Abstract
Advanced renal cell carcinoma (RCC) is an invariably fatal cancer. Currently, small-molecule inhibitors that target cell-growth, angiogenesis, or nutrient-sensing pathways represent the primary pharmacological interventions for this disease, but these inhibitors only delay tumor progression and are not curative. The cytokine interferon (IFN)-γ showed the potential to provide lasting remission in several phase I/II trials for advanced RCC, but subsequent trials, including a multi-center phase III study using IFN-γ as a monotherapy for RCC, were less promising. Notably, these trials were designed to exploit the indirect immune-modulatory effects of IFN-γ, while its direct anti-tumor properties – including its ability to trigger programmed cell death in tumors - remain mostly untapped. Here, we show that the proteasome inhibitor bortezomib (PS-341, Velcade) sensitizes otherwise-resistant RCC cells to direct necrotic death by IFN-γ. Mechanistically, we demonstrate that bortezomib functions at least in part by inhibiting pro-survival NF-κB signaling. In the absence of this signal, IFN-γ triggers programmed necrosis (or ‘necroptosis’) dependent on the kinase RIP1. When taken together with the observation that NF-κB signaling is elevated in RCC, these results provide rationale for the combined use of IFN-γ and bortezomib in the treatment of metastatic RCC.
Keywords: interferon, bortezomib, NF-κB, RIP1, necroptosis, necrosis
Introduction
Renal cell carcinoma (RCC), the 13th most-common malignancy worldwide, kills ~116,000 people annually (1). While early-stage RCC is treatable by surgical and other interventions, the metastatic form of this malignancy is chemotherapy-resistant and lethal (2). RCC comprises several distinct histological varieties, of which Clear Cell (cc)RCC represents the dominant subtype and accounts for up to 85% of all RCC cases (3). Over the past decade, small-molecule therapies that target growth factor-, angiogenesis-, and nutrient-sensing pathways (e.g. the tyrosine kinase inhibitors sunitinib and sorafenib) have become the frontline treatment options for advanced RCC (2, 4). Although most patients will derive some benefit from these agents, virtually all will experience significant side-effects, eventually develop resistance, and ultimately succumb to metastatic disease within five years (5, 6). Treatment of advanced RCC is therefore still a significant therapeutic challenge.
Prior to the introduction of small-molecule targeted agents, cytokine-based immunotherapy with either IFN-α or high-dose interleukin-2 was the most common treatment option for metastatic RCC (7). Based on its similarity to IFN-α, the cytokine IFN-γ was evaluated in several clinical trials for RCC (8). In particular, two key phase I/II trials employing low-dose IFN-γ produced overall-response rates of 15–30% and complete-response rates of up to 10% in metastatic RCC, highlighting the ability of this cytokine to provide long-term remission even after termination of treatment (9, 10). Based on these encouraging findings, a large multi-center phase III trial employing IFN-γ as a monotherapy for RCC was conducted, but this trial found no significant difference between IFN-γ and placebo in overall response rates, time to disease progression, or median survival (11). As a result, IFN-γ was not pursued much further as a potential anti-RCC biotherapeutic.
It is noteworthy that the trials described above – including the disappointing phase III trial – were designed primarily to exploit the indirect immune-modulatory effects of IFN-γ (i.e. its capacity to stimulate the immune response to RCC, without necessarily acting on the tumor itself). We suggest that a major advantage of IFN-γ over current small-molecule approaches is its pleiotropic nature: IFN-γ is not only a powerful activator of the anti-tumor immune response, but is also anti-angiogenic and directly tumoricidal to susceptible cells. Emphasizing the immune-modulatory effects of IFN-γ at the expense of its other direct anti-tumor properties (for example, its anti-angiogenic and growth-suppressive effects) may have contributed to the failure of the phase III clinical trial. We are therefore focused on resurrecting IFN-γ as an anti-RCC therapeutic by exploiting its direct anti-neoplastic properties, and, specifically, its capacity to selectively kill tumor cells. To this end, we have recently shown that the transcription factor NF-κB activates a survival program that protects mammalian cells from IFN-γ (12). In the absence of this survival program, we found that IFN-γ activates a novel process of caspase-independent necrotic cell death [sometimes termed ‘necroptosis’ (13)], mediated by the kinase RIP1 (12). As NF-κB drives a well-described survival program in many tumors – including RCC (14–16), and as dividing cells were found to be especially susceptible to IFN-γ-induced necrosis (12), these discoveries readily lend themselves to exploitation for the treatment of RCC.
One mechanism by which the small-molecule proteasome inhibitor bortezomib (PS-341, Velcade) functions as an anti-neoplastic agent is by inhibiting NF-κB (17), and studies have shown that blocking NF-κB with bortezomib in RCC cells (i) sensitizes them to the pro-apoptotic effects of TNF-α and TRAIL (18–20); (ii) synergistically potentiates the tumoricidal capacity of EGFR inhibitors (21); and (iii) increases susceptibility to oncolysis by encephalomyocarditis virus (22). In this study, we took advantage of the NF-κB-inhibitory capacity of bortezomib to test if blocking NF-κB signaling in RCC rendered them susceptible to IFN-γ-induced necrosis.
Using a panel of patient-derived ccRCC cell-lines, we report that inhibiting NF-κB by bortezomib renders RCC cells selectively susceptible to IFN-γ-induced necrosis. IFN-γ-triggered necrotic death was found to be independent of von Hippel-Lindau (VHL) gene or protein status and required the kinase RIP1. All RCC cells contained readily-detectable basal NF-κB activity, comprising chiefly of canonical RelA:P50 dimers, and bortezomib sensitized these cells to IFN-γ in part by inhibiting the pro-survival signaling activity of these NF-κB complexes. NF-κB signaling components and pro-survival target genes displayed elevated expression in samples from RCC tumors, compared to surrounding normal renal tissue, suggesting that RCC cells are more reliant on NF-κB survival signaling than their normal counterparts. In agreement, IFN-γ – in the setting of NF-κB inhibition by bortezomib – selectively induced necrosis in a panel of ATCC-derived RCC cell lines at doses that were largely non-toxic to normal kidney epithelial cells. Collectively, these results suggest that the combination of IFN-γ and bortezomib will have therapeutic benefit in RCC.
Materials and Methods
Cell lines
Clear Cell RCC cell lines HRC31, HRC45 and HRC63 were established at the Fox Chase Cancer Center (FCCC) as described previously (23). These cells were maintained in IIA medium (DMEM/F12 supplemented with 1.2 g/ml NaHCO3, 1.6 μM FeSO4, 50 nM sodium selenite, 25 μg/ml insulin, 200 nM hydrocortisone, 10 g/ml transferrin, 1 nM triiodothyronine, 10 μU/ml vasopressin, 10 nM cholesterol, 10 ng/ml epidermal growth factor, and 15% FBS). ACHN, Caki-1, 786-O and RenCa were obtained from the ATCC and cultured in DMEM/10% FBS (ACHN and Caki-1) or RPMI 1640/10% FBS (786-O and RenCa). Normal kidney epithelium-derived cell populations NKC1 and NKC2 were generated from minced renal tissue obtained from FCCC patients, and cultured in ACL-4 medium (DMEM/F12 supplemented with 0.02 mg/ml insulin, 0.01 mg/ml transferrin, 25 nM sodium selenite, 50 nM hydrocortisone, 1 ng/ml epidermal growth factor, 0.01 mM ethanolamine, 0.01 mM phosphorylethanolamine, 100 pM triiodothyronine, 0.5% (w/v) bovine serum albumin, 0.5 mM sodium pyruvate, 2mM L-glutamine, and 15% FBS). ATCC cell lines were not authenticated in-house, but were used within six months of resuscitation.
Reagents
Cytokines and chemicals were from the following sources: human IFN-γ (Pestka Biomedical Laboratories), Necrostatin-1 (Enzo Life Sciences), bortezomib (Millenium), IMD-0354 (EMD Biosciences). Oligonucleotides used to prepare radiolabeled wild-type and mutant NF-κB probes for EMSA were purchased from Santa Cruz, while oligonucleotides used to make the GAS probe were custom-synthesized at the Fox Chase Cancer Center. All other reagents were from Sigma-Aldrich, unless otherwise mentioned.
Antibodies
Primary antibodies for use in immunohistochemical and immunoblotting studies were purchased from Santa Cruz (pVHL, IRF-1, I-κBα, IKKβ, P50, P52, RelA, c-Rel), Upstate (STAT1, pSTAT1 Y701, pSTAT1 S727), BD Biosciences (HIF-1α, RIP1), Ventana (cytokeratin, RCC marker), Dako (vimentin), Novocastra (CD10), and Novus (HIF-2α).
Electrophoretic mobility shift assay (EMSA)
EMSA reactions were performed as described previously (24). Briefly, cells (2×106/condition) were lysed hypotonically and nuclear extracts prepared by high-salt extraction. 10 μg nuclear protein was incubated with a radiolabeled NF-κB probe derived from the HIV-1 Long Terminal Repeat (5′-AGTTGAGGGGACTTTCCCAGGC-3′) (25), mutant NF-κB probe (5′-AGTTGAGGCGACTTTCCCAGGC-3′), or GAS probe derived from the human IRF1 gene (5′-GATCGATTTCCCCGAAAT-3′), and reactions resolved by 5% non-denaturing PAGE. Gels were then vacuum-dried and subjected to autoradiography. For antibody supershift experiments, antibodies (1 μg) were added to nuclear extracts 15 minutes prior to incubation with radiolabeled oligonucleotide.
RNAi
RCC cells (6×104/well) seeded into six-well dishes were transfected with pools of four distinct proprietary siRNAs (SMARTpool, Dharmacon) to RIP1 at 20nM using Oligofectamine (Invitrogen) as a transfection reagent. As controls, non-targeting siRNA duplexes (Dharmacon) were employed. Cells were used in experiments 48–72 hr post-transfection.
Real-time quantitative PCR
Cells (2 × 106/condition) were harvested in TRI Reagent (Applied Biosystems), and total RNA was extracted by phase separation in bromochloropropane (Molecular Research Center). RNA was reverse transcribed into cDNA according to the manufacturer’s protocol (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems). Real-time quantitative (q) PCR was performed on an ABI7000 System using the Fast Start Universal Probe Master Mix (Roche), with probe and primer sets designed and supplied by the Roche Universal Probe Library System.
Cell viability
Cell viability was measured by Trypan Blue exclusion analysis. As necessary, necrosis was established by rescue of viability with Nec-1 (50 μM) pre-treatment.
Statistical analysis
Student’s T-test was used for comparison between two groups, and P-values <0.05 were considered significant.
Results
Characterization of three patient-derived ccRCC cell lines
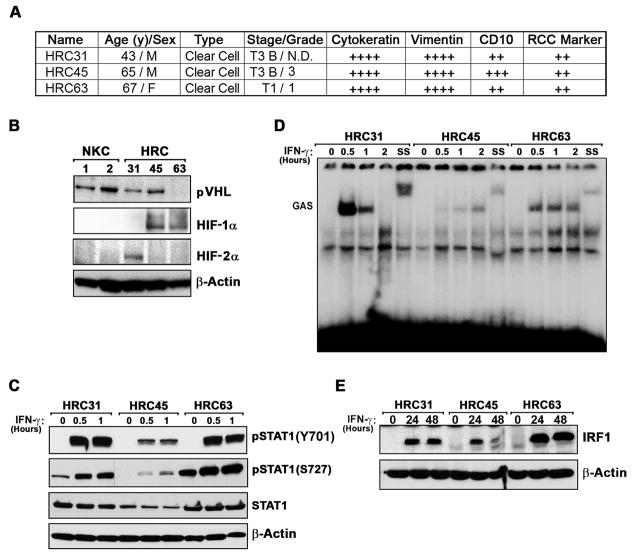
Three ccRCC cell lines, designated HRC31, HRC45 and HRC63, were established from tumor biopsies of patients undergoing surgery at the Fox Chase Cancer Center (23, 26). Each cell line stained positive in immunohistochemical studies with antibodies to cytokeratin, vimentin, CD10 and RCC marker, confirming ccRCC diagnosis (Fig 1A).
Figure 1. Characterization of ccRCC cell lines.
(A) Three FCCC patient-derived ccRCC cell lines, designated HRC31, HRC45 and HRC63, were histologically analyzed by staining with anti-pan-cytokeratin, anti-vimentin, anti-CD10 and anti-RCC marker antibodies. Staining was arbitrarily graded as weak (+), moderate (++), strong (+++), or very strong (++++) in at least 80% of individual cells in a representative field. (B) Whole-cell extracts from HRC cell lines were examined for pVHL, HIF-1α, and HIF-2α expression by immunoblotting. As normal controls, extracts from two populations of cultured kidney epithelial cells (NKC1 and NKC2) were used. (C) Whole-cell extracts from HRC cell lines were examined for phospho-STAT1 Y701, pSTAT1 S727, or total STAT1 after IFN-γ (100U/ml) treatment. (D) Kinetics of STAT1 binding to a GAS element were examined by EMSA of nuclear lysates from HRC cells. SS = supershift with an anti-STAT1 antibody. (E) Induction of the ISG product IRF-1 following treatment of HRC cell lines by IFN-γ (100U/ml) was determined by immunoblotting.
The VHL gene, a key tumor suppressor in ccRCC (27), was not mutated in any of the three cell lines (not shown), and its product – pVHL - was expressed in two out of three cell lines (HRC31 and HRC45) at levels equivalent to those seen in two independent populations of cells cultured from normal renal epithelium (herein designated NKC1 and NKC2; Fig. 1B). Although VHL was unmutated in HRC63, these cells did not express detectable pVHL protein, suggestive of VHL inactivation in this cell line by epigenetic or post-transcriptional mechanisms (Fig. 1B). It is noteworthy that VHL mutations on average are found to only occur in approximately half of all ccRCC samples (with a range of ~20% to >70%, depending on the study), so unmutated VHL in all three RCC cell lines is not a statistical improbability (28–31).
Interestingly, although HRC31 and HRC45 each display normal pVHL levels (Fig. 1B) and two copies of chromosome 3p (where VHL resides), both cell lines showed loss of heterozygosity (LOH) for all or part of chromosome 3p, based on allele profiling (Supplementary Fig. 1), indicative of uniparental isodisomy, a mechanism for copy-neutral LOH (32). The LOH included the region 3p21-p25, which, besides VHL, harbors several other tumor suppressor loci implicated in ccRCC tumorigenesis, including PBRM1, SETD2, and BAP1 (33). Such LOH may unmask recessive mutations in the remaining allele of one or more of these tumor suppressor genes, providing a possible explanation for the normal VHL status of these two cell lines.
The VHL gene product (pVHL) - in its best-described role - functions by controlling the protein levels of Hypoxia Inducible Factor (HIF)-α, a master regulator of the cellular response to hypoxia (34). There are three HIF-α family members, HIF-1α, HIF-2α and HIF-3α, of which HIF-1α and HIF-2α are particularly important in renal cell carcinogenesis (27). HIF-1α accumulated to robust levels in two out of three cell lines (HRC45, HRC63), and HIF-2α in the third cell line (HRC31) even under normoxic conditions, in concordance with their roles as drivers of ccRCC tumorigenesis. Thus, all three HRC cell lines display elevated HIF-α expression despite unmutated VHL and (at least in the cases of HRC31 and HRC45) detectable pVHL protein.
As the exploitation of the direct tumoricidal properties of IFN-γ in RCC is dependent on the capacity of RCC cells to support signaling downstream of the IFN-γ receptor, we next tested key aspects of classical IFN-γ signaling in the three ccRCC cell lines. The major IFN-γ-initiated signaling axis proceeds via Janus kinase (Jak)1/2-mediated phosphorylation of the transcription factor STAT1, which then translocates to the nucleus and transactivates expression of interferon-stimulated genes (ISGs) containing gamma-activated sequence (GAS) elements in their promoters (35). In all three cell lines, IFN-γ induced robust phosphorylation of STAT1 on the key Y701 and S727 residues within 30 minutes of treatment (Fig. 1C), and a subsequent EMSA experiment revealed that nuclei from all three HRC cell lines contained IFN-γ-inducible activity capable of binding GAS elements, albeit with cell line-specific differences in kinetics and magnitude (Fig. 1D). Supershifting the IFN-γ-activated band with an anti-STAT1 antibody demonstrated that the IFN-γ-inducible activity contains STAT1. In accordance with these observations, IFN-γ treatment upregulated protein levels of the prototypic ISG product IRF-1 in each HRC cell line (Fig. 1E). HRC45 displayed somewhat reduced GAS activity and IRF-1 induction, consistent with the modestly-lower levels of total STAT1 protein seen in these cells. Together, these findings indicate that all three HRC cell lines are capable of mounting a STAT1-dependent transcriptional response following IFN-γ stimulation, a prerequisite for activation of necrosis by this cytokine.
Bortezomib and IKK inhibition sensitizes ccRCC cell lines to IFN-γ-induced cell death
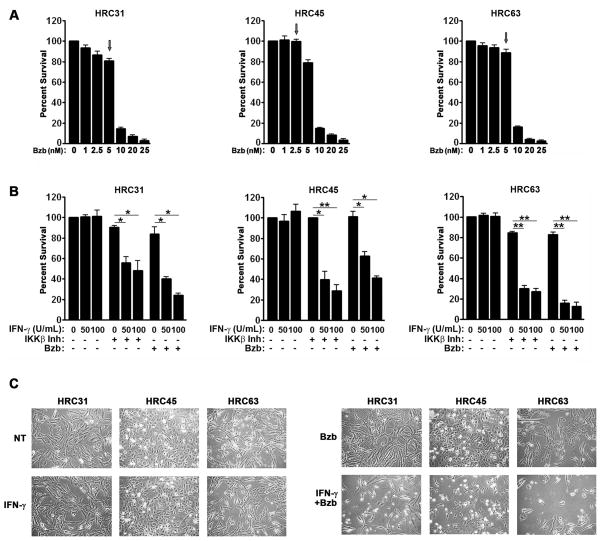
We had previously reported that neutralizing NF-κB signaling sensitized dividing cells to IFN-γ-triggered necrotic death (12). As a start to exploiting this observation for the treatment of RCC, we took advantage of the fact that a major mechanism by which the proteasome inhibitor bortezomib exerts its anti-tumor effects is via inhibiting NF-κB activation (17, 36). We first conducted dose-finding studies to identify doses of bortezomib that, on their own, were relatively non-toxic to each cell line (i.e. >85% cell viability in the continuous presence of that dose over 72 h, Fig. 2A). We designated the highest such dose of bortezomib as the maximum tolerated dose (MTD, typically in the 2.5–5 nM range) for the purposes of these studies, and used that dose in subsequent experiments (Fig. 2A, arrows).
Figure 2. Bortezomib and IKK inhibition sensitize ccRCC cells to IFN-γ-induced cell death.
(A) HRC cell lines were treated with the indicated dose-range of bortezomib (Bzb) and cell viability was measured 72 h post-treatment to determine the MTD (arrow). (B) HRC cell lines were pre-treated with their respective MTDs of bortezomib or, as a positive control, the IKKβ inhibitor IMD-0354 (500 nM) for 1 h, following which they were treated with the indicated concentrations of human IFN-γ. Viability was measured 72 h post-IFN-γ treatment. (C) HRC cell lines that were either untreated (NT), treated with IFN-γ alone (100U/ml), bortezomib alone (MTD) or the combination of IFN-γ and bortezomib (at 100U/ml and MTD, respectively) were photographed 72 h post-treatment. Error bars represent mean +/− S.D., n=3. * = p<0.05; ** = p<0.005
We next treated each HRC cell line with its MTD of bortezomib, following which we incubated them with increasing amounts of IFN-γ [up to 100 U/ml, well within the therapeutic range for this cytokine (37)]. In control experiments, we treated parallel populations of HRC cells with the IKKβ inhibitor IMD-0354, before exposing them to these same doses of IFN-γ. IFN-γ alone was largely non-toxic to HRC cells, but induced progressive, dose-dependent cytotoxicity in the presence of either bortezomib or IKKβ inhibitor (Fig. 2B). In HRC31 and HRC63, the combination of IFN-γ and bortezomib was cytotoxic to over 80% of cells within 72 h, while HRC45 was somewhat more resistant (~60% loss of viability in the same time frame; Fig. 2B,C), perhaps as a result of its modestly-poorer responsiveness to IFN-γ stimulation (see Fig. 1). Of note, ~100% of all three cell lines eventually succumbed to this combination 96–120 h post-treatment, but the interpretation of these results are somewhat confounded by issues of toxicity arising from such prolonged exposure to bortezomib alone (data not shown). Collectively, these results show that bortezomib sensitizes RCC cells to IFN-γ-induced cell death at therapeutically-achievable doses for both agents.
IFN-γ induces RIP1 kinase-dependent necrosis in ccRCC cells when NF-κB is inhibited
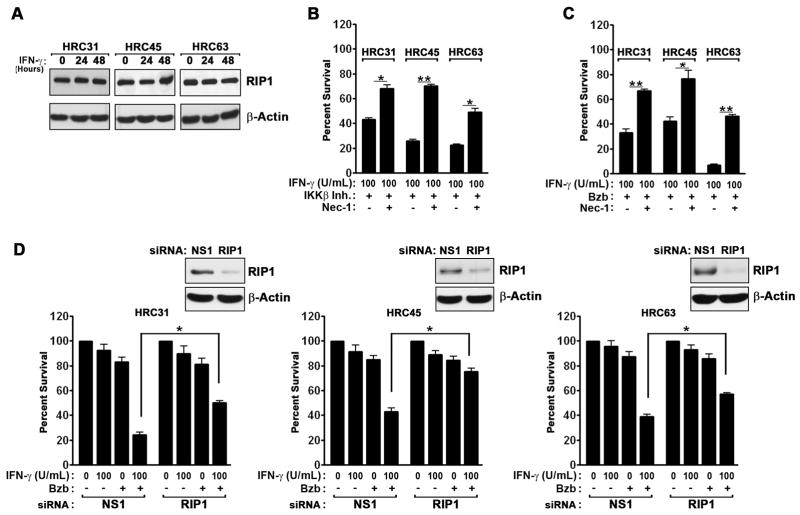
Necrosis in NF-κB-deficient MEFs exposed to IFN-γ resulted from accumulation of ROS in mitochondria, and required the kinase RIP1 (12). Mechanistically, we have found that IFN-γ transcriptionally activated RIP1, which then impinged on mitochondria to hyperactivate ROS production and induce respiratory failure (12). To test if a similar RIP1-dependent necrotic mechanism accounted for IFN-γ-triggered cytotoxicity in IKKβ inhibitor- or bortezomib-treated ccRCC cells, we first verified that each HRC cell line expressed RIP1 protein (Fig. 3A). We then incubated the HRC cell lines with the potent, specific RIP1 kinase inhibitor Necrostatin-1 [Nec-1; (38)], before exposing them IFN-γ in the presence of either the IKKβ inhibitor (Fig. 3B) or bortezomib (Fig. 3C). Nec-1 afforded significant protection against IFN-γ induced cell death in all three cell lines, rescuing each of them by at least 30% (Fig. 3B,C). Of note, the caspase inhibitor zVAD provided modest protection (up to 15%, depending on the cell line) against the combination of bortezomib and IFN-γ, suggesting that both necrotic and caspase-dependent apoptotic mechanisms contribute to IFN-γ-triggered cytotoxicity in these cells (data not shown).
Figure 3. IFN-γ induces RIP1 kinase-dependent necrosis in the presence of IKKβ inhibition or bortezomib.
(A) Immunoblot analysis of RIP1 protein expression in HRC cell lines after IFN-γ (100U/ml) treatment. (B) HRC cell lines were pre-treated with the IKKβ inhibitor IMD-0354 (500 nM) for 1 hr, following which they were exposed to IFN-γ (100U/ml) in the presence or absence of the specific RIP1 kinase inhibitor Necrostatin-1 (Nec-1, 50μM). Cell viability was measured 72 h post IFN-γ treatment. (C) HRC cell lines were pre-treated with bortezomib (MTD) for 1 hr, following which they were exposed to IFN-γ (100U/ml) in the presence or absence Nec-1 (50μM). Cell viability was measured 72 h post IFN-γ treatment. (D) HRC cell lines were transfected with the indicated siRNAs for 24 h, following which they were treated with either IFN-γ (100U/ml), bortezomib (MTD), or the combination of IFN-γ and bortezomib at these doses for a further 48 h, at which time cell viability was measured. Knockdown of RIP1 was confirmed by immunoblotting (inset). Error bars represent mean +/− S.D., n=3. * = p<0.05; ** = p<0.005.
To confirm the role of RIP1 in IFN-γ-driven cell death, we used RNAi to silence RIP1 expression by >75% in each of the three HRC cell lines (Fig. 3D, inset), following which we treated these cells with IFN-γ in the presence or absence of bortezomib. In all three cell lines, RIP1 RNAi – like Nec-1 pre-treatment – provided significant protection against the combination of IFN-γ and bortezomib (Fig. 3D). These data demonstrate that RIP1-dependent necrosis is a dominant mechanism of cell death induced by IFN-γ in the presence of bortezomib.
Bortezomib inhibits NF-κB in ccRCC cells
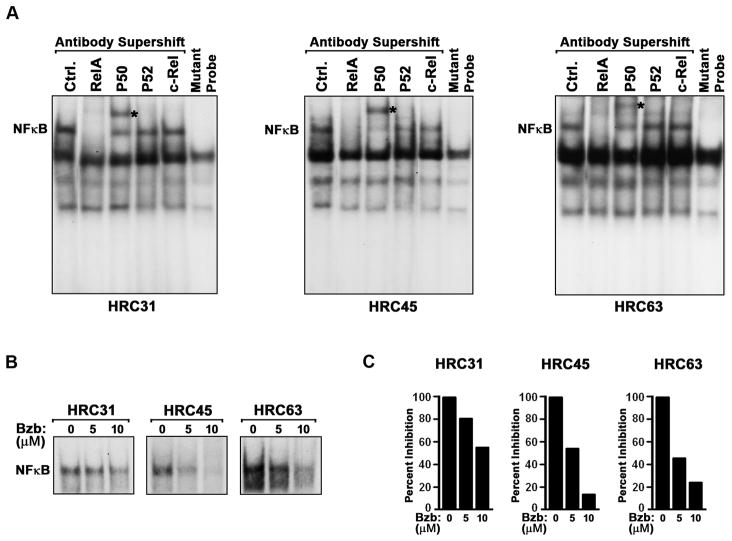
These results demonstrate that bortezomib and NF-κB inhibition can both sensitize ccRCC cells to necrosis induced by IFN-γ, suggesting that bortezomib – a known NF-κB inhibitor – functions, at least in part, by blocking constitutive NF-κB activity in ccRCC cells. To test if bortezomib inhibited NF-κB in HRC cells, we first determined by EMSA that all three HRC cell lines displayed detectable basal NF-κB activity (Fig. 4A). Supershift studies using antibodies to NF-κB subunits revealed that the primary NF-κB complexes in all three cell lines comprised chiefly of canonical RelA:P50 heterodimers, although dimeric combinations of other sub-units (e.g., P52- and c-Rel-containing complexes in HRC45) may also exist in lesser amounts (Fig. 4A).
Figure 4. Bortezomib inhibits NF-κB in HRC cells.
(A) Nuclear extracts from unstimulated HRC cell lines were pre-incubated with the indicated anti-NF-κB subunit antibodies and supershift of NF-κB complexes was examined by EMSA using radiolabeled NF-κB consensus or mutant oligonucleotides. (Ctrl. = control extract without antibody) Visibly-supershifted P50-containing complexes are indicated with an asterisk. The anti-RelA–supershifted complex is not seen in these autoradiograms as it can often be too large to enter the gel (12). (B) HRC cell lines were treated with bortezomib for 12 h, and nuclear extracts prepared from these cells were examined for binding to a radiolabeled NF-κB probe by EMSA. (C) NF-κB bands in the EMSA autoradiograms shown in (B) were densitometrically quantified by Image J analysis.
Next, we incubated each HRC cell line with bortezomib for 12 h, following which we subjected nuclear extracts from these cells to NF-κB EMSA. As shown in Fig. 4B, bortezomib treatment significantly reduced basal NF-κB activity in all three HRC cell lines. By densitometry, we determined that bortezomib treatment reduced NF-κB DNA binding activity by ~50% (HRC31), ~90% (HRC45) and ~80% (HRC63) in the three HRC cell lines (Fig. 4C). DNA binding to a control Oct-1 probe (not shown), or RelA/P50 protein levels (not shown) were not significantly affected by bortezomib treatment, demonstrating that bortezomib specifically inhibits NF-κB activity in HRC cells. In agreement with these findings, bortezomib efficiently prevented degradation of I-κBα following acute exposure of HRC63 cells to the well-established NF-κB stimulator TNF-α (Supplementary Figure 2). As a consequence, bortezomib was as effective as an IKKβ inhibitor in sensitizing HRC63 cells to TNF-α-induced cell death (Supplementary Figure 2).
We note, however, that while bortezomib can inhibit constitutive NF-κB signaling in HRC cells, the degree to which it does so (particularly in HRC31) does not fully correlate with its ability to sensitize cells to IFN-γ. Thus, bortezomib likely engages multiple proteasome-sensitive mechanisms to sensitize HRC cells to IFN-γ.
NF-κB pro-survival target gene expression is inhibited by bortezomib and elevated in ccRCC
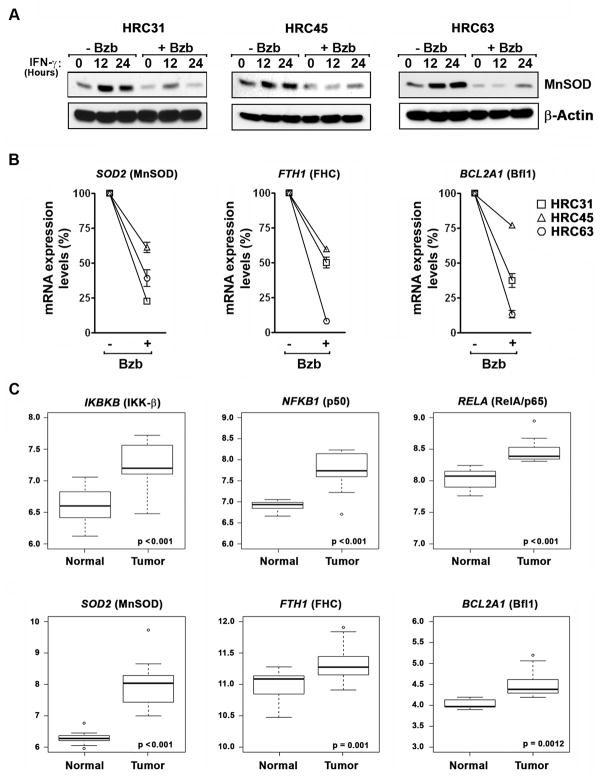
We have previously reported that the NF-κB target SOD2 (encoding the antioxidant enzyme MnSOD) is necessary for protection against IFN-γ-induced necrosis (12). To test if bortezomib inhibited MnSOD expression in the three HRC cell lines, we treated these cells with IFN-γ and examined MnSOD levels in lysates from these cells. As shown in Fig. 5A, we found that bortezomib inhibited both basal and IFN-γ-induced expression of MnSOD in all three HRC cell lines.
Figure 5. . NF-κB pro-survival targets are down-regulated by bortezomib and show elevated expression in ccRCC tumors.
(A) Immunoblot analysis of MnSOD protein expression in HRC cell lines after exposure to IFN-γ (100U/ml) in the presence or absence of bortezomib (5μM). (B) mRNA expression levels of SOD2, BCL2A1, and FTH1 after bortezomib (5 μM, 12h) treatment of HRC cell lines, compared to untreated samples (arbitrarily set to 100% in each cell line), after normalization to an internal GAPDH control. Error bars represent mean +/− S.D, n=3. (C) Box plots showing gene expression profiles of NF-κB signaling components (upper panels) or downstream survival targets (lower panels) in paired normal and tumor samples from 21 ccRCC patients (44). Y-axis displays the Robust Multi-array Average (RMA)- normalized expression level (on log2 scale) of each mRNA. Open circles represent outliers in that particular comparison. The solid line within each box is the median, and distance between box and whiskers indicate interquartile ranges.
To extend these observations, we analyzed the mRNA expression profiles of SOD2, as well as of two additional mitochondrial pro-survival targets of NF-κB - the Bcl-2 family member Bfl-1 (encoded by BCL2A1) and the antioxidant enzyme Ferritin Heavy Chain (FHC, encoded by FTH1) - following treatment of HRC cell lines with bortezomib. Like MnSOD, both Bfl-1 and FHC have been reported to protect cells from cytokine-driven mitochondrial damage and cell death (39, 40). We found that bortezomib significantly down-regulated the expression of each of these mRNAs in all three cell lines (Fig. 5B).
While these results support the idea that bortezomib will sensitize ccRCC cells to IFN-γ-induced necrotic death by blocking NF-κB survival signaling (e.g. by downregulating MnSOD levels) in vivo, a precondition for the specific killing of ccRCC – but not normal – cells by the combination of IFN-γ and bortezomib (or other NF-κB blockers) is that ccRCC cells, compared to normal kidney tissues, must selectively display an increased reliance on NF-κB-dependent survival mechanisms. Data suggest that established ccRCC cell lines do indeed display constitutively-elevated NF-κB activity (16, 19, 21, 41, 42), and that increased NF-κB activity correlates with progression of disease in patients (43).
Elevated NF-κB activity, however, is not necessarily indicative of increased pro-survival gene induction, and whether dedicated NF-κB pro-survival targets are upregulated in ccRCC has not been determined. We therefore examined NF-κB pro-survival gene expression in publicly-available datasets where ccRCC and normal renal tissues were assessed for transcriptomic changes. In particular, we focused on raw expression data generated by Copland and colleagues (44) (made available through GEO), as these data paired ccRCC and normal renal cortical tissue obtained from the same patients. Analysis by Linear Models for Microarray Data (LIMMA) (45) of the expression levels of the primary NF-κB signaling components (genes encoding IKKβ, P50 and RelA) and key downstream survival targets (genes encoding MnSOD, Bfl-1, and FHC) in this dataset revealed that all these genes were significantly overexpressed in ccRCC samples, compared to their paired normal controls, strongly suggesting that elevated NF-κB survival signaling is a common feature of ccRCC (Fig. 5C). Notably, very similar results were obtained when the larger (albeit less-robustly controlled) dataset generated by Libermann and colleagues (46) was analyzed (Supplementary Fig. 3).
Bortezomib sensitizes established RCC cell lines to IFN-γ-induced necrosis
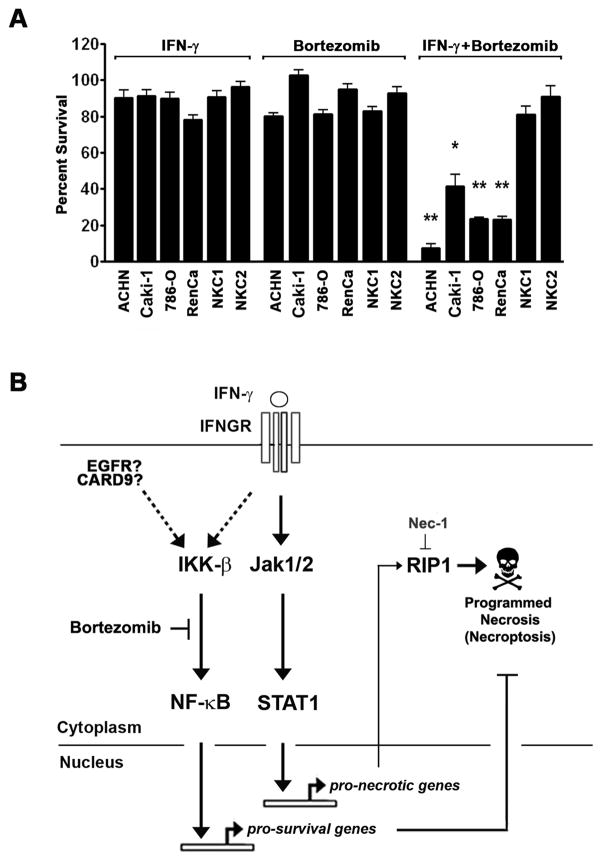
The elevated expression of NF-κB pro-survival targets – and, in particular antioxidant-encoding genes like SOD2 and FTH1 - in RCC samples suggests that RCC cells possess mitochondria that are metabolically more active than those found in surrounding normal tissue, and are therefore more reliant on ROS-quenching antioxidant enzymes like MnSOD and FHC for their survival. As IFN-γ-driven necrosis is a ROS-driven process, RCC cells, compared to normal kidney epithelial cells, will be expected to display selective susceptibility to IFN-γ when NF-κB is disabled (i.e. RCC cells are ‘addicted’ to elevated NF-κB signaling). To test this prediction, we treated a panel of four ATCC-derived human (ACHN, CAKI-1 and 786-O) and murine (RenCa) RCC cell lines with the combination of IFN-γ and bortezomib. In parallel, we also treated two independent populations of cells from normal renal epithelium (NKC1 and NKC2) with the same combination. In line with the expectation that RCC cells are selectively addicted to elevated NF-κB activity for their survival, both NKC populations displayed undetectable basal NF-κB activity (not shown) and remained resistant to the combination of IFN-γ and bortezomib at concentrations of each agent (50–100U/ml IFN-γ and 5nM bortezomib) that induced potent necrosis in all RCC cell lines tested (Fig. 6A). In fact, both NKC1 and NKC2 remained mostly viable (<10% cell death) for at least 72 hours in the continued presence of up to ten-fold higher concentrations of IFN-γ (i.e.1000U/ml) or 2.5-fold higher concentrations of bortezomib (12.5 nM) in subsequent dose-escalation experiments, when the concentration of one agent was held constant while that of the other was increased (not shown).
Figure 6. Bortezomib sensitizes established RCC cell lines to IFN-γ-induced necrosis.
(A) The ATCC-derived RCC cell lines ACHN, CAKI-1, 786-O and RenCa were pre-treated with bortezomib (5nM) for 1 hr, following which they were exposed to either 100U/ml of human IFN-γ (ACHN, CAKI-1, 786-O) or murine IFN-γ (RenCa) for 72 h. In parallel, two populations of normal kidney epithelial cells (NKC1 and NKC2) were also treated with identical concentrations of bortezomib and IFN-γ for 72 h. Error bars represent mean +/− S.D., n=3. * = p<0.05; ** = p<0.005. (B) Schematic of IFN-γ pro-necrotic and NF-κB pro-survival pathways. IFN-γ activates Jak/STAT signaling downstream of its tetrameric receptor (IFNGR) to induce expression of several hundred genes, include putative pro-necrotic targets. In parallel, RCC cells also display elevated levels of NF-κB activity (ostensibly as a result of constitutive EGFR or CARD9 activity) that drives expression of pro-survival genes like SOD2 to quench mitochondrial ROS. When NF-κB is inactivated (for example, by bortezomib), then IFN-γ induces necrosis via RIP1-mediated ROS accrual.
Collectively, these data demonstrate that elevated NF-κB activity can be exploited to selectively sensitize RCC cells to the pro-necrotic effects of IFN-γ, and provide strong rationale for the combined use of bortezomib (or other NF-κB inhibitors) with IFN-γ for the treatment of this malignancy.
Discussion
The ideal IFN-γ-based anti-RCC therapy will not only take advantage of the immune-modulatory activity of IFN-γ, but will also exploit this cytokine’s other anti-neoplastic properties - including its anti-angiogenic and direct tumoricidal capacity – and do so with minimal toxic side-effects. With this goal in mind, we have identified NF-κB as one druggable mechanism by which RCC cells are resistant to the direct tumoricidal properties of IFN-γ. We had previously shown that when NF-κB is disabled, IFN-γ transcriptionally activates a novel form of necrotic cell death (sometimes termed necroptosis) dependent on the kinase RIP1 (12). Here, we exploited these findings to demonstrate that the small molecule proteasome inhibitor bortezomib sensitizes RCC cells to IFN-γ-induced necrotic death, at least in part by blocking NF-κB.
How does NF-κB protect RCC cells from IFN-γ? Results from our previous work suggests that IFN-γ utilizes RIP1 to boost mitochondrial activity and ATP biogenesis – for example, to fuel ATP-dependent anti-microbial enzymes – during physiological immune responses (12). An unavoidable consequence of increased mitochondrial activity is the generation of toxic reactive oxygen species (ROS), typically as a byproduct of leaky electron transport during the process of oxidative phosphorylation (47). Scavenging of ROS and other free radicals is a primary task of the NF-κB pro-survival program, as evidenced by the facts that (i) ROS accumulates to toxic levels following cytokine stimulation of NF-κB-deficient cells, and (ii) overexpression of the NF-κB targets MnSOD and FHC quenches ROS after cytokine exposure without need for NF-κB activity (12, 40, 48). When ROS is allowed to accrue in mitochondria (for example, following exposure to IFN-γ in settings of NF-κB inhibition), mitochondrial membrane potential is lost, respiratory failure occurs, and the cell dies by caspase-independent necrosis (12).
Although the process of tumorigenesis often results in a shift in ATP production from oxidative phosphorylation to aerobic glycolysis, oxidative phosphorylation is still thought to generate a significant fraction (from less than half to over 90%, depending on the malignancy) of the tumor cell’s ATP needs (49, 50). We propose that the increased demand for ATP during tumorigenesis makes tumor cells more reliant on NF-κB-activated free radical scavenging mechanisms - both constitutively, and in response to acute stimuli such as IFN-γ - to ensure mitochondrial integrity and cell survival. Indeed, several observations lend support to the idea that tumor cells (including RCC) have co-opted NF-κB pro-survival signaling to sustain their own viability. These include the findings that (i) high constitutive NF-κB activity in a frequent occurrence in diverse cancers; (ii) activating mutations in genes encoding NF-κB subunits have been reported in several tumor types; and (iii) tumor cells are often ‘addicted’ to NF-κB survival signaling, and succumb when NF-κB signaling is inhibited or ablated (14, 51).
Established RCC cell lines and patient tumor samples – as a result of CARD9 activation and/or chronic EGF receptor signaling - also display constitutively-elevated NF-κB pro-survival activity (22, 26, 28). In agreement, our in silico analyses of published patient data reveal that NF-κB signaling components and downstream pro-survival target genes are chronically upregulated in RCC tumors (Fig. 5). It is particularly noteworthy that the NF-κB targets SOD2 (encoding manganese superoxide dismutase, or MnSOD) and FTH1 (encoding ferritin heavy chain, FHC) were elevated in ccRCC, as both these gene products are antioxidant enzymes critical for NF-κB-dependent buffering of mitochondria against cytokines such as IFN-γ and TNF-α (12, 40, 48). Thus, when NF-κB is blocked in RCC (e.g. by bortezomib), antioxidant enzymes like MnSOD and FHC are not induced, and consequent accrual of ROS and other free radicals following IFN-γ stimulation likely results in mitochondrial toxification, respiratory failure and eventual necrotic death (schematically depicted in Fig. 6B).
Collectively, these observations suggest that the combination of IFN-γ and NF-κB inhibition will have therapeutic benefit in RCC. NF-κB, however, is a master transcription factor with multiple functions, and agents that inhibit NF-κB therefore have numerous toxic side-effects (51). The pleiotropic nature of the NF-κB transcriptional response is a major limitation to the development of new NF-κB antagonists, and – to the best of our knowledge - no dedicated NF-κB inhibitor has yet been approved for anti-tumor therapy (51). Although bortezomib mediates its anti-tumor effects at least in part by NF-κB blockade, it almost certainly sensitizes RCC cells to IFN-γ by mechanisms additional to simple inhibiton of NF-κB pro-survival signaling, such as, for example, stabilization of pro-apoptotic proteins or induction of ER stress (17). We therefore suggest that determining how NF-κB specifically mediates cell survival (i.e. identifying druggable pro-survival targets of NF-κB) is essential to the development of next-generation NF-κB-targeted anti-tumor agents. Such agents, by selectively blocking NF-κB pro-survival signaling without affecting other aspects of NF-κB signaling, will be expected to minimize toxicity issues impacting current NF-κB inhibitors.
Supplementary Material
Acknowledgments
We thank Sharon Howard and the Fox Chase Cell Culture Facility for generating and maintaining the FCCC patient-derived NKC and HRC cell lines. We also thank Kerry Campbell for helpful comments and Holly Gillin for administrative assistance.
Grant Support. This work was supported by an ACS Research Scholar Grant (RSG-09-195-01-MPC) to S. Balachandran. Additional funds were provided by the F.M Kirby Foundation, and via institutional support of the Kidney Cancer Keystone Program. R.J. Thapa was supported by an NIH Postdoctoral Training Grant (T32 CA00903534).
Footnotes
Conflicts of Interest: The authors declare that no conflicts of interest exist.
Author Contributions. RJT and PC performed most of the experiments, with assistance from SN. MC, JP and JRT conducted cytological and genetic analyses of HRC cell lines shown in Figure 1 and Supplementary Figure 1. SP analysed NF-κB gene expression profiles in data from RCC patient samples (Figure 5). SB designed the experiments, interpreted data and, together with RJT, PC, SP, and JRT, wrote the manuscript.
References
- 1.Ljungberg B, Campbell SC, Choi HY, Jacqmin D, Lee JE, Weikert S, et al. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60:615–21. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 2.Singer EA, Gupta GN, Srinivasan R. Targeted therapeutic strategies for the management of renal cell carcinoma. Curr Opin Oncol. 2012;24:284–90. doi: 10.1097/CCO.0b013e328351c646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, Delahunt B, et al. The Heidelberg classification of renal cell tumours. J Pathol. 1997;183:131–3. doi: 10.1002/(SICI)1096-9896(199710)183:2<131::AID-PATH931>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 4.Pirrotta MT, Bernardeschi P, Fiorentini G. Targeted-therapy in advanced renal cell carcinoma. Curr Med Chem. 2011;18:1651–7. doi: 10.2174/092986711795471293. [DOI] [PubMed] [Google Scholar]
- 5.Pal SK, Figlin RA. Targeted therapies for renal cell carcinoma: understanding their impact on survival. Target Oncol. 2010;5:131–8. doi: 10.1007/s11523-010-0145-6. [DOI] [PubMed] [Google Scholar]
- 6.Eisen T, Sternberg CN, Robert C, Mulders P, Pyle L, Zbinden S, et al. Targeted therapies for renal cell carcinoma: review of adverse event management strategies. J Natl Cancer Inst. 2012;104:93–113. doi: 10.1093/jnci/djr511. [DOI] [PubMed] [Google Scholar]
- 7.Rosenblatt J, McDermott DF. Immunotherapy for renal cell carcinoma. Hematol Oncol Clin North Am. 2011;25:793–812. doi: 10.1016/j.hoc.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Miller CH, Maher SG, Young HA. Clinical Use of Interferon-gamma. Ann N Y Acad Sci. 2009;1182:69–79. doi: 10.1111/j.1749-6632.2009.05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aulitzky W, Gastl G, Aulitzky WE, Herold M, Kemmler J, Mull B, et al. Successful treatment of metastatic renal cell carcinoma with a biologically active dose of recombinant interferon-gamma. J Clin Oncol. 1989;7:1875–84. doi: 10.1200/JCO.1989.7.12.1875. [DOI] [PubMed] [Google Scholar]
- 10.Ellerhorst JA, Kilbourn RG, Amato RJ, Zukiwski AA, Jones E, Logothetis CJ. Phase II trial of low dose gamma-interferon in metastatic renal cell carcinoma. J Urol. 1994;152:841–5. doi: 10.1016/s0022-5347(17)32587-9. [DOI] [PubMed] [Google Scholar]
- 11.Gleave ME, Elhilali M, Fradet Y, Davis I, Venner P, Saad F, et al. Interferon gamma-1b compared with placebo in metastatic renal-cell carcinoma. Canadian Urologic Oncology Group. N Engl J Med. 1998;338:1265–71. doi: 10.1056/NEJM199804303381804. [DOI] [PubMed] [Google Scholar]
- 12.Thapa RJ, Basagoudanavar SH, Nogusa S, Irrinki K, Mallilankaraman K, Slifker MJ, et al. NF-kappaB protects cells from gamma interferon-induced RIP1-dependent necroptosis. Mol Cell Biol. 2011;31:2934–46. doi: 10.1128/MCB.05445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–14. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–23. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 15.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 16.Morais C, Gobe G, Johnson DW, Healy H. The emerging role of nuclear factor kappa B in renal cell carcinoma. Int J Biochem Cell Biol. 2011;43:1537–49. doi: 10.1016/j.biocel.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 17.McConkey DJ, Zhu K. Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Resist Updat. 2008;11:164–79. doi: 10.1016/j.drup.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Brooks AD, Jacobsen KM, Li W, Shanker A, Sayers TJ. Bortezomib sensitizes human renal cell carcinomas to TRAIL apoptosis through increased activation of caspase-8 in the death-inducing signaling complex. Mol Cancer Res. 2010;8:729–38. doi: 10.1158/1541-7786.MCR-10-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell. 2007;28:15–27. doi: 10.1016/j.molcel.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oya M, Ohtsubo M, Takayanagi A, Tachibana M, Shimizu N, Murai M. Constitutive activation of nuclear factor-kappaB prevents TRAIL-induced apoptosis in renal cancer cells. Oncogene. 2001;20:3888–96. doi: 10.1038/sj.onc.1204525. [DOI] [PubMed] [Google Scholar]
- 21.An J, Rettig MB. Epidermal growth factor receptor inhibition sensitizes renal cell carcinoma cells to the cytotoxic effects of bortezomib. Mol Cancer Ther. 2007;6:61–9. doi: 10.1158/1535-7163.MCT-06-0255. [DOI] [PubMed] [Google Scholar]
- 22.Roos FC, Roberts AM, Hwang, Moriyama EH, Evans AJ, Sybingco S, et al. Oncolytic targeting of renal cell carcinoma via encephalomyocarditis virus. EMBO Mol Med. 2010;2:275–88. doi: 10.1002/emmm.201000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thrash-Bingham CA, Greenberg RE, Howard S, Bruzel A, Bremer M, Goll A, et al. Comprehensive allelotyping of human renal cell carcinomas using microsatellite DNA probes. Proc Natl Acad Sci U S A. 1995;92:2854–8. doi: 10.1073/pnas.92.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basagoudanavar SH, Thapa RJ, Nogusa S, Wang J, Beg AA, Balachandran S. Distinct roles for the NF-kappa B RelA subunit during antiviral innate immune responses. J Virol. 2011;85:2599–610. doi: 10.1128/JVI.02213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbeau B, Bernier R, Dumais N, Briand G, Olivier M, Faure R, et al. Activation of HIV-1 long terminal repeat transcription and virus replication via NF-kappaB-dependent and -independent pathways by potent phosphotyrosine phosphatase inhibitors, the peroxovanadium compounds. J Biol Chem. 1997;272:12968–77. doi: 10.1074/jbc.272.20.12968. [DOI] [PubMed] [Google Scholar]
- 26.Thrash-Bingham CA, Salazar H, Freed JJ, Greenberg RE, Tartof KD. Genomic alterations and instabilities in renal cell carcinomas and their relationship to tumor pathology. Cancer Res. 1995;55:6189–95. [PubMed] [Google Scholar]
- 27.Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–73. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 28.Schraml P, Struckmann K, Hatz F, Sonnet S, Kully C, Gasser T, et al. VHL mutations and their correlation with tumour cell proliferation, microvessel density, and patient prognosis in clear cell renal cell carcinoma. J Pathol. 2002;196:186–93. doi: 10.1002/path.1034. [DOI] [PubMed] [Google Scholar]
- 29.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44:17–9. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 31.Duns G, Hofstra RM, Sietzema JG, Hollema H, van Duivenbode I, Kuik A, et al. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Hum Mutat. 2012;33:1059–62. doi: 10.1002/humu.22090. [DOI] [PubMed] [Google Scholar]
- 32.Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, et al. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet. 1988;42:217–26. [PMC free article] [PubMed] [Google Scholar]
- 33.Duns G, Hofstra RMW, Sietzema JG, Hollema H, van Duivenbode I, Kuik A, et al. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Hum Mutat. 2012;33:1059–62. doi: 10.1002/humu.22090. [DOI] [PubMed] [Google Scholar]
- 34.Sufan RI, Jewett MA, Ohh M. The role of von Hippel-Lindau tumor suppressor protein and hypoxia in renal clear cell carcinoma. Am J Physiol Renal Physiol. 2004;287:F1–6. doi: 10.1152/ajprenal.00424.2003. [DOI] [PubMed] [Google Scholar]
- 35.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 36.Chen D, Frezza M, Schmitt S, Kanwar J, QPD Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11:239–53. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Younes HM, Amsden BG. Interferon-gamma therapy: evaluation of routes of administration and delivery systems. J Pharm Sci. 2002;91:2–17. doi: 10.1002/jps.10007. [DOI] [PubMed] [Google Scholar]
- 38.Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–21. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 1999;13:382–7. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pham CG, Bubici C, Zazzeroni F, Papa S, Jones J, Alvarez K, et al. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 2004;119:529–42. doi: 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 41.An J, Rettig MB. Mechanism of von Hippel-Lindau protein-mediated suppression of nuclear factor kappa B activity. Mol Cell Biol. 2005;25:7546–56. doi: 10.1128/MCB.25.17.7546-7556.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An J, Sun Y, Fisher M, Rettig MB. Maximal apoptosis of renal cell carcinoma by the proteasome inhibitor bortezomib is nuclear factor-kappaB dependent. Mol Cancer Ther. 2004;3:727–36. [PubMed] [Google Scholar]
- 43.Oya M, Takayanagi A, Horiguchi A, Mizuno R, Ohtsubo M, Marumo K, et al. Increased nuclear factor-kappa B activation is related to the tumor development of renal cell carcinoma. Carcinogenesis. 2003;24:377–84. doi: 10.1093/carcin/24.3.377. [DOI] [PubMed] [Google Scholar]
- 44.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–9. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 45.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 46.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–9. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 47.Kowaltowski AJ, de Souza-Pinto NC, Castilho RF, Vercesi AE. Mitochondria and reactive oxygen species. Free Radic Biol Med. 2009;47:333–43. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Sakon S, Xue X, Takekawa M, Sasazuki T, Okazaki T, Kojima Y, et al. NF-kappaB inhibits TNF-induced accumulation of ROS that mediate prolonged MAPK activation and necrotic cell death. Embo J. 2003;22:3898–909. doi: 10.1093/emboj/cdg379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathupala SP, Ko YH, Pedersen PL. The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim Biophys Acta. 1797:1225–30. doi: 10.1016/j.bbabio.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moreno-Sanchez R, Rodriguez-Enriquez S, Saavedra E, Marin-Hernandez A, Gallardo-Perez JC. The bioenergetics of cancer: is glycolysis the main ATP supplier in all tumor cells? Biofactors. 2009;35:209–25. doi: 10.1002/biof.31. [DOI] [PubMed] [Google Scholar]
- 51.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.