Abstract
A hexamer of the bacteriophage T4 tail terminator protein, gp15, attaches to the top of the phage tail stabilizing the contractile sheath and forming the interface for binding of the independently assembled head. Here we report the crystal structure of the gp15 hexamer, describe its interactions in T4 virions that have either an extended tail or a contracted tail, and discuss its structural relationship to other phage proteins. The neck of T4 virions is decorated by the “collar” and “whiskers”, made of fibritin molecules. Fibritin acts as a chaperone helping to attach the long tail fibers to the virus during the assembly process. The collar and whiskers are environment-sensing devices, regulating the retraction of the long tail fibers under unfavorable conditions, thus preventing infection. Cryo-electron microscopy analysis suggests that twelve fibritin molecules attach to the phage neck with six molecules forming the collar and six molecules forming the whiskers.
Keywords: bacteriophage T4; fibritin; gpwac; T4 collar and whiskers; tail terminator protein, gp15
Introduction
Bacteriophage T4 has long served as a model system for molecular and structural biology. Its structure and assembly have been extensively studied using biochemical methods, cryo-electron microscopy (cryo-EM), and X-ray crystallography (for recent reviews, see Refs. 1–3). T4 is a lytic phage that uses Escherichia coli as a host and belongs to the Myoviridae family, characterized by contractile tails. The T4 virion (Fig. 1) has a 1200-Å-long and 860-Å-wide head, and a 1000-Å-long tail containing a rigid central tube surrounded by a contractile sheath.4–6 At the distal end of the tail, there is a multiprotein baseplate7 to which six long and six short tail fibers are attached. A different set of fibers attach to the neck region of T4 virions, near the head-to-tail interface, forming the phage collar and whiskers.8,9
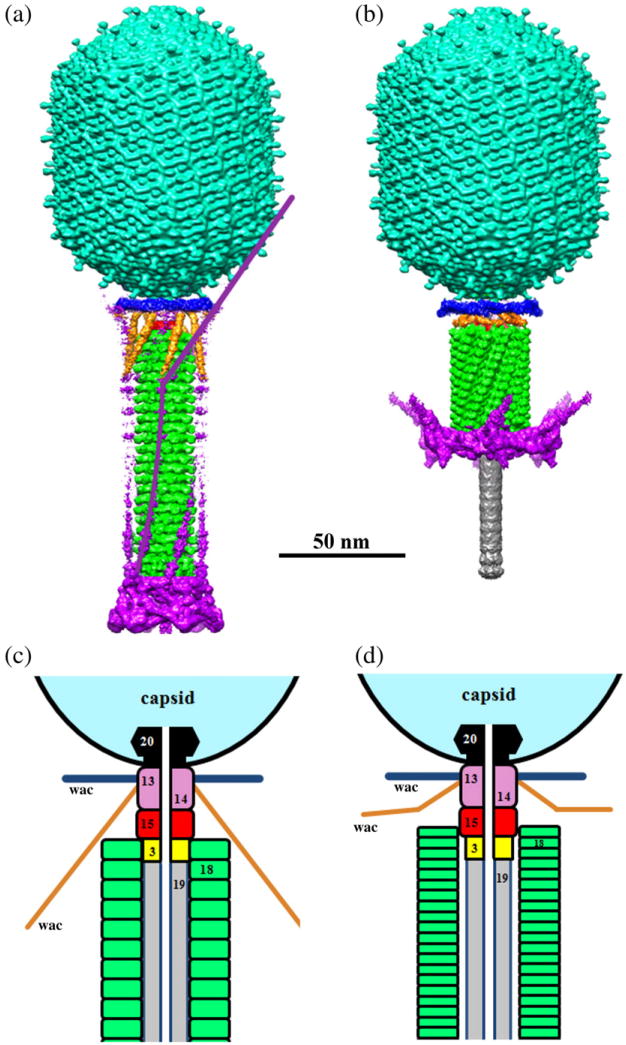
Fig. 1.
Structure of bacteriophage T4. (a and b) Surface-shaded views of T4 virions with (a) extended and (b) contracted tails. The figures are based on the cryo-EM reconstructions of the T4 capsid,4 extended tail,6 and contracted tail. The capsid, tail sheath, and tail tube are colored cyan, green, and gray, respectively. The phage collar is colored blue and whiskers are colored gold. Density corresponding to the baseplate and long tail fibers is colored magenta. One retracted long tail fiber is outlined in (a) by the magenta continuous line. (c and d) Schematic representations of the bacteriophage T4 neck and a part of the tail in the extended (c) and contracted (d) forms. Regions occupied by different proteins are in different colors. The portal protein gp20 is in black; the tail tube, formed by gp19, is in gray; the tail tube terminator protein, gp3, is in yellow; the discs of the tail sheath protein, gp18, are in green; the tail terminator protein, gp15, is in red; gp13 and gp14 are in pink.
The T4 head and tail are assembled via independent pathways. Assembly of the T4 head is a complex process that includes a number of intermediate stages.2,10,11 The head assembly is initiated by the dodecamer of the portal protein, gp20 (gp, gene product). First, a head precursor, called the prohead, is assembled, which is subsequently processed by a scaffold-associated protease. Then the phage genomic DNA is packaged into the capsid through the portal vertex by an ATP-driven motor composed of five gp17 molecules.12 Upon completion of the DNA packaging, the head assembly is finalized by attachment of several copies of the gp13 and gp14 proteins to the portal vertex.9,13 Monomers of gp13 and gp14 have a size of 309 and 256 amino acid (aa) residues, respectively. The gp13–gp14 complex seals the portal vertex and creates a site for attachment of the independently assembled tail. Mutant phages lacking these proteins produce heads that are unable to bind tails and lose their DNA.14
The T4 tail assembly begins with the baseplate formation and proceeds with polymerization of the tail tube and the contractile sheath. The tail tube is formed by gp19 molecules (163 aa residues) and has external and internal diameters of 90 Å and 40 Å, respectively. The length of the tube is controlled by a mechanism involving the “tape-measure protein”, gp29. The elongation of the tail tube is terminated by attachment of the hexamer of the 175-residue tail tube terminator protein, gp3, which binds to the last row of gp19 subunits (probably also to gp29) and stabilizes the tail tube.15,16 The tail tube assembly in the Myoviridae phages is similar to that of the Siphoviridae phages that have long non-contractile tails. The tail tube proteins of the Myoviridae phages have a fold that has similarity to the tail tube proteins of Siphoviridae phages and to the tube proteins of the bacterial secretion system VI.17–19 The T4 tail tube is used as a scaffold for the polymerization of the contractile sheath. The gp18 sheath molecules (659 aa residues) assemble around the tube in the form of a six-start helix. The assembled sheath has 138 gp18 molecules arranged into 23 hexameric rings, stacked onto each other. Each ring isrotatedby17.2° and translated by 40.6 Å relative to the previous ring. The structure of the gp18M deletion mutant that consisted of three out of four protein domains was determined by X-ray crystallography.20 The deletion mutant did not contain the C-terminal domain that interacts with the tail tube. However, X-ray structures of two sheath proteins LIN1278 [Protein Data Bank (PDB) ID: 3LML] and DSY3957 (PDB ID: 3HXL) encoded in prophages of bacteria Listeria innocua and Desulfitobacterium hafniense were reported later.21 The structures are homologous to gp18 and include the C-terminal domains.
