Abstract
Lipid secretions from algae pose a great opportunity for engineering biofueler feedstocks. The lipid exudates could be interesting from a process engineering perspective because lipids could be collected directly from the medium without harvesting and disrupting cells. We here report on the extracellular secretions of algal metabolites from the strain UTEX 2341 (Chlorella minutissima) into the culture medium. No detailed analysis of these lipid secretions has been performed to date. Using multiple mass spectrometric platforms, we observed around 1000 compounds and were able to annotate 50 lipids by means of liquid chromatography coupled to accurate mass quadrupole time-of-flight mass spectrometry (LC-QTOF), direct infusion with positive and negative electrospray ion trap mass spectrometry and gas chromatography coupled to mass spectrometry (GC–MS). These compounds were annotated by tandem mass spectral (MS/MS) database matching and retention time range filtering. We observed a series of triacylglycerols (TG), sulfoquinovosyldiacylglycerols (SQDG), phosphatidylinositols and phosphatidylglycerols, as well as betaine lipids diacylglyceryl-N,N,N-trimethylhomoserines (DGTS).
Keywords: Biofuel, Algae, Lipids, LC–MS, GC–MS
1. Introduction
Algae secrete small molecules such as fatty acids, polysaccharides, small peptides and amino acids as well as exotoxins into their environment [1]. Understanding such secretions is important for deeper insights into laboratory microecosystems of algae, metabolism studies of algae and algal biofuel research. Pratt observed such extracellular secretions from Chlorella algae diffuse through cell walls and leak into the culture medium [2]. The algal exudate (Chlorellin) had inhibitory effects on bacteria and algae. The mixture was later analyzed to contain free fatty acids which are cytotoxic to cell membranes [3]. Isotopic tracer experiments were used to postulate that the excretion of a series of amino acids is synchronized to the light cycle of algal photosynthesis [4,5]. Bell and Mitchel observed that bacteria belonging to the genus Spirillum were attracted within 10–15 min to algal exudates from the diatomaceous algae Skeletonema [6]. Bacteria can grow in algal exudates such as amino acids and sugars without additional carbon sources [7]. It also has been discussed that macroalgae release chemical compounds in defense of herbivores that graze on such algae [8] or predators that actively evolved to detect chemical signals released from algae [9]. Toxic algal secretions including domoic acid or brevetoxins occur during harmful algal blooms (HAB) and are threatening to other wildlife species [10]. It has been hypothesized that algae and bacteria release mucoid substances as protection against ultraviolet radiation [11]. On diatom algae (phytoplankton) the extracellular mucilages contain a complex mixture of proteoglycans and sulfated polysaccharides [12].
In this research paper we investigate the extracellular lipid secretions (see Fig. 1) from the unicellular green algae UTEX 2341 (annotated as Chlorella minutissima [13,14]). Such extracellular secretions were reported previously by Gladu et al. where an scanning electron microscope (SEM) image shows mucoid substances outside the cell walls [14]. UTEX 2341 is a promising candidate for biofuel and biomass production, however the molecular composition of lipid secretions has not been reported in detail. The analysis of such material at the molecular level and the complexity calls for a series of analytical platforms and separation techniques to be used (see Fig. 2). Fast fingerprint infusion spectra can be obtained with the direct analysis in real time (DART) ion source coupled to mass spectrometers [15]. Fatty acids analysis can be performed best by converting free fatty acids into their methyl esters (FAME) and analyzing them with GC–MS. Using a preseparation technique such as thin-layer chromatography (TLC) enhances the information content of FAME analysis [16].Also matrix-assisted laser desorption/ionization coupled to accurate mass time-of-flight instruments (MALDI-TOF-MS) were used for the analysis of algal lipids in the past [17]. A detailed fragmentation analysis using tandem mass spectrometry analysis software (MS/MS) is possible by nanoelectrospray (nanoESI) chip based direct infusion [18]. Using ultra-high resolving power Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FT-ICR-MS) it is possible to analyze multiple lipid classes and especially generate unique elemental compositions for lipid species [19]. A detailed separation of multiple lipid species can be obtained with high-performance liquid chromatography coupled to accurate mass quadrupole time-of-flight mass spectrometers (LC-QTOF) [20,21] or LC-Orbitrap mass spectrometry [22].
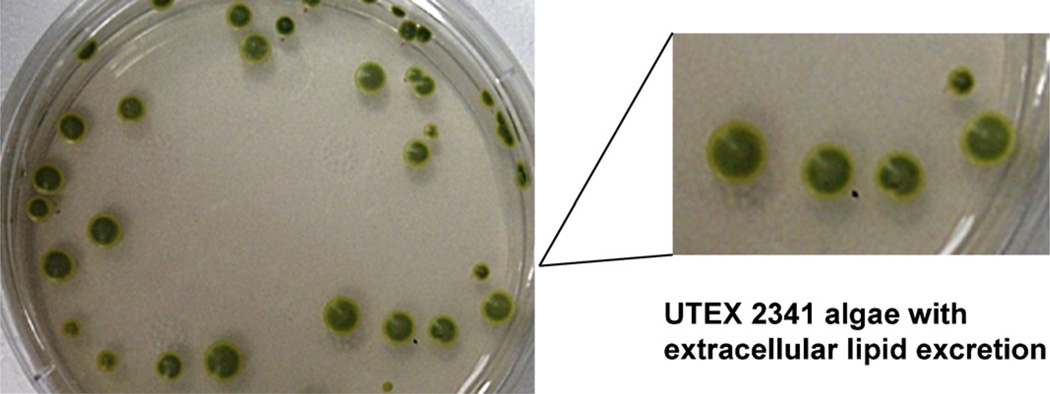
Fig. 1.
The algae UTEX 2341 (Chlorella minutissima) secretes lipid material into the surrounding media. The photo shows a circular yellow secretion around the green cell body. The exudate was collected, extracted and analyzed with different analytical techniques. Bacterial contaminations were not observed by means of a microscopy method.
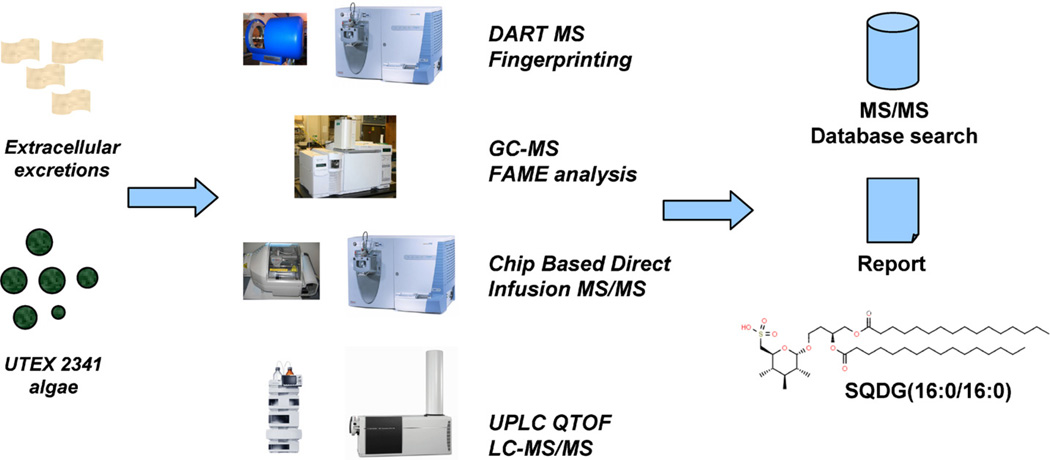
Fig. 2.
Analytical platform to investigate extracellular lipid secretions from algae. Four different mass spectrometric platforms were used to analyze a complex mixture of lipids.
2. Methods
2.1. Algal growth and separation of algal secretions
The algal strain UTEX 2341 (assigned as C. minutissima, also assigned to the Nannochloropsis genus [14] and other species [13]) was obtained from the culture collection of algae at the University of Texas at Austin (UTEX). Chlorella UTEX 2341 was maintained in N8Y medium, prepared by adding 0.1% yeast extract to N8 medium at 25 °C and irradiated with fluorescent light at ∼2000 lx on a 16:8 h light/dark cycle. For extracellular matter production UTEX 2341 was grown on solid ATCC medium #5 (sporulation agar) in Petri dishes. A control plate (no growth) was included for comparison. After one week a yellow-white secretion from individual UTEX 2341 colonies was observed (see Fig. 1). The extracellular matter was separated and extracted with a modified Folch method [23]. To do this, the secreted matter was first washed from the algal colony using 2 ml methanol and filtered through a 0.22 µm filter into a 50 ml Falcon tube. The total volume was adjusted to 5 ml with methanol and 10 ml chloroform was added and vortexed. Additionally 3 ml water was added and vortexed. The mixture was centrifuged at 1000 × g for 2 min, and the upper phase, interface and lower phase were collected and lyophilized for 48 h.
2.2. Direct analysis in real time (DART) analysis
A direct analysis in real time (DART) ion source (IonSense Inc., Saugus, MA) was coupled to a ThermoFinnigan LTQ linear iontrap (ThermoFinnigan, San Jose, CA) operating in full-scan mode from 200 to 2000 Da. Spectra were recorded with the Xcalibur software. Infusion mass spectra were summed over abundant scans. Solvent blanks were measured to exclude peaks from contaminations. The infusion time was around 5–10 s. The DART 100-CE was operated with an effective helium flow rate of 2.0l/min. The heater control was set to 250 °C, the grid electrode voltage was 250 V, the discharge electrode voltage was 150 V and needle voltage was set to 3000 V.
2.3. GC–MS FAME analysis
In order to analyze fatty acids in the GC–MS, their methyl ester analogs (FAME) had to be prepared. The dried sample was dissolved in 200 µl hexane and transferred to another glass vial and mixed with 50 µl toluene, and then 100 µl BCl3-methanol was added (Sigma–Aldrich; Supelco 33353; boron trichloride in methanol 12%). The mixture was heated in a water bath at 75 °C for 10 min. After cooling, 200 µl distilled water was added. The organic and aqueous phases were then separated and 100 µl of the upper organic phase containing the fatty esters was used for further GC–MS analysis. All solvents and plastic Eppendorf tubes were screened to determine the level of contamination (solvent blanks and method blanks). The injector was an Apex ProSep 800 precolumn separation inlet (Apex Technologies, Inc., NJ). The precolumn mode program was set to splitless mode for 0.17 min, and then changed to GC split for 2.5 min. The precolumn oven of the ProSep was held at 50 °C for 0.10 min and then programmed to 250 °C at 75 °C/min and held for 0.10 min, finally got to 350 °C at 50 °C/min and held for 32 min. The precolumn method here was for FAME analysis.
The samples were analyzed with an Agilent 6890 gas chromatograph coupled to an Agilent 5973 MSD (Agilent, Santa Clara, CA). 5 µl sample was injected at different split ratios of 10:1, 20:1 and 100:1 by an Agilent 7683B series autosampler (Agilent, Santa Clara, CA). The GC column was an Agilent HP5-MS capillary column bonded 5% phenyl, 95% dimethylpolysiloxane (30 m × 250 µm i.d., 0.25 µm film thickness). The temperature program was started at 60 °C for 0.5 min. Then the temperature was ramped to 320 °C at 10 °C/min, and then held at 325 °C for 10 min. Helium was used as carrier gas with constant flow rate of 1.0 ml/min. The solvent delay was 6.35 min. The MS filament source temperature was set to 230 °C; the quadrupole temperature was set to 150 °C and the transfer line temperature was 280 °C. Spectra were acquired in positive (70 eV) full scan mode from 50 to 650 m/z at 2 spectra/s scan speed.
2.4. Nanoelectrospray chip-based infusion into a linear iontrap mass spectrometer
A chip based nano-electrospray infusion (Advion Nanomate) was coupled to a LTQ linear iontrap mass spectrometer (Thermo Fisher Scientific). 10 µg of the dried Folch extract was resuspended with 100 µl methanol/chloroform (9/1, v/v) (degassed with nitrogen). The sample was vortexed and centrifuged for 2 min at 14,000 rpm. Then 10 µl is diluted with 90 µl methanol/chloroform (9/1, v/v) containing 7.5 mM ammonium acetate and is transferred to the well plates. The plates were covered with aluminum foil to prevent evaporation of the solvent. The infusion of the extracted lipid samples was performed using with an Advion “C” chip, the Nanomate cooling plate was set to 10 °C, the Nanomate gas pressure to 0.4 psi and the voltage to 2.0 kV. 10 µl is infused with an aspiration delay of 3 s. A data–dependent MS/MS method collected tandem mass spectra in positive and negative mode over a range of 20 min infusion time. The covered m/z range in the method was split from 350 to 450 Da, 450 to 750 Da and 750 to 1200 Da in order to increase the number of MS/MS spectra for individual components. Each file contained between 100 and 150 MS/MS precursor ions and the related product ion spectra that averaged between 1 and 30 scans each. Solvent blanks and method blanks were measured to analyze existing background contaminants from plastic material and extraction solvents.
2.5. Chromatographic and mass spectral settings on the Q-TOF instrument
Instrumentation consisted of an Agilent 1200 LC stack (binary pump SL, degasser, autosampler, thermostat, column oven) interfaced to an Agilent 6530 accurate–mass quadrupole time-of-flight (Q-TOF) with a JetStream ESI source. Due to the low solubility of the excretion material in water/methanol, 5 µl of the Nanomate infusion solvent was used for LC–MS analysis (see method above). The material was injected on an Acquity 1.7 µm BEH HILIC 2.1 mm × 150 mm column (Waters Corporation, Milford, MA). Column temperature was kept at 40 °C. The mobile phase consisted of water with 5 mM ammonium acetate and 0.2% acetic acid (A) and 9:1 acetonitrile/water with 5 mM ammonium acetate and 0.2% acetic acid (B). The gradient method was: 0–4 min − 100% B, 4–12 min – linear gradient to 45% B, and 12–20 min − 45% B. The column was re-equilibrated for 20 min following the separation of each sample, and the flow rate was constant at 0.25 ml/min throughout the gradient method and re-equilibration. MS and MS/MS data were collected with a 0.25 s scan rate in both profile and centroid modes, and mass calibration was maintained by constant infusion of reference ions at 121.0509 and 922.0098 m/z. MS/MS data was generated utilizing data-dependent MS/MS triggering with dynamic exclusion. Precursor ions, with a minimum 1 k signal intensity were isolated with a 4 m/z isolation width (medium setting), and a variable collision energy was applied based on precursor ion m/z (10 eV + 0.03 eV× ion m/z). Ions were excluded from data dependent MS/MS analysis for 30 s following acquisition of two spectra. Data were exported into the open exchange format mzXML Samples were measured in negative and positive mode. Additionally blank runs were acquired to determine the level of background contaminations.
2.6. Data processing and MS/MS database search
The identification of compounds was based on authentic reference MS/MS spectra and an in-house developed MS/MS database (LipidBlast).The QTOF-LC–MS/MS spectra were extracted with Agilent MassHunter software and exported as MGF format (10 counts abundance minimum). For the LTQ instrument the Thermo RAW files were converted using the Thermo ExtractMSn software and the freely available DeconMSn program [24]. The MGF format is a container format that holds multiple MS/MS scans with their referenced precursor ion masses. In order to create consensus MS/MS scans and to reduce the number of spectra all similar MS/MS scans with the same precursor ions were clustered using the MSCluster program [25].
For low-resolution MS/MS search the freely available NIST MS Search GUI program [26] and the NIST MSPepsearch GUI [27] were used. A precursor search accuracy of ±0.4 Da and a product ion tolerance to ±0.8 Da was set for the low resolution iontrap and ±0.05 Da precursor accuracy and ±0.4 Da product ion tolerance was applied in case of the high resolution QTOF instrument. A precursor accuracy of 0.008 Da was used for the direct lookup of masses from the QTOF in data-tables containing accurate adduct masses.
All calculations were performed on a Monarch Computer Dual Opteron 254 (2.8 GHz) with an ARECA-1120 Raid-6 array using WD Raptor and Samsung hard disks (max hard disk burst read/write transfer rate 500 MB/s) equipped with 2.8 GB RAM running a 32-bit Windows XP. An additional RamDisk (QSoft Ramdisk Enterprise) was used for file based operations allowing burst read-write rates of 1000 MB/s.
3. Results
3.1. Analysis of algal secretions with DART-linear iontrap mass spectrometry
The DART analysis allows the fast acquisition of mass spectra without any sample preparation or preseparation, because the sample is directly infused by holding a glass capillary into the open DART ion source. Such fingerprinting methods are preferable in order to perform classifications in a high-throughput mode. A complex mixture was detected from the mass spectrum (see Supplement) and based on this observation additional analysis steps were performed as discussed below.
3.2. Analysis of algal free fatty acid secretions with GC–MS (FAME method)
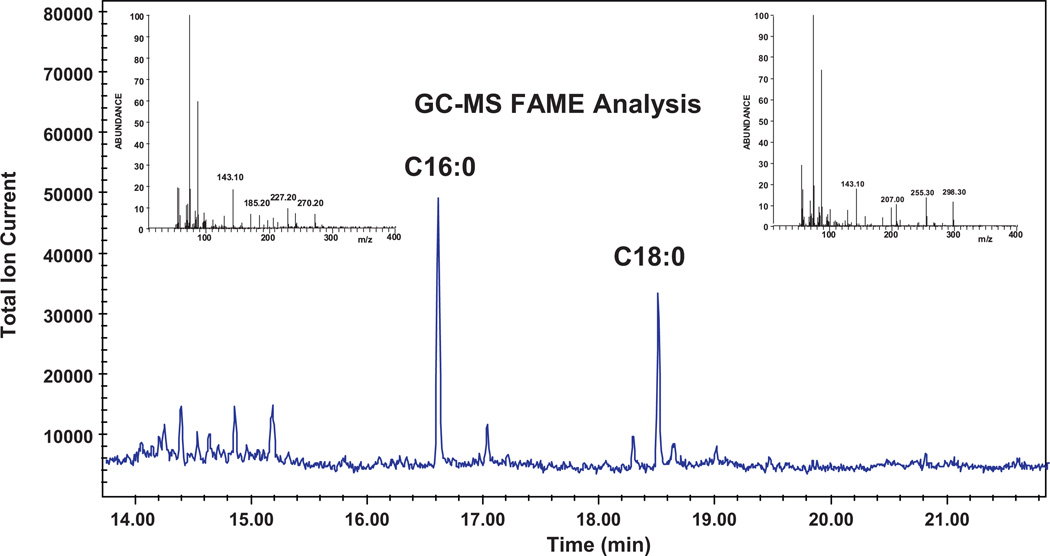
Two main free fatty acids components were observed in the excretion by FAME analysis. C16:0 (hexadecanoic acid methyl ester; methyl palmitate) and C18:0 (octadecanoid acid methyl ester; methyl stearate) were annotated using authentic FAME reference standards with retention time and mass spectral matching. Fig. 3 shows the chromatogram and the mass spectra with the characteristic FAME marker ions m/z 74, 87, 143 and molecular ions m/z 270 (C16:0) and m/z 298 (C18:0). No other free fatty acids as FAMES were observed in the excretion using these characteristic m/z markers.
Fig. 3.
A GC–MS fatty acid methyl ester (FAME) analysis revealed two major peaks in the secretions. These peaks in the chromatogram were identified as C16:0 (hexadecanoic acid methyl ester) and C18:0 (octadecanoid acid methyl ester) using retention time and mass spectral matching.
3.3. Analysis of algal secretions with chip based nanoESI direct infusion
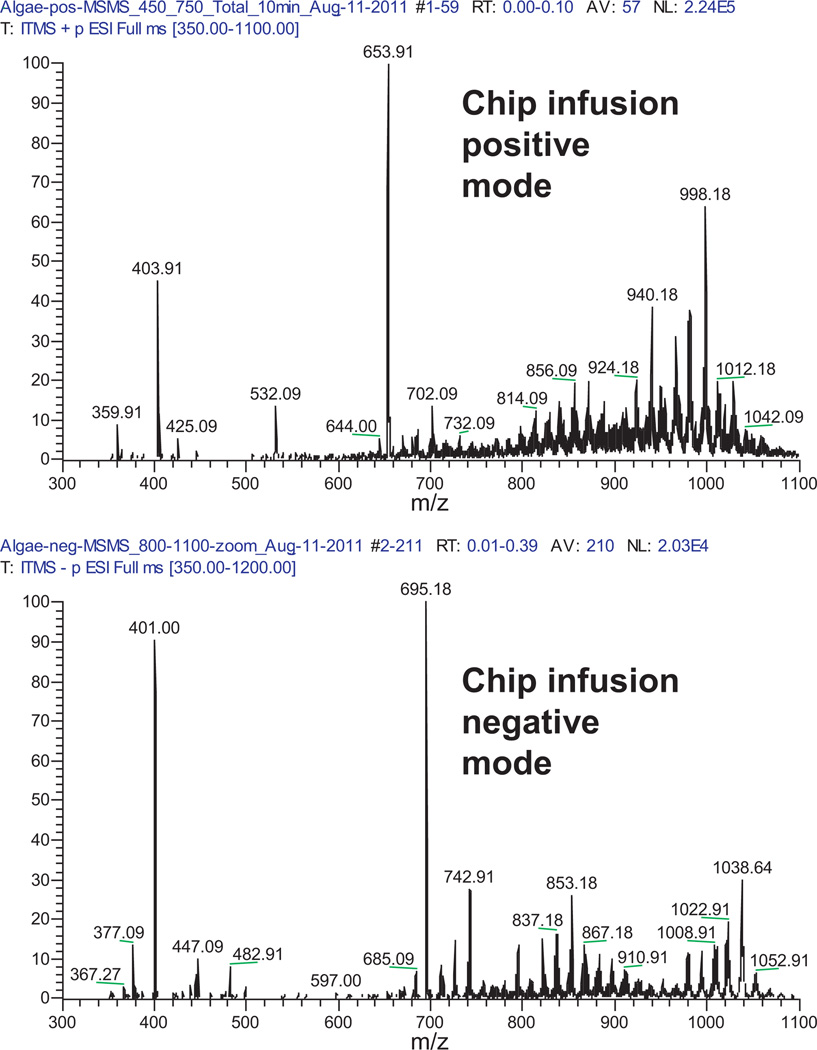
The chip-based nanoelectrospray direct infusion coupled to a low resolution iontrap platform utilized data-dependent MS/MS scans in positive and negative mode (see Fig. 4). Because of the extended infusion times it is possible to collect MS/MS scans over the range of minutes. Around 2500 MS/MS precursors were fragmented in positive mode and around 1400 in negative mode. Compounds were identified based on precursor search and subsequent MS/MS match. Among the annotated compounds are triacylglycerols, phosphatidylcholines and DGTS betaine lipids in positive mode and sulfoquinovosyldiacylglycerols (SQDG) and phosphatidylinositols in negative mode (see Table 1). Due to the complexity of the exudate and same mass isobaric overlaps many of the tandem mass spectra represented mixed compounds.
Fig. 4.
Chip based nanoelectrospray infusion allows for long direct infusion and data dependent MS/MS. Complex lipids were observed in the algal secretions. The tandem mass spectra are then searched in MS/MS libraries.
Table 1.
Identified compounds in UTEX 2341 lipid secretions using LC–QTOF-MS/MS and direct infusion nanoESI-iontrap-MS/MS. All compounds were confirmed with MS/MS library search. The reverse dot product represents the level of confidence. Compound abbreviations see text. Compound annotations without retention time were obtained from the low resolution direct infusion method. Doublet identifications from multiple adducts or platforms were removed.
| Num | RT [min] | Prec m/z experiment | Prec m/z library | Delta m/z | Rev-Dot library | Library MS/MS match | Platform |
|---|---|---|---|---|---|---|---|
| 1 | 1.51 | 793.51996 | 793.51358 | −0.00638 | 996 | SQDG 32:0; [M−H]−; SQDG(16:0/16:0) | LC–MS, Inf-MS/MS |
| 2 | 1.54 | 765.48730 | 765.48228 | −0.00502 | 821 | SQDG 30:0; [M−H]−; SQDG(14:0/16:0) | LC–MS, Inf-MS/MS |
| 3 | 2.13 | 900.80153 | 900.80153 | 0.00000 | 806 | TG 54:4; [M+NH4]+; TG(18:1/18:1/18:2) | LC–MS/MS |
| 4 | 2.13 | 900.80153 | 900.80153 | 0.00000 | 706 | TG 54:4; [M+NH4]+; TG(18:0/18:2/18:2) | LC–MS/MS |
| 5 | 2.13 | 900.80153 | 900.80153 | 0.00000 | 768 | TG 54:4; [M+NH4]+; TG(18:0/18:1/18:3) | LC–MS/MS |
| 6 | 2.16 | 874.78714 | 874.78589 | −0.00125 | 837 | TG 52:3; [M+NH4]+; TG(16:0/18:1/18:2) | LC–MS/MS |
| 7 | 2.16 | 874.78714 | 874.78589 | −0.00125 | 823 | TG 52:3; [M+NH4]+; TG(16:1/18:1/18:1) | LC–MS/MS |
| 8 | 2.16 | 874.78714 | 874.78589 | −0.00125 | 431 | TG 52:3; [M+NH4]+; TG(16:0/18:0/18:3) | LC–MS/MS |
| 9 | 2.19 | 902.81833 | 902.81717 | −0.00116 | 662 | TG 54:3; [M+NH4]+; TG(18:0/18:1/18:2) | LC–MS/MS |
| 10 | 2.38 | 848.77054 | 848.77025 | −0.00029 | 812 | TG 50:2; [M+NH4]+; TG(16:0/16:1/18:1) | LC–MS/MS |
| 11 | 2.38 | 848.77054 | 848.77025 | −0.00029 | 787 | TG 50:2; [M+NH4]+; TG(16:0/16:0/18:2) | LC–MS/MS |
| 12 | 2.38 | 848.77054 | 848.77025 | −0.00029 | 642 | TG 50:2; [M+NH4]+; TG(16:1/16:1/18:0) | LC–MS/MS |
| 13 | 2.55 | 835.53741 | 835.53368 | −0.00373 | 380 | PI 34:1; [M−H]−; GPIns(16:0/18:1) | LC–MS, Inf-MS/MS |
| 14 | 2.58 | 833.52075 | 833.51800 | −0.00275 | 509 | PI 34:2; [M−H]−; GPIns(16:0/18:2) | LC–MS/MS |
| 15 | 2.62 | 850.78668 | 850.78589 | −0.00079 | 687 | TG 50:1; [M+NH4]+; TG(16:0/16:0/18:1) | LC–MS/MS |
| 16 | 2.62 | 850.78668 | 850.78589 | −0.00079 | 621 | TG 50:1; [M+NH4]+; TG(16:0/16:1/18:0) | LC–MS/MS |
| 17 | 2.62 | 850.78668 | 850.78589 | −0.00079 | 776 | TG 50:1; [M+NH4]+; TG(14:0/18:0/18:1) | LC–MS/MS |
| 18 | 2.98 | 876.80280 | 876.80153 | −0.00127 | 667 | TG 52:2; [M+NH4]+; TG(16:1/18:0/18:1) | LC–MS/MS |
| 19 | 2.98 | 876.80280 | 876.80153 | −0.00127 | 772 | TG 52:2; [M+NH4]+; TG(16:0/18:0/18:2) | LC–MS/MS |
| 20 | 3.32 | 904.83202 | 904.83281 | 0.00079 | 602 | TG 54:2; [M+NH4]+; TG(16:0/18:1/20:1) | LC–MS/MS |
| 21 | 3.32 | 904.83202 | 904.83281 | 0.00079 | 745 | TG 54:2; [M+NH4]+; TG(18:0/18:1/18:1) | LC–MS/MS |
| 22 | 3.32 | 904.83202 | 904.83281 | 0.00079 | 897 | TG 54:2; [M+NH4]+; TG(16:0/18:2/20:0) | LC–MS/MS |
| 23 | 3.32 | 904.83202 | 904.83281 | 0.00079 | 641 | TG 54:2; [M+NH4]+; TG(18:0/18:0/18:2) | LC–MS/MS |
| 24 | 3.32 | 904.83202 | 904.83281 | 0.00079 | 531 | TG 54:2; [M+NH4]+; TG(16:1/18:1/20:0) | LC–MS/MS |
| 25 | 3.32 | 904.83202 | 904.83281 | 0.00079 | 552 | TG 54:2; [M+NH4]+; TG(16:1/18:0/20:1) | LC–MS/MS |
| 26 | 3.32 | 904.83202 | 904.83281 | 0.00079 | 339 | TG 54:2; [M+NH4]+; TG(16:0/18:0/20:2) | LC–MS/MS |
| 27 | 3.47 | 902.81833 | 902.81717 | −0.00116 | 678 | TG 54:3; [M+NH4]+; TG(18:1/18:1/18:1) | LC–MS/MS |
| 28 | 3.47 | 902.81833 | 902.81717 | −0.00116 | 919 | TG 54:3; [M+NH4]+; TG(16:1/18:0/20:2) | LC–MS/MS |
| 29 | 3.47 | 902.81833 | 902.81717 | −0.00116 | 329 | TG 54:3; [M+NH4]+; TG(16:1/18:1/20:1) | LC–MS/MS |
| 30 | 3.72 | 876.80280 | 876.80153 | −0.00127 | 809 | TG 52:2; [M+NH4]+; TG(16:0/18:1/18:1) | LC–MS/MS |
| 31 | 3.72 | 876.80280 | 876.80153 | −0.00127 | 827 | TG 52:2; [M+NH4]+; TG(16:1/16:1/20:0) | LC–MS/MS |
| 32 | 3.11 | 736.60925 | 736.60912 | −0.00013 | 983 | DGTS 34:2; [M+H]+; DGTS(16:0/18:2) | LC–MS/MS |
| 33 | 3.16 | 738.62463 | 738.62477 | 0.00014 | 986 | DGTS 34:1; [M+H]+; DGTS(16:0/18:1) | LC–MS/MS |
| 34 | 3.17 | 764.63788 | 764.64042 | 0.00254 | 990 | DGTS 36:2; [M+H]+; DGTS(18:1/18:1) | LC–MS/MS |
| 35 | 3.22 | 762.62387 | 762.62477 | 0.00090 | 987 | DGTS 36:3; [M+H]+; DGTS(18:1/18:2) | LC–MS/MS |
| 36 | 9.31 | 786.60181 | 786.60126 | −0.00055 | 614 | PC 36:2; [M+H]+; GPCho(18:1/18:1) | LC–MS/MS |
| 37 | 9.34 | 784.58606 | 784.58564 | −0.00042 | 600 | PC 36:3; [M+H]+; GPCho(18:1/18:2) | LC–MS/MS |
| 38 | 9.35 | 760.58679 | 760.58564 | −0.00115 | 606 | PC 34:1; [M+H]+; GPCho(16:0/18:1) | LC–MS/MS |
| 39 | 9.42 | 758.56970 | 758.56995 | 0.00025 | 504 | PC 34:2; [M+H]+; GPCho(16:0/18:2) | LC–MS/MS |
| 40 | 11.14 | 522.35629 | 522.35596 | −0.00033 | 318 | LPC 18:1; [M+H]+; PC(18:1(11E)/0:0) | LC–MS/MS |
| 41 | NA | 814.950 | 814.633 | −0.317 | 995 | PC 38:2; [M+H]+; GPCho(18:1/20:1) | Inf-MS/MS |
| 42 | NA | 837.320 | 837.549 | 0.229 | 761 | PI 34:0; [M−H]−; GPIns(16:0/18:0) | Inf-MS/MS |
| 43 | NA | 851.420 | 851.565 | 0.145 | 488 | PI 35:0; [M−H]−; GPIns(16:0/19:0) | Inf-MS/MS |
| 44 | NA | 865.412 | 865.581 | 0.169 | 161 | PI 36:0; [M−H]−; GPIns(16:0/20:0) | Inf-MS/MS |
| 45 | NA | 881.427 | 881.518 | 0.091 | 246 | PI 38:6; [M−H]−; GPIns(16:0/22:6) | Inf-MS/MS |
| 46 | NA | 737.060 | 737.451 | 0.391 | 853 | SQDG 28:0; [M−H]−; SQDG(12:0/16:0) | Inf-MS/MS |
| 47 | NA | 752.130 | 752.640 | 0.510 | 780 | DGTS 35:1; [M+H]+; DGTS(16:0/19:1) | Inf-MS/MS |
| 48 | NA | 754.260 | 754.656 | 0.396 | 773 | DGTS 35:0; [M+H]+; DGTS(16:0/19:0) | Inf-MS/MS |
| 49 | NA | 896.060 | 896.770 | 0.710 | 913 | TG 54:6; [M+NH4]+; TG(18:2/18:2/18:2) | Inf-MS/MS |
| 50 | NA | 895.320 | 894.755 | −0.565 | 987 | TG 54:7; [M+NH4]+; TG(18:2/18:2/18:3) | Inf-MS/MS |
3.4. Analysis of algal secretions with LC–QTOF-MS/MS
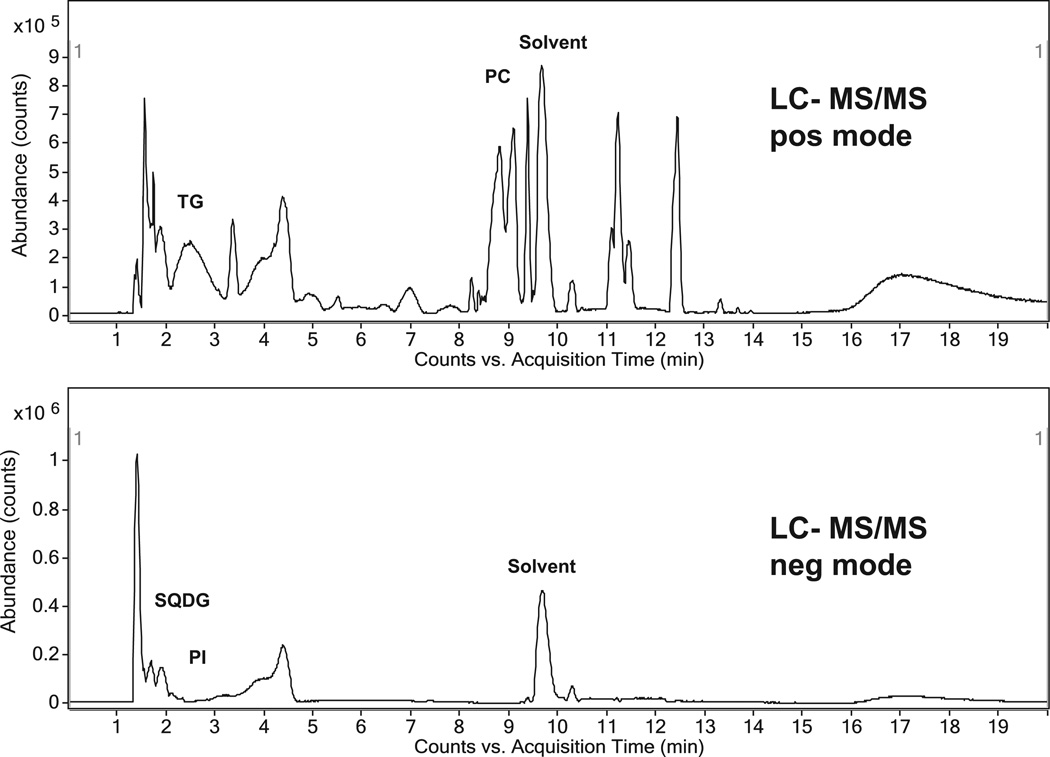
The high-resolution accurate mass LC–MS was performed in positive mode and negative mode (see Fig. 5). Around 3000 signals (m/z-retention time pairs) were observed in positive mode and 1300 m/z-RT pairs in negative mode. 1400 MS/MS scans in positive and negative mode were acquired. Half of these MS/MS scans were redundant, because they were obtained from the same precursor ion. The positive ionization mode allowed the annotation of triacylglycerols, phosphatidylcholines and diacylglyceryl-N,N,N-trimethylhomoserines (betaine lipids), whereas in negative mode several phosphatidylinositols and sulfoquinovosyldiacylglycerols were observed (see Table 1). All annotations were performed via accurate mass precursor search, retention time matching and additional MS/MS search or confirmation of product ion matches. Because of the high complexity many compounds co-eluted, however MS/MS product ion search and fatty acyl losses allowed for confident annotation of compound classes and fatty acyl carbon chain length and degree of unsaturation. The method did not allow for assignment of stereochemistry and regiospecific or positional analysis of fatty acyl chains. Using negative ion mode a series of fatty acyl chains were observed including m/z 249.153 (C16:3), 255.233 (C16:0), 281.248 (C18:1), 283.264 (C18:0), 295.229 (C18:2(OH)), 297.155 and 297.243 (epoxy/OH/cyclo FA), 311.170 (C20:0), 321.175, 325.185 and 339.200 (epoxy/OH/cyclo FA). The complete elucidation of these fatty acyls requires a more detailed analysis with picolinyl, TMS or DMOX derivates [28]. In addition, an exhaustive query on matching accurate precursor ion masses against lipid databases was performed yielding a high number of tentative lipid annotations (see supplemental data). However, these tentative annotations lack confirmatory MS/MS product spectra and therefore such annotations potentially have a higher false-positive and false negative-rate.
Fig. 5.
Hydrophilic interaction chromatography (HILIC-UPLC) coupled to high resolution QTOF-MSMS reveals a complex mixture of compounds. A series of lipids are annotated using MS/MS scans including triacylglycerols (TG), sulfoquinovosyl-diacylglycerols (SQDG), phosphatidylinositols (PI) and phosphatidylcholines (PC).
4. Discussion
4.1. Taxonomic classification of algae is still a challenging process
Correct taxonomic classifications are important for the compound annotations process. If taxonomy–metabolite relationships are known, such target compound lists can be assembled from databases and a targeted analytical screening process can be performed [29]. The problem is that even in major strain collections, taxonomy annotations may be wrong [13]. The classifications found in these strain collections can be based on microscopic observations, chemotaxonomy data such as the content of alkanes, chlorophyll or carotenoids or they are purely based on genomic information. The algal strain UTEX 2341 (C minutissima) was assigned to the Nannochloropsis species in one publication [14]. This information is also maintained at the UTEX website (http://www.sbs.utexas.edu/utex/). A phylogenetic comparison based on the 18S ribosomal RNA subunit placed UTEX 2341 (named as Chlorella multissima) into the Trebouxiophyceae class, within close proximity to Botryococcaceae and Chlorellaceae as well as Nanochlorum eukaryotum (Nannochloris species) [30]. A very recent sequence comparison based on the 23 s rRNA genomic sequence [31], shows a cladogram with UTEX 2341 in near proximity to other Chlorella species [32]. A final correct classification may be obtained after all 4000 Chlorophyta species (green algae) will be fully sequenced.
4.2. Algae contain and secrete diverse compound mixtures during their life cycle
Algae contain a series of lipid-like compound classes including free fatty acids, sterols, carotenes, phospholipids, galactolipids, betaine lipids and glycerolipids [33]. The main phospholipid compound classes are phosphatidylcholine (PC), phosphatidylethanolamines (PE), phosphatidic acids (PA), phosphatidylinositols (PI), and phosphatidylglycerols (PG). The main glycolipids include monogalactosyldiacylglycerol (MGDG), digalac-tosyldiacylglycerols (DGDG) and sulfoquinovosyldiacylglycerols (SQDG). Nonpolar lipids include monoacylglycerols (MG), diacylglycerols (DG) and triacylglycerols (TG). Many algae and bacteria also substitute nitrogen for phosphorus and form phosphate-free betaine and ornithine lipids [34]. These betaine lipids include diacylglyceryl-N,N,N-trimethylhomoserine (DGTS), diaclyglycerylhydroxymethyl-N,N,N-trimethyl-beta-alanine (DG-TA) and 1,2-diacylglyceryl-3-O-carboxyhydroