x. [DOI] [PubMed] [Google Scholar]
- Digilio MC, Pacifico C, Tieri L, Marino B, Giannotti A, Dallapiccola B. Audiological findings in patients with microdeletion 22q11 (di George/velocardiofacial syndrome) Br J Audiol. 1999;33:329–333. doi: 10.3109/03005369909090116. [DOI] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132:785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Brauer C, Chen YJ, Brauer PM, Pimkina J, Raychaudhuri P. ARF stimulates XPC to trigger nucleotide excision repair by regulating the repressor complex of E2F4. EMBO Rep. 2009;10:1036–1042. doi: 10.1038/embor.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DA, Budarf ML, Emanuel BS. A genetic etiology for DiGeorge syndrome: consistent deletions and microdeletions of 22q11. Am J Hum Genet. 1992;50:924–933. [PMC free article] [PubMed] [Google Scholar]
- Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, Khochbin S, Gazzeri S. p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol Cell Biol. 2006;26:4339–4350. doi: 10.1128/MCB.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg DE, Rigas SH, Russak J, Gai W, Kaplow M, Osman I, Turner F, Randerson-Moor JA, Houghton A, Busam K, Timothy BD, Bastian BC, Newton-Bishop JA, Polsky D. Frequent p16-independent inactivation of p14ARF in human melanoma. J Natl Cancer Inst. 2008;100:784–795. doi: 10.1093/jnci/djn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelli S, Beri S, Drabkin HA, Gambini C, Gregorio A, Fiorio P, Zuffardi O, Gemmill RM, Giorda R, Gimelli G. The tumor suppressor gene TRC8/RNF139 is disrupted by a constitutional balanced translocation t(8;22)(q24.13;q11.21) in a young girl with dysgerminoma. Mol Cancer. 2009;8:52. doi: 10.1186/1476-4598-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Nimmakayalu MA, Jalali GR, Hacker AM, Vorstman J, Conforto DD, Medne L, Emanuel BS. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Shaikh TH, Budarf ML, Rhodes CH, Emanuel BS. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum Mol Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg F, Crowder WE, Paschall V, Colon-Linares J, Lubianski B, Ledbetter DH. Familial DiGeorge syndrome and associated partial monosomy of chromosome 22. Hum Genet. 1984;65:317–319. doi: 10.1007/BF00291554. [DOI] [PubMed] [Google Scholar]
- Guo T, McDonald-McGinn D, Blonska A, Shanske A, Bassett AS, Chow E, Bowser M, Sheridan M, Beemer F, Devriendt K, Swillen A, Breckpot J, Digilio MC, Marino B, Dallapiccola B, Carpenter C, Zheng X, Johnson J, Chung J, Higgins AM, Philip N, Simon TJ, Coleman K, Heine-Suner D, Rosell J, Kates W, Devoto M, Goldmuntz E, Zackai E, Wang T, Shprintzen R, Emanuel B, Morrow B. Genotype and cardiovascular phenotype correlations with TBX1 in 1,022 velo-cardio-facial/DiGeorge/22q11.2 deletion syndrome patients. Hum Mutat. 2011;32:1278–1289. doi: 10.1002/humu.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha L, Ichikawa T, Anver M, Dickins R, Lowe S, Sharpless NE, Krimpenfort P, DePinho RA, Bennett DC, Sviderskaya EV, Merlino G. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci U S A. 2007;104:10968–10973. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland M, Taylor CF, Chambers PA, Kukalizch K, Randerson-Moor JA, Gruis NA, de Snoo FA, ter Huurne JA, Goldstein AM, Tucker MA, Bishop DT, Bishop JA. A mutation hotspot at the p14ARF splice site. Oncogene. 2005;24:4604–4608. doi: 10.1038/sj.onc.1208678. [DOI] [PubMed] [Google Scholar]
- Hewitt C, Lee WC, Evans G, Howell A, Elles RG, Jordan R, Sloan P, Read AP, Thakker N. Germline mutation of ARF in a melanoma kindred. Hum Mol Genet. 2002;11:1273–1279. doi: 10.1093/hmg/11.11.1273. [DOI] [PubMed] [Google Scholar]
- Holmes SE, Riazi MA, Gong W, McDermid HE, Sellinger BT, Hua A, Chen F, Wang Z, Zhang G, Roe B, Gonzalez I, Donald-McGinn DM, Zackai E, Emanuel BS, Budarf ML. Disruption of the clathrin heavy chain-like gene (CLTCL) associated with features of DGS/VCFS: a balanced (21;22)(p12;q11) translocation. Hum Mol Genet. 1997;6:357–367. doi: 10.1093/hmg/6.3.357. [DOI] [PubMed] [Google Scholar]
- Huh SH, Ornitz DM. Beta-catenin deficiency causes DiGeorge syndrome-like phenotypes through regulation of Tbx1. Development. 2010;137:1137–1147. doi: 10.1242/dev.045534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS3, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types [see comments] Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Kato T, Kurahashi H, Emanuel BS. Chromosomal translocations and palindromic AT-rich repeats. Curr Opin Genet Dev. 2012 doi: 10.1016/j.gde.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui H, Ueda T, Emmert S, Schneider TD, Kraemer KH. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res. 2002;30:3624–3631. doi: 10.1093/nar/gkf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2001a;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet. 2001b;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Inagaki H, Hosoba E, Kato T, Ohye T, Kogo H, Emanuel BS. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh T, Takata M, Toda T, Emanuel BS. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J Hum Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- Ledbetter DH, Rich DC, O'Connell P, Leppert M, Carey JC. Precise localization of NF1 to 17q11.2 by balanced translocation. Am J Hum Genet. 1989;44:20–24. [PMC free article] [PubMed] [Google Scholar]
- Li MM, Nimmakayalu MA, Mercer D, Andersson HC, Emanuel BS. Characterization of a cryptic 3.3 Mb deletion in a patient with a "balanced t(15;22) translocation" using high density oligo array CGH and gene expression arrays. Am J Med Genet A. 2008;146:368–375. doi: 10.1002/ajmg.a.32116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, Epstein JA, Brown MC, Adams J, Morrow BE. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13:1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Linggi B, Muller-Tidow C, van de Locht L, Hu M, Nip J, Serve H, Berdel WE, van der Reijden B, Quelle DE, Rowley JD, Cleveland J, Jansen JH, Pandolfi PP, Hiebert SW. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- Magro PG, Russo AJ, Li WW, Banerjee D, Bertino JR. p14ARF expression increases dihydrofolate reductase degradation and paradoxically results in resistance to folate antagonists in cells with nonfunctional p53. Cancer Res. 2004;64:4338–4345. doi: 10.1158/0008-5472.CAN-03-1045. [DOI] [PubMed] [Google Scholar]
- Merlino G. The ARFul truth about melanoma susceptibility genes. Pigment Cell Melanoma Res. 2008;21:503–504. doi: 10.1111/j.1755-148X.2008.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmakayalu MA, Gotter AL, Shaikh TH, Emanuel BS. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum Mol Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- Ohtani I, Schuknecht HF. Temporal bone pathology in DiGeorge's syndrome. Ann Otol Rhinol Laryngol. 1984;93:220–224. doi: 10.1177/000348948409300306. [DOI] [PubMed] [Google Scholar]
- Oskarsdottir S, Vujic M, Fasth A. Incidence and prevalence of the 22q11 deletion syndrome: a population-based study in Western Sweden. Arch Dis Child. 2004;89:148–151. doi: 10.1136/adc.2003.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenne P, Eymin B, Brambilla E, Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer. 2010;127:2239–2247. doi: 10.1002/ijc.25511. [DOI] [PubMed] [Google Scholar]
- Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet 12 Spec No. 2003;1:R37–R44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- Padilla-Nash HM, Barenboim-Stapleton L, Difilippantonio MJ, Ried T. Spectral karyotyping analysis of human and mouse chromosomes. Nat Protoc. 2006;1:3129–3142. doi: 10.1038/nprot.2006.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerson-Moor JA, Harland M, Williams S, Cuthbert-Heavens D, Sheridan E, Aveyard J, Sibley K, Whitaker L, Knowles M, Bishop JN, Bishop DT. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10:55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- Reyes MR, LeBlanc EM, Bassila MK. Hearing loss and otitis media in velo-cardio-facial syndrome. Int J Pediatr Otorhinolaryngol. 1999;47:227–233. doi: 10.1016/s0165-5876(98)00180-3. [DOI] [PubMed] [Google Scholar]
- Rizos H, Darmanian AP, Holland EA, Mann GJ, Kefford RF. Mutations in the INK4a/ARF melanoma susceptibility locus functionally impair p14ARF. J Biol Chem. 2001a;276:41424–41434. doi: 10.1074/jbc.M105299200. [DOI] [PubMed] [Google Scholar]
- Rizos H, Puig S, Badenas C, Malvehy J, Darmanian AP, Jimenez L, Mila M, Kefford RF. A melanoma-associated germline mutation in exon 1beta inactivates p14ARF. Oncogene. 2001b;20:5543–5547. doi: 10.1038/sj.onc.1204728. [DOI] [PubMed] [Google Scholar]
- Sarkar-Agrawal P, Vergilis I, Sharpless NE, DePinho RA, Runger TM. Impaired processing of DNA photoproducts and ultraviolet hypermutability with loss of p16INK4a or p19ARF. J Natl Cancer Inst. 2004;96:1790–1793. doi: 10.1093/jnci/djh307. [DOI] [PubMed] [Google Scholar]
- Scambler PJ. 22q11 deletion syndrome: a role for TBX1 in pharyngeal and cardiovascular development. Pediatr Cardiol. 2010;31:378–390. doi: 10.1007/s00246-009-9613-0. [DOI] [PubMed] [Google Scholar]
- Scambler PJ, Kelly D, Lindsay E, Williamson R, Goldberg R, Shprintzen R, Wilson DI, Goodship JA, Cross IE, Burn J. Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet. 1992;339:1138–1139. doi: 10.1016/0140-6736(92)90734-k. [DOI] [PubMed] [Google Scholar]
- Seroussi E, Kedra D, Pan HQ, Peyrard M, Schwartz C, Scambler P, Donnai D, Roe BA, Dumanski JP. Duplications on human chromosome 22 reveal a novel Ret Finger Protein-like gene family with sense and endogenous antisense transcripts. Genome Res. 1999;9:803–814. doi: 10.1101/gr.9.9.803. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, Donald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, O'Connor RJ, Pierpont ME, McGrath J, Hacker AM, Nimmakayalu M, Geiger E, Emanuel BS, Saitta SC. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan MB, Kato T, Haldeman-Englert C, Jalali GR, Milunsky JM, Zou Y, Klaes R, Gimelli G, Gimelli S, Gemmill RM, Drabkin HA, Hacker AM, Brown J, Tomkins D, Shaikh TH, Kurahashi H, Zackai EH, Emanuel BS. A palindrome-mediated recurrent translocation with 3:1 meiotic nondisjunction: the t(8;22)(q24.13;q11.21) Am J Hum Genet. 2010;87:209–218. doi: 10.1016/j.ajhg.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Bertwistle D, DEN BW, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ. Velo-cardio-facial syndrome: 30 Years of study. Dev Disabil Res Rev. 2008;14:3–10. doi: 10.1002/ddrr.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon R, Stone G. Phylogenomic analysis by chromosome sorting and painting. Methods Mol Biol. 2008;422:13–29. doi: 10.1007/978-1-59745-581-7_2. [DOI] [PubMed] [Google Scholar]
- Swillen A, Devriendt K, Legius E, Prinzie P, Vogels A, Ghesquiere P, Fryns JP. The behavioural phenotype in velo-cardio-facial syndrome (VCFS): from infancy to adolescence. Genet Couns. 1999;10:79–88. [PubMed] [Google Scholar]
- Tapia-Paez I, Kost-Alimova M, Hu P, Roe BA, Blennow E, Fedorova L, Imreh S, Dumanski JP. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- Udayakumar D, Mahato B, Gabree M, Tsao H. Genetic determinants of cutaneous melanoma predisposition. Semin Cutan Med Surg. 2010;29:190–195. doi: 10.1016/j.sder.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli F, Viola A, Morishima M, Pramparo T, Baldini A, Lindsay E. TBX1 is required for inner ear morphogenesis. Hum Mol Genet. 2003;12:2041–2048. doi: 10.1093/hmg/ddg216. [DOI] [PubMed] [Google Scholar]
- Wang Y, DiGiovanna JJ, Stern JB, Hornyak TJ, Raffeld M, Khan SG, Oh KS, Hollander MC, Dennis PA, Kraemer KH. Evidence of ultraviolet type mutations in xeroderma pigmentosum melanomas. Proc Natl Acad Sci U S A. 2009;106:6279–6284. doi: 10.1073/pnas.0812401106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Strom SS, Wang LE, Guo Z, Qiao Y, Amos CI, Spitz MR, Duvic M. Repair of UV light-induced DNA damage and risk of cutaneous malignant melanoma. J Natl Cancer Inst. 2003;95:308–315. doi: 10.1093/jnci/95.4.308. [DOI] [PubMed] [Google Scholar]
- Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.