The T4 tail assembly is completed by the hexamer of the tail terminator protein, gp15 (the monomer is 272 aa residues long), which binds to the top† of the tail. The gp15 hexamer interacts with the gp3 hexamer and, presumably, with the C-terminal domains of gp18 molecules located in the last ring of the contractile sheath (Fig. 1c). Mutant tails lacking gp3 also lack gp15 and sheaths of the gp15-lacking tails are unstable.22,23
Contraction of the tail during infection is associated with a substantial rearrangement of the gp18 subunits and results in shortening of the sheath to less than one-half of its original length. The contracted sheath is also a six-start helix with a pitch of 16.4 Å and a twist of 32.9°. After the tail contraction, the C-terminal domains of gp18 do not interact with the tail tube. However, the top gp18 ring of the sheath presumably remains in contact with the gp15 hexamer.
The gp15 protein also creates the binding site for attachment of the capsid. Mutant phage tails lacking gp15 are unable to join the heads. The tail binds to the head via interactions of gp15 with gp14 (or with both gp13 and gp14).
At a late stage of phage assembly, the collar and whiskers,9 made of fibritin molecules, attach to the phage neck via the gp13–gp14 complex (Fig. 1). Each 500-Å-long fibritin molecule is a trimer of the 486-residue product of the late gene wac (whisker antigen control).8 Sequence and structural analyses show that most of the fibritin structure consists of coiled-coil segments.24 Biochemical data suggest that the N-terminal domain of fibritin binds to the phage.25 Structures of the N- and C-terminal parts of fibritin, containing globular domains, were determined using X-ray crystallography.26–28 The collar and whiskers serve as molecular chaperons facilitating attachment of the long tail fibers to the phage.29 Fibritin also serves as an environment-sensing device that controls the retraction of the long tail fibers under adverse conditions (e.g., low temperature, low ionic strength), thus preventing undesirable infection.30
Here we report the X-ray structure of the gp15 hexamer and describe its interactions with the T4 neck and tail proteins. We also present a cryo-EM reconstruction of the contracted T4 tail containing the collar and whiskers. The reconstruction suggests that twelve fibritin molecules bind to the phage neck via their N-terminal domains with six molecules forming the collar and six molecules forming the whiskers. The new interpretation of the structure of the collar–whisker complex is different from the previously proposed model6 that assumed six fibritin molecules per virion.
Results and Discussion
Structure of gp15 and its relationship to other phage proteins
The structure of the hexamer of the gp15 protein, which terminates the T4 tail assembly and stabilizes the contractile sheath, has been determined using X-ray crystallography (Fig. 2 and Table 1) to 2.7 Å resolution. The hexamer has a shape resembling a hexagonal nut with a 40-Å pore through which the genomic DNA exits during the infection. The wall of the pore is formed by six 4-stranded antiparallel β-sheets and six α-helices coming from different gp15 monomers. The central pore and the side surface of the gp15 hexamer are negatively charged whereas the top and the bottom surfaces contain positively charged areas (Fig. 3a–c). The top and bottom parts of the hexamer contain flexible regions and loops, some of which (residues 88–104, residues 179–182, and the C-terminal region following residue 234) are disordered in the crystals.
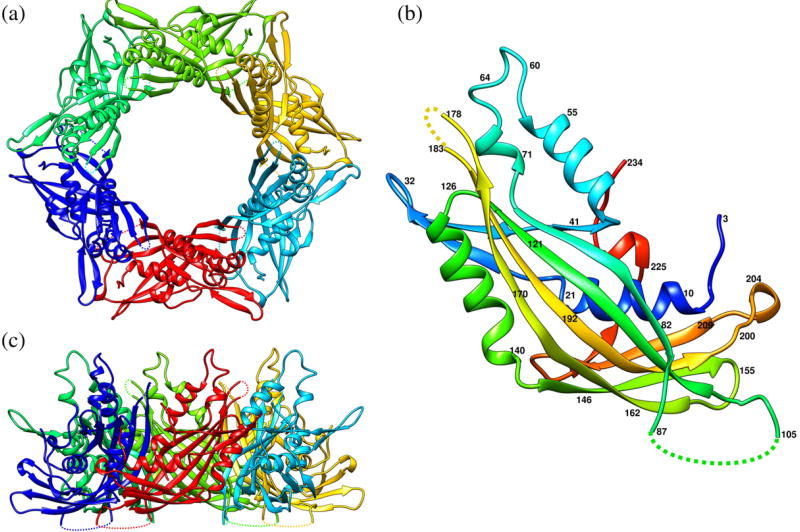
Fig. 2.
Ribbon diagrams of the gp15 protein. (a) and (c) represent the top and side views of the gp15 hexamer, respectively. Different protein chains are shown in different colors. (b) gp15 monomer represented in rainbow color running from N terminus (blue) to C terminus (red). The broken lines indicate protein regions that are disordered in the crystals.
Table 1. X-ray data and refinement statistics.
| gp15 C-terminal truncation mutant (residues 1–261) | ||||
|---|---|---|---|---|
|
|
||||
| Full length gp15 | Triclinic, Pt derivative | |||
|
|
|
|||
| Trigonal, native | Trigonal, native | Peak | Inflection point | |
| X-ray wavelength (Å) | 1.0 | 0.97941 | 1.07195 | 1.07228 |
| Space group | P32 | P1 | P1 | |
| Cell dimensions | ||||
| a, b, c (Å) | 100.68, 100.68, 155.891 | 65.87, 76.31, 93.89 | 65.89, 77.01, 94.16 | |
| α, β, γ (°) | 90, 90, 120 | 109.04, 104.62, 89.30 | 109.01, 104.43, 88.94 | |
| Resolutiona (Å) | 2.7 (2.75–2.7) | 3.2 (3.31–3.2) | 3.6 (3.73–3.6) | 3.6 (3.73–3.6) |
| Rmergea | 0.078 (0.27) | 0.077 (0.21) | 0.076 (0.17) | 0.073 (0.27) |
| I/σa | 20.0 (3.3) | 18.7 (5.9) | 15.9 (5.6) | 17.3 (3.6) |
| Completenessa (%) | 93 (85) | 92 (72) | 79 (58) | 77(51) |
| Redundancya | 5.5 (2.3) | 3.8 (3.5) | 2.3 (2.2) | 2.3 (2.1) |
|
| ||||
| Refinement statistics | ||||
|
| ||||
| Crystal form | Trigonal, native | Triclinic, native | ||
| Resolution (Å) | 2.7 | 3.2 | ||
| No. of reflections | 45,176 | 25,229 | ||
| No. of atoms | ||||
| Protein | 10,452 | 10,145 | ||
| Water | 72 | 0 | ||
| Rwork/Rfree | 0.207/0.252 | 0.171/0.202 | ||
| Root-mean-square deviations | ||||
| Bond length (Å) | 0.010 | 0.009 | ||
| Bond angles (°) | 1.30 | 1.22 | ||
| Average B-factor (Å2) | 64 | 62 | ||
| Ramachandran plot (%) | ||||
| Favored | 90 | 90 | ||
| Outliers | 1.9 | 2.0 | ||
Numbers in parentheses represent values in the highest-resolution shell.
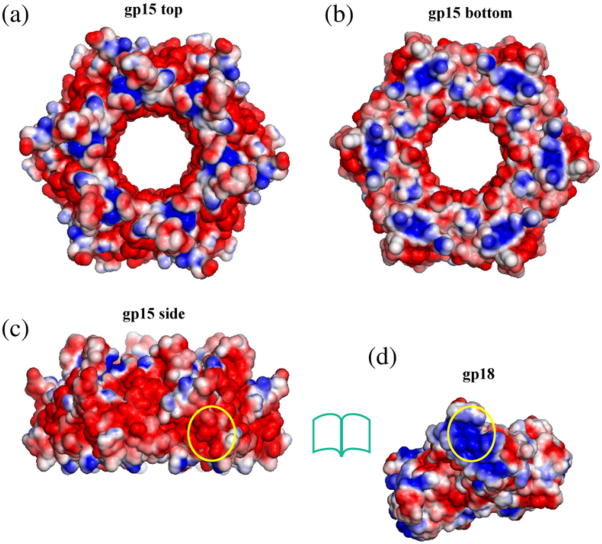
Fig. 3.
Solvent-accessible surfaces of gp15 and gp18 colored according to the electrostatic potential. The blue color corresponds to a potential of 2 kT/e. The red color corresponds to a potential of -2 kT/e. (a), (b), and (c) represent the top, bottom, and side views of the gp15 hexamer, respectively. One region on the side of the gp15 hexamer that faces gp18 in the contracted tail is outlinedby the yellow oval. (d) shows the surface of the gp18 model. The region of the gp18 molecule that faces gp15 in the contracted tail is outlined by the yellow oval. The electrostatic potential was calculated using the APBS software31 and visualized with PyMOL.64
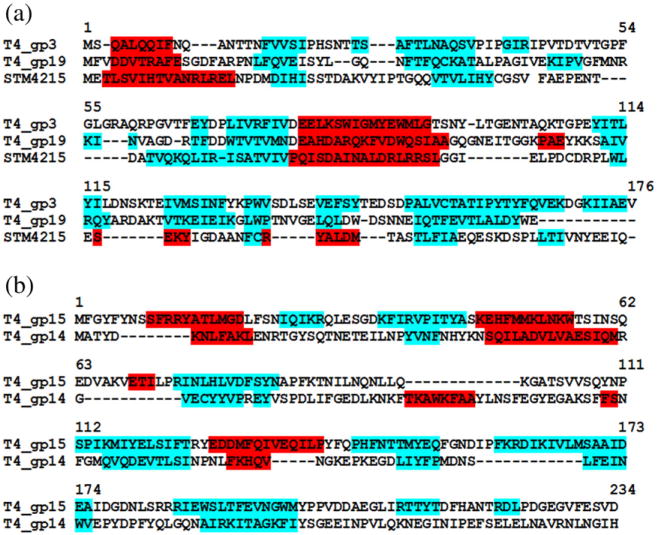
Comparison of the gp15 structure with known structures using the Dali algorithm32 showed that the core of the gp15 molecule is similar to the hypothetical protein STM4215 from Salmonella typhimurium (PDB ID: 2GJV), the gpU protein33 from bacteriophage λ (PDB ID: 3FZ2), and the hypothetical phage-related protein NP_888769.1 from Bordetella bronchiseptica34 (PDB ID: 2L25). Molecules of gp15 and STM4215 can be superimposed with a root-mean-square deviation of 2.4 Å between 107 equivalenced Cα atoms (Fig. 4), although these proteins have rather different sizes (272 and 151 aa residues, respectively) and only 8% sequence identity. The gp15 protein contains large insertions and an additional C-terminal region. Like gp15, the STM4215 and gpU proteins form crystals that have hexameric arrangements of molecules. The relative orientations of monomers in the gp15 hexamer are analogous to those in the STM4215 and gpU hexamers, although the similarity between gp15 and STM4215 hexamers is greater. The structure of the central part of the gp15 hexamer is rather similar to the STM4215 hexamer; however, the gp15 hexamer has additional structural elements in its periphery (Fig. 4b). The gpU protein (131 aa residues) terminates the assembly of the tail tube in the Siphoviridae phage λ. Since the STM4215 and NP_888769.1 molecules (151 and 140 aa residues, respectively) have structures similar to that of gpU, they most likely correspond to the tail tube terminator proteins.35 The STM4215 protein is encoded in a prophage-related region of the Salmonella genome that also contains genes encoding proteins of a contractile tail.33,36 Therefore, STM4215 probably represents a Myoviridae tail tube terminator protein, whereas gpU and NP_888769.1 correspond to the tail tube terminators of the Siphoviridae phages.35
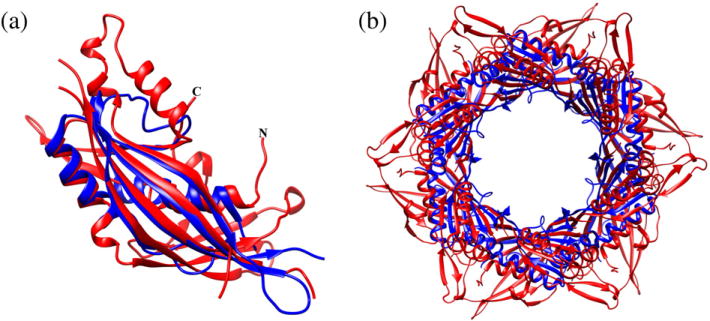
Fig. 4.
Superposition of the STM4215 protein (blue) onto gp15 (red). (a) The STM4215 monomer was superimposed onto the gp15 monomer. (b) Superposition of the gp15 and STMP4215 hexamers.
The size and putative function of the STM4215 protein correspond to those of the T4 tube terminator, gp3 (175 aa residues), which is located in the phage particle between the molecules of the tail tube protein, gp19, and the gp15 hexamer (Fig. 1c). The secondary structure elements of the STM4215 protein align reasonably well with those predicted for gp3 (Fig. 5a). The size of the gp3 hexamer observed by electron microscopy is comparable to that of the STM4215 hexamer.16 Taking into account the structural similarity of gp15 and STM4215, the structure of the gp3 hexamer should be closely related to that of the STM4215 hexamer despite low sequence similarity between these proteins.
Fig. 5.
Comparison of observed and predicted secondary structure elements (helices are colored red, and β-strands are colored cyan). (a) Predicted secondary structure elements for gp3 and gp19 compared to the observed secondary structure elements in the STM4215 protein. (b) Predicted secondary structure elements for gp14 compared to the secondary structure elements observed in the gp15 structure (region 1–234).
In bacteriophage λ, the N-terminal domain of the tail tube protein gpV17 has structural similarity to the gpU protein.33 The T4 tail tube protein, gp19, has size and predicted secondary structure comparable to those of the gp3 and STM4215 proteins (Fig. 5a), suggesting that gp19 structure is probably similar to that of STM4215 and gp3. This assumption is consistent with the hypothesis of common ancestry of the proteins of the phage tail tubes, baseplate hubs, and tubes of the bacterial secretion system VI.1,37 The gp19, gp3, and gp15 proteins probably evolved from an ancestral tail tube protein by gene duplication and diverged to acquire their specific functions. During the evolution process, these proteins diverged substantially in their amino acid sequences, whereas the gp15 protein also gained additional structural elements required for stabilization of the contractile sheath. These proteins must be similar enough to be incorporated into the growing tail tube structure yet be able to perform their specific functions.
Interactions of gp15 with the tail proteins gp3 and gp18
The structure of gp15 was fitted into the cryo-EM reconstructions of the extended and contracted T4 tails (Fig. 6).5,6 The fitting suggests that the periphery of the gp15 hexamer interacts with the C-terminal domain of the tail sheath protein gp18. To examine the interface between the gp18 and gp15 proteins, we created a model of the entire gp18 molecule based on the crystal structure of the gp18 truncation mutant20 (PDB ID: 3FOA) lacking the C-terminal domain and the prophage tail sheath protein LIN1278, encoded in the L. innocua genome (PDB ID: 3LML), which includes the C-terminal domain. Although the C-terminal domains of the gp18 and LIN1278 proteins have only 14% sequence identity, the fitting of the gp15 and gp18 structures into the cryo-EM reconstructions (Fig. 6) made it possible to identify the regions that are probably involved in the gp15–gp18 interactions. In the extended T4 tail, region 572–583 of gp18 probably interacts with region 152–159 of gp15. During the tail contraction, the gp18 molecules dramatically change their positions and detach from the tail tube.5,20 However, the cryo-EM density5 suggests that, after the tail contraction, the C-terminal domains of the gp18 molecules belonging to the topmost gp18 hexamer ring still interact with the gp15 hexamer. Therefore, gp15 helps to maintain the integrity of the contracted tail by keeping the top of the gp18 sheath attached to the rest of the phage particle. The interface between gp18 and gp15 molecules in the contracted tail is different from that of the extended tail. In the contracted tail, regions 153–156 and 203–208 of the gp15 molecule probably interact with regions 578–582 and 606–619 of gp18. The gp15 surface involved in the binding interface is negatively charged whereas the corresponding gp18 surface has positive charge (Fig. 3c and d). Although there could be some uncertainty in the atomic positions, the exact positions of charges are not critical in establishing the nature of the interactions between gp18 and gp15. The resultant electrostatic interaction will help to keep the sheath attached to the phage particle after the sheath has contracted during genome ejection. In the crystal structure of the DSY3957 protein, which is homologous to gp18, 25 N-terminal residues are involved in the formation of a β-sheet within the C-terminal domain of a symmetry-related molecule. The 20 amino-terminal residues of the gp18M protein were disordered in the crystal structure. Leiman and Shneider38 suggested that, in the phage particle, the N-terminal regions of the gp18 molecules may interact with the C-terminal domains of the gp18 molecules located immediately above. Similarly, the N-terminal regions of the gp18 molecules located in the topmost gp18 ring may be involved in the interaction with gp15.
Fig. 6.
Relative positions of the gp15 and gp18 molecules in the extended and contracted T4 tails. The gp15 hexamer is shown in red and the gp18 molecules belonging to the topmost ring of the contractile sheath are shown in green. (a) and (b) show the gp15 and gp18 molecules fitted into the cryo-EM reconstructions of the (a) extended and (b) contracted tails,5,6 represented by the semitransparent surfaces. (c and d) Side view of the gp15 hexamer and gp18 molecules. (e and f) The gp15 and gp18 molecules viewed from the top of the phage.
The changes in the gp15–gp18 interaction during the tail contraction may cause conformational changes in the gp15 hexamer, which in turn may induce changes in the gp13–gp14 complex and the portal gp20 protein, allowing escape of the DNA. Although the comparison of the extended and contracted tail cryo-EM reconstructions suggests that the gp15 pore becomes wider after the tail contraction,6 the rearrangement of the protein subunits cannot be reliably assessed with the present resolution of the cryo-EM reconstructions.
The electrostatic forces probably also play an important role in the binding of the gp15 hexamer to the gp3 hexamer. The bottom part of the gp15 hexamer that interacts with gp3 is mainly positively charged. gp3 is a negatively charged protein and the homology modeling of the gp3 hexamer based on the structure of the STM4215 protein suggests that the top part of the gp3 hexamer that interacts with gp15 contains many negatively charged residues.
Stoichiometry of the gp13–gp14 complex and its interactions with gp15 and the portal
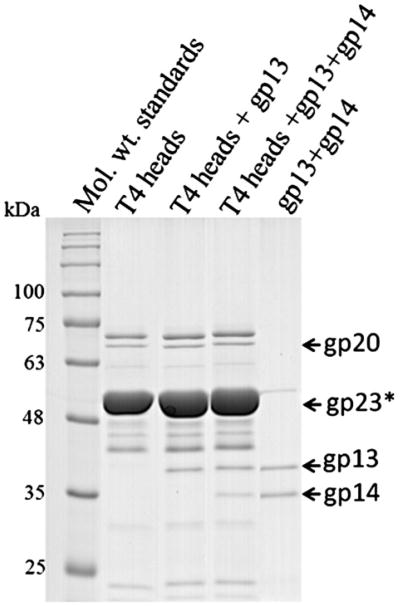
The gp15 hexamer provides the surface for capsid attachment. The top part of the hexamer interacts with the gp13–gp14 complex attached to the portal vertex of the capsid. The exact number of the gp13 and gp14 molecules in the phage is not known. These proteins are monomers in solution and they do not interact with each other under normal physiological conditions.13 However, these proteins can form a complex in vitro when treated with ammonium sulfate.13 Ultracentrifugation analysis suggested that the complex contains ten gp13 and five gp14 molecules. However, the complex assembled on the phage most likely has the stoichiometry (gp13)12–(gp14)6 because, within the phage, it is located between the 12-fold symmetric portal protein, gp20, and the gp15 hexamer. Binding experiments show that the gp13 and gp14 proteins attach in vitro to T4 heads with approximately twelve and six copies per head, respectively (Fig. 7). The cryo-EM reconstruction of the contracted phage5 suggests that the neck region located just below the gp20 portal has a 12-fold symmetry. Therefore, in the virus, the gp13 protein likely forms a dodecamer located just below the gp20 dodecameric portal. Secondary structure prediction and circular dichroism spectra13 show that the gp13 protein has high α-helical content. The Siphoviridae phages HK97, SPP1, and the Podoviridae phage P22 also contain dodecamers of α-helical proteins attached to the portal,39–41 although these proteins have a much smaller size compared to T4 gp13, supporting the conclusion that there are twelve gp13 molecules per phage.
Fig. 7.
Copy number determination of gp13 and gp14 bound to phage T4 heads. Purified gp13 and gp14 were bound to purified phage T4 heads prepared as described earlier.14 See Experimental Procedures for details of binding experiment.
In bacteriophages λ and SPP1, the proteins possessing the head-to-tail connector function, equivalent to that of T4 gp14, have a structural similarity to the tail tube terminator proteins, tail tube proteins,37 and, consequently, to gp15 of T4. The gp14 and gp15 proteins are similar in size and are encoded by consecutive genes in the T4 genome. The secondary structure elements of gp15 are comparable to those predicted for gp14 (Fig. 5b). Therefore, the gp14 protein is likely to have a fold similar to that of the gp15 protein. In the T4 virion, the top surface of the gp15 hexamer has positively charged areas that interact with the large negative charge (estimated as -16 electrons at pH 7) on the gp14 molecule. Therefore, the electrostatic forces also play an important role in attachment of the head to the tail.
Structure of T4 collar and whiskers
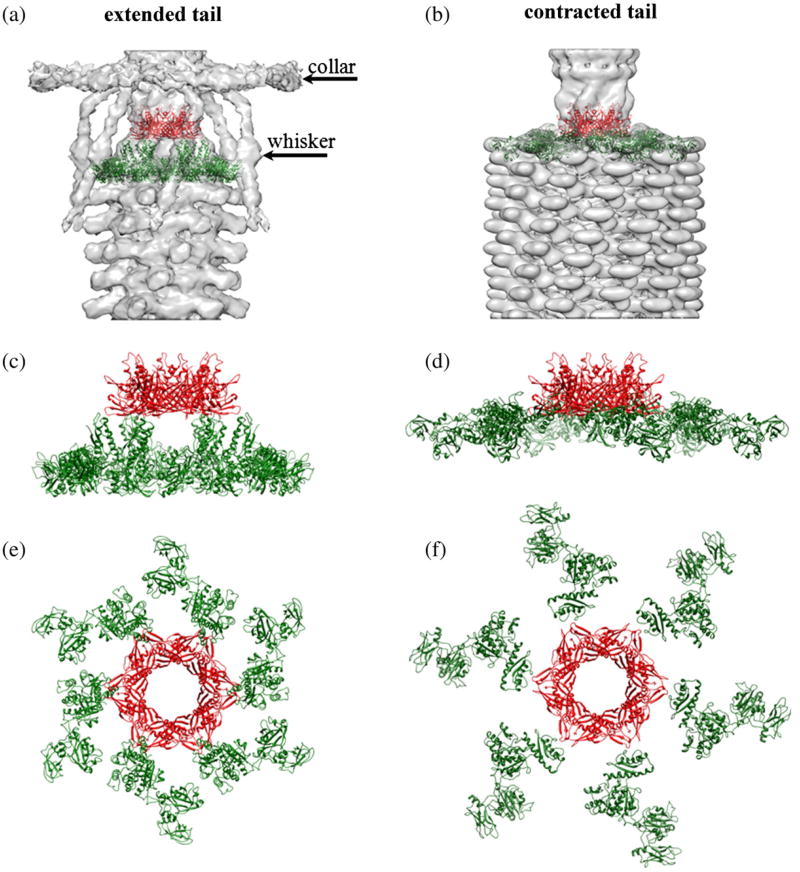
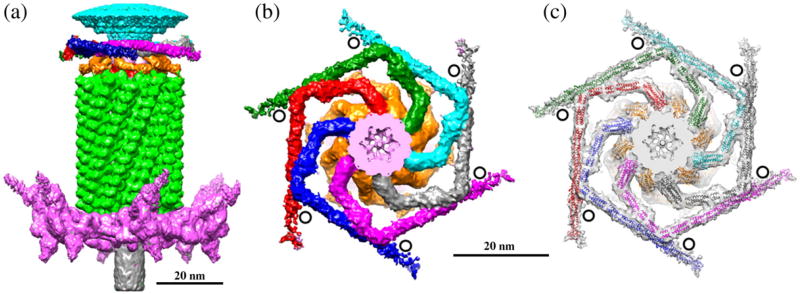
The neck of T4 is decorated with the collar and whiskers formed by fibritin molecules9 that consist of gpwac trimers. The structure of the contracted T4 tail containing the attached collar and whiskers was determined by cryo-EM (Fig. 8) and compared with the cryo-EM reconstruction of the extended tail and the attached collar and whiskers.6 The collar has a similar structure independent of whether the tail is extended or contracted, whereas the whiskers change their conformation during the tail contraction because of steric hindrances between the whiskers and contracted tail sheath. Only a part of the whiskers is visible in the contracted tail reconstruction because of their flexibility. The previous model of the collar and whiskers, based on the cryo-EM reconstruction of the extended T4 sheath, assumed that the virus contained six molecules of fibritin6 with the N-terminal parts of fibritin molecules located in the phage collar and the C-terminal parts forming the whiskers. In the previous model, a part of the cryo-EM density in the collar region remained uninterpreted. The new reconstruction of the contracted tail with the collar and whiskers suggests that the T4 phage contains twelve fibritin molecules. Six fibritin molecules are located approximately in one plane and form the phage collar, whereas the other six molecules stretch down and form the whiskers (Fig. 8). This interpretation is consistent with earlier estimation of the gpwac copy number using gel densitometry.9 Fibritin molecules bind to the neck where gp13 molecules are presumably located, suggesting that twelve fibritin molecules attach to twelve molecules of gp13. Genetic experiments24,25 showed that the N-terminal domain of fibritin binds to the phage. In support of the new interpretation of the collar and whiskers structure, the structure of the minifibritin protein,26 containing the 80 aminoterminal residues of fibritin, fits well into the cryo-EM density with the N-terminal domain attached to the phage neck.
Fig. 8.
Structure of the T4 collar and whiskers. (a) Side view of the cryo-EM reconstruction of the contracted T4 tail. The baseplate, tail sheath, and tail tube are colored magenta, green, and gray, respectively. The phage whiskers are colored gold. Only parts of the whiskers are visible in the reconstruction because of their flexibility. (b) Top view of the collar and whisker structure. Surfaces of six different fibritin molecules forming the phage collar are colored green, cyan, gray, magenta, blue, and red. The black circles show the positions where the retracted long tail fibers interact with the collar (in virions with extended tails). (c) The cryo-EM map is represented as a semitransparent surface. Models of the fibritin molecules fitted into the reconstruction are shown as ribbon diagrams. Fibritin molecules forming the collar are shown in different colors. Fibritin molecules forming the whiskers are colored gold. The figure was prepared using the program Chimera.63 The Chimera “hide dust” tool was used to remove small unconnected blobs of the cryo-EM density.
The gp13 dodecamer contains twelve binding sites for fibritin, which separate into two 6-fold symmetric subgroups. One subgroup of sites attaches the whiskers whereas the other binds the fibritin molecules that form the collar. The collar and whiskers help to attach six long tail fibers to the phage particle during the virus assembly and control the retractions of the long tail fibers under conditions unfavorable for infection.29,30 In the retracted conformation, the elbow region of each long tail fiber interacts with one whisker and the distal part of the fibers (formed by gp37) interacts with the collar.1,6 Such interactions require the proper orientation on the collar and whiskers with respect to the tail baseplate to which the long tail fibers are attached. Therefore, the separation of the fibritin molecules into the whiskers and collar should be correlated with the orientation of the tail. The correlation between the orientations of the tail, collar, and whiskers is probably achieved via interactions between the gp13 dodecamer and the hexamer of gp14, which break the 12-fold symmetry of gp13 and result in separation of the fibritin binding sites into two subgroups that have somewhat different orientations. The binding sites corresponding to the collar allow six fibritin molecules to attach almost perpendicular to the phage axis. Such orientation favors the interactions between the fibritin molecules and results in the formation of the collar disk.
Conclusion
Bacteriophages probably represent the most abundant biological entities on Earth with the estimated number of phage particles to be in the order of 1032 (Refs. 42 and 43). The number of phages outnumber about 10-fold the number of bacteria.44 As phages are a major cause of bacterial mortality, they have a considerable impact on the Earth's ecology. Phages also play an important role in evolution as they participate in the horizontal transfer of genes. Furthermore, phages have potential for treatment of bacterial infections and for control of food-borne pathogens. T4 is one of the most studied phages and by far the most studied phage with a contractile tail. Crystal structures of 14 out of more than 40 proteins present in the mature T4 virion have been completely or partially determined using X-ray crystallography. Although the protein composition of the T4 neck, a critical part for virus assembly, had been studied earlier using electron microscopy and biochemical methods, no atomic structure of the T4 neck proteins have been available until now. The similarity of gp15 to other phage proteins, together with the model of the T4 collar and whiskers, greatly enhance knowledge on bacterio-phage assembly and evolution.
Experimental Procedures
Expression and purification of the full-length gp15 protein and its C-terminal truncation mutant containing residues 1–261
The vector for the expression of the full-length gp15 and the protein over-expression in E. coli cells were described previously.16 Ammonium sulfate was added to the supernatant of gp15 over-expressed cell lysate at a concentration of 25% saturation and gp15 was fractionated by centrifugation. The gp15 precipitates were resuspended and solubilized in 25 mM Tris–HCl (pH 8.0) and subsequently applied to HiTrap Q HP (5 ml; GE Healthcare) anion-exchange column. Proteins were eluted with a 0- to 1-M NaCl linear gradient using the same buffer. The fractions containing gp15 were pooled and applied to Superdex 200 PG (120 ml; GE Healthcare) gel-filtration column that was equilibrated with 25 mM Tris–HCl (pH 8.0) and 0.2 M NaCl. Purified gp15 fractions were pooled and dialyzed against 10 mM Tris–HCl (pH 8.0).
The gene encoding the C-terminal truncation mutant of gp15 was constructed by PCR amplification of the coding sequence corresponding to amino acids 1–261 using appropriate primers and purified phage T4 DNA as the template. The amplified DNA was digested with NheI and BamHI restriction enzymes and ligated to the pET-28b plasmid DNA that had been linearized with the same restriction enzymes. This resulted in fusion at the 5′ end of the gene 15 to a vector sequence containing a hexahistidine-tag. The ligated DNA was transformed into E. coli XL10 Gold cells and colonies containing the correctly oriented inserted DNA were selected. The accuracy of the cloned DNA was confirmed by sequencing the entire insert. The plasmid DNA containing the gene of the gp15 C-terminal truncation mutant was then transformed into the expression strain E. coli BL21 (DE3) RIPL.
The E. coli cells carrying the plasmid were induced with 1 mM IPTG at 30 °C for 3 h. Cells were harvested by centrifugation at 8200g for 12 min at 4 °C and were lysed using an Aminco French press. The cell lysate was centrifuged at 34,000g for 20 min at 4 °C to separate the soluble fraction from the pellet. The His-tagged gp15 C-terminal truncation mutant was purified by HisTrap HP affinity chromatography (AKTA-PRIME; GE Healthcare) followed by gel filtration on a HiLoad Superdex 200 column (AKTA-FPLC; GE Healthcare) in a buffer containing 20 mM Tris–HCl (pH 8) and 100 mM NaCl. The His-tag was cleaved off with thrombin. The Superdex-purified protein was incubated with thrombin in the cleavage buffer [20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 20 mM imidazole, and 2.5 mM CaCl2] overnight at 8 °C. The cleaved protein was then separated from the His-tag, thrombin, and any uncleaved protein by HisTrap chromatography. The protein recovered in the flow-through fraction was concentrated and purified by gel filtration on the Superdex 200 column.
Crystallization of the gp15 proteins
Purified full-length gp15 was concentrated to about 20 mg/ml by Ultra free centrifugal filter units (50 kDa cutoff; Millipore). Initial screening for crystallization conditions was performed by the hanging-drop method at 28 °C. Crystals appeared under condition 41 of the Crystal Screen Cryo Kit from Hampton Research, which contained 8.5% isopropanol, 0.085 M Na Hepes (pH 7.5), 17% polyethylene glycol (PEG) 4000, and 15% glycerol. After optimization, the best crystals were obtained using 0.15 M ammonium sulfate, 10.5% isopropanol, 0.085 M Na Hepes (pH 7.5), 20% PEG 4000, and 15% glycerol. The gp15 C-terminal truncation mutant was concentrated to 5 mg/ml using Ultra free centrifugal filter units with a 10-kDa molecular mass cutoff. Initial crystallization trials were performed using the hanging-drop method at 20 °C with several commercially available crystallization screens from Hampton Research and Emerald Biosystems. The best crystals were grown under condition 23 of the PEG Ion Screen from Hampton Research, which contained 20% (w/v) PEG 3350 and 0.2 M ammonium formate. Platinum derivative of the gp15 C-terminal truncation mutant was obtained by soaking the crystals for 1.5 h in the crystal growth solution containing platinum potassium thiocyanate at 6 mM concentration.
X-ray data collection and structure determination
X-ray data for the crystals of the gp15 C-terminal truncation mutant containing residues 1–261 (Table 1) were collected at the GM/CA-CAT beamline 23-ID-B of the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Prior to data collection, crystals were placed for a few seconds into a cryoprotectant solution and flash-frozen in the nitrogen stream. The cryoprotectant solution was obtained from the crystal growth solution by adding glycerol to obtain a 20% (v/v) concentration. Data for the full-length gp15 protein were collected at the BioCars beamline 14-BM-C at the Advanced Photon Source (Table 1).
Crystals of the gp15 C-terminal truncation mutant belonged to the space group P1 with six molecules in the asymmetric unit whereas the crystals of the full-length gp15 protein belonged to space group P32 with six molecules in the asymmetric unit. Both crystal forms contained layers of gp15 hexamers. In the crystals belonging to the P32 space group, the planes of the layers were perpendicular to the c-axis of the crystals. Interactions between gp15 molecules within the layers were stronger than contacts between layers, resulting in diffraction anisotropy.
The X-ray data sets were processed using the program HKL2000.45 Structure determination was performed using the PHENIX software suite46 with the help of the AutoSol wizard.47 Correction for the diffraction anisotropy was performed using the program Xtriage.48 Determination of the platinum positions and subsequent phasing of the triclinic crystal form was performed using SIRAS (single isomorphous replacement with anomalous scattering) with the help of the program SOLVE.49 The phases were improved by density modification with the program RESOLVE50 using the 6-fold non-crystallographic symmetry. An atomic model was built into the electron density using the program Coot.51 The model was subjected to several rounds of crystallographic refinement using the program phenix.refine52 with manual rebuilding in Coot51 and automatic rebuilding using PHENIX AutoBuild.53 The structure of the trigonal P32 crystal form was determined using Molecular Replacement54 with the program Phaser.55 The structure was subjected to several cycles of manual fitting and refinement using the programs Coot51 and phenix.refine.52 The structure refined in the trigonal crystal form was placed into the P1 unit cell and subjected to refinement with phenix.refine.52
Fitting the gp15 structure into the cryo-EM density
The gp15 crystal structure was fitted into the cryo-EM reconstructions of the extended and contracted T4 tails5,6 using the program EMFIT.56,57 The gp15 hexamer was placed such that its axis coincided with the axis of the phage particle. Systematic two-dimensional search was performed varying the rotation angle of the gp15 hexamer around the phage axis and the position of the hexamer center along the phage axis. The criterion for the goodness of fit was the value of Sumf.56,57 The angular and translational steps were 3° and 2 Å, respectively. The gp15 hexamers fitted in the reconstructions of the extended6 and contracted5 tails had similar orientations with respect to the phage particle. Therefore, it was assumed that the hexamer does not change its orientation during the tail contraction.
Model of the full gp18 molecule
The HHpred server58 was used to align the sequences of T4 gp18 and the LIN1278 protein from L. innocua. These two proteins had ∼14% sequence identity in their C-terminal domains. A homology model of the gp18 C-terminal domain was created based on the LIN1278 structure21 (PDB ID: 3LML) using the program MODELLER.59 A model of the entire gp18 molecule was then created by superposition of the crystal structure of the gp18M truncation mutant (lacking the C-terminal domain) and the homology model of the gp18 C-terminal domain onto the structure of the LIN1278 protein. Molecules of gp18 were fitted into the cryo-EM reconstructions using the same orientation of the gp18M fragments with respect to the phage particle as described by Aksyuk et al.20 (PDB IDs: 3FOH and 3FOI).
Sequence alignments and comparison of secondary structure elements
Alignment of the gp3, gp19, and STM4215 sequences using the secondary structure information was performed using the PRALINE server.60‡ The secondary structure of the gp14 protein was predicted using PSIPRED.61 Alignment of the gp14 and gp15 sequences using the secondary structure was performed with the program YAP.§
Binding of the gp13 and gp14 proteins to T4 heads
Purified phage T4 heads (2×1011 particles)14 were incubated with purified gp13 and gp14 proteins at a 1:100 molar ratio of the portal protein gp20 to gp13 or gp14 in 500 μl of binding buffer [50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, and 100 mM NaCl] for 30 min at room temperature. The heads were sedimented by centrifugation at 34,000g for 45 min and the pellets were washed twice with 500 μl of the binding buffer. The pellets were then resuspended in 10 μl SDS sample buffer and analyzed by SDS-PAGE (Fig. 7). The density of the gp13, gp14, and gp20 bands were determined for each lane separately and the number of gp13 molecules per head was calculated using the known copy number of gp20 (12 molecules per head).
Preparation of phages with contracted tails, collar, and whiskers for cryo-EM
A solution of wild-type T4 phage with a titer of about 1011 plaque-forming units per milliliter was dialyzed overnight into 4 M urea in buffer A, containing 50 mM Tris–HCl at pH 8.0, 0.2 M NaCl, and 8 mM MgCl2 at 4 °C. Subsequently, DNase I was added to the sample to reach a final concentration of 40 μg/ml in order to remove DNA from broken phage particles. The phage was then pelleted by centrifugation for 1 h at 100,000g and resuspended in buffer A without urea. In this procedure, urea was not added directly into the sample, in contrast to the protocol used by Leiman et al.,5 thus possibly explaining the preservation of fibritin around the neck of the phage.
Cryo-EM data collection and three-dimensional reconstruction
Images of frozen-hydrated particles were recorded on Kodak film at a magnification of 39,100× with an FEI CM200 electron microscope using the radiation dose of 16 e−/Å2 and a defocus range from 1.5 to 3 μm. Images were digitized with a Nikon scanner and binned to the final pixel size of 3.57 Å per pixel. Contracted tails of 2727 individual phage particles were boxed using the program boxer from the EMAN software package.62 The contrast transfer function parameters of boxed particles were determined and the phases were flipped using the program ctfit from EMAN.62 The three-dimensional (3D) image reconstruction was performed with the EMAN package using the projection matching technique. The reconstruction of the contracted T4 tail5 lacking gpwac was used as the initial model. Thirty cycles of image reconstruction were performed while imposing the 6-fold symmetry. During the reconstruction process, the reference projections were calculated from the current model at 1.5° angular intervals. Orientations of individual particles were determined from projection angles of best-matching projections of the 3D map obtained in the previous reconstruction cycle. 3D reconstructions were calculated from the boxed particles with the make3d program from the EMAN package. Contrary to default EMAN procedure, class averages were not used in the reconstruction procedure. The resolution of the final reconstruction was estimated to be 25 Å using a Fourier shell correlation cutoff of 0.5. The structure of the collar region was similar in the reconstructions of the extended6 and contracted tails. The collar region was used to determine relative orientations of these two reconstructions with the program Chimera.63
Model of the fibritin structure
The model of fibritin was composed of the crystal structure of its N-terminal part26 (residues 1–80), the crystal structure of the C-terminal part27 (residues 371– 483), and the central region, modeled with segments of triple-helical coiled coil.6,24 The parts of the fibritin structure were fitted into the cryo-EM density using the program Chimera.63
Accession codes
The atomic coordinates and structure factors of T4 gp15 have been deposited in the PDB, with accession codes 4HUD and 4HUH. The cryo-EM map of the contracted T4 tail containing the collar and whiskers has been deposited in the Electron Microscopy Data Bank with the accession code EMD-5528. The model of fibritin fitted into the cryo-EM map has been deposited in PDB with the accession code 3J2O. The gp15 structure and gp18 model fitted into the cryo-EM reconstructions of the extended and contracted T4 tails have been deposited in PDB with accession codes 3J2M and 3J2N, respectively.
Acknowledgments
We thank Sheryl Kelly for help in the preparation of the manuscript. Use of the Advanced Photon Source (sectors 23 and 14) was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Support for this project was provided to M.G.R. by the National Institutes of Health (AI081726) and, for early work, by the National Science Foundation (MCB-0443899). Figures were prepared using the program Chimera63 and PyMOL.64
Abbreviations used
- cryo-EM
cryo-electron microscopy
- PDB
Protein Data Bank
- PEG
polyethylene glycol
- 3D
three-dimensional
Footnotes
Author Contributions. A.F., Z.Z., S.K., A.A.A., F.A., V.B.R., and M.G.R. designed research. A.F., Z.Z., S.K., V.D.B., and A.A.A. performed research. A.F., V.B.R., and M.G.R. wrote the paper.
Conflict of Interest Statement. The authors declare that they have no conflict of interest.
The terms “top” and “bottom” are used in accordance with the orientation of the T4 virions shown in Fig. 1.
References
- 1.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao VB, Black LW. Structure and assembly of bacteriophage T4 head. Virol J. 2010;7:356. doi: 10.1186/1743-422X-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black LW, Rao VB. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv Virus Res. 2012;82:119–153. doi: 10.1016/B978-0-12-394621-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci USA. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 6.Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- 7.Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 8.Dewey MJ, Wiberg JS, Frankel FR. Genetic control of whisker antigen of bacteriophage T4D. J Mol Biol. 1974;84:625–634. doi: 10.1016/0022-2836(74)90120-x. [DOI] [PubMed] [Google Scholar]
- 9.Coombs DH, Eiserling FA. Studies on the structure, protein composition and aseembly of the neck of bacteriophage T4. J Mol Biol. 1977;116:375–405. doi: 10.1016/0022-2836(77)90076-6. [DOI] [PubMed] [Google Scholar]
- 10.Black LW, Showe MK, Steven AC. In: Morphogenesis of the T4 head. Karam JD, editor. American Society for Microbiology; Washington, DC: 1994. pp. 218–258. [Google Scholar]
- 11.Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Akhter T, Zhao L, Kohda A, Mio K, Kanamaru S, Arisaka F. The neck of bacteriophage T4 is a ring-like structure formed by a hetero-oligomer of gp13 and gp14. Biochim Biophys Acta. 2007;1774:1036–1043. doi: 10.1016/j.bbapap.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Kottadiel VI, Vafabakhsh R, Dai L, Chemla YR, Ha T, Rao VB. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 2011;9:e1000592. doi: 10.1371/journal.pbio.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vianelli A, Wang GR, Gingery M, Duda RL, Eiserling FA, Goldberg EB. Bacteriophage T4 self-assembly: localization of gp3 and its role in determining tail length. J Bacteriol. 2000;182:680–688. doi: 10.1128/jb.182.3.680-688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, Kanamaru S, Chaidirek C, Arisaka F. P15 and P3, the tail completion proteins of bacteriophage T4, both form hexameric rings. J Bacteriol. 2003;185:1693–1700. doi: 10.1128/JB.185.5.1693-1700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage λ major tail protein structure reveals a common evolution for longtailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aksyuk AA, Leiman PG, Kurochkina LP, Schneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aksyuk AA, Kurochkina LP, Fokine A, Forouhar F, Mesyanzhinov VV, Tong L, Rossmann MG. Structural conservation of the Myoviridae phage tail sheath protein fold. Structure. 2011;19:1885–1894. doi: 10.1016/j.str.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968;32:231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- 23.King J. Bacteriophage T4 tail assembly: four steps in core formation. J Mol Biol. 1971;58:693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- 24.Letarov A, Manival X, Desplats C, Krisch HM. gpwac of the T4-type bacteriophages: structure, function, and evolution of a segmented coiledcoil protein that controls viral infectivity. J Bacteriol. 2005;187:1055–1066. doi: 10.1128/JB.187.3.1055-1066.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efimov VP, Nepluev IV, Sobolev BN, Zurabishvili TG, Schulthess T, Lustig A, et al. Fibritin encoded by bacteriophage T4 gene wac has a parallel triple-stranded a-helical coiled-coil structure. J Mol Biol. 1994;242:470–486. doi: 10.1006/jmbi.1994.1595. [DOI] [PubMed] [Google Scholar]
- 26.Boudko SP, Strelkov SV, Engel J, Stetefeld J. Design and crystal structure of bacteriophage T4 mini-fibritin NCCF. J Mol Biol. 2004;339:927–935. doi: 10.1016/j.jmb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 28.Strelkov SV, Tao Y, Shneider MM, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin M: a troublesome packing arrangement. Acta Crystallogr, Sect D: Biol Crystallogr. 1998;54:805–816. doi: 10.1107/s0907444997018878. [DOI] [PubMed] [Google Scholar]
- 29.Terzaghi BE, Terzaghi E, Coombs D. The role of the collar/whisker complex in bacteriophage T4D tail fiber attachment. J Mol Biol. 1979;127:1–14. doi: 10.1016/0022-2836(79)90454-6. [DOI] [PubMed] [Google Scholar]
- 30.Conley MP, Wood WB. Bacteriophage T4 whiskers: a rudimentary environment-sensing device. Proc Natl Acad Sci USA. 1975;72:3701–3705. doi: 10.1073/pnas.72.9.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pell LG, Liu A, Edmonds L, Donaldson LW, Howell PL, Davidson AR. The X-ray crystal structure of the phage λ tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J Mol Biol. 2009;389:938–951. doi: 10.1016/j.jmb.2009.04.072. [DOI] [PubMed] [Google Scholar]
- 34.Atia-tul-Wahab, Serrano P, Geralt M, Wuthrich K. NMR structure of the Bordetella bronchiseptica protein NP_888769.1 establishes a new phage-related protein family PF13554. Protein Sci. 2011;20:1137–1144. doi: 10.1002/pro.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:115–142. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- 36.Edmonds L, Liu A, Kwan JJ, Avanessy A, Caracoglia M, Yang I, et al. The NMR structure of the gpU tail-terminator protein from bacteriophage lambda: identification of sites contributing to Mg(II)-mediated oligomerization and biological function. J Mol Biol. 2007;365:175–186. doi: 10.1016/j.jmb.2006.09.068. [DOI] [PubMed] [Google Scholar]
- 37.Cardarelli L, Pell LG, Neudecker P, Pirani N, Liu A, Baker LA, et al. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc Natl Acad Sci USA. 2010;107:14384–14389. doi: 10.1073/pnas.1005822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 39.Lhuillier S, Gallopin M, Gilquin B, Brasilès S, Lancelot N, Letellier G, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA. 2009;106:8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, et al. The crystal structure of bacteriophage HK97 gp6: defining a large family of head–tail connector proteins. J Mol Biol. 2010;395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 42.Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Ackermann HW. 5500 phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 44.Brüssow H, Hendrix RW. Phage genomics: small is beautiful. Cell. 2002;108:13–16. doi: 10.1016/s0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter Charles W., Jr, editor. Methods in Enzymology. Vol. 276. Academic Press; New York, NY: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr, Sect D: Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwart PH, Grosse-Kunstleve RW, Adams PD. Xtriage and Fest: automatic assessment of X-ray data and substructure structure factor estimation. CCP4 Newsletter 43, contribution 7 2005 [Google Scholar]
- 49.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr, Sect D: Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr, Sect D: Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix. refine Acta Crystallogr, Sect D: Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr, Sect D: Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossmann MG. The Molecular Replacement Method. Gordon and Breach; New York, NY: 1972. [Google Scholar]
- 55.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X-ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 57.Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallogr, Sect D: Biol Crystallogr. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- 58.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 60.Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 62.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 63.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 64.DeLano WL. DeLano Scientific; San Carlos, CA: 2002. The PyMOL Molecular Graphics System. http://www.pymol.org. [Google Scholar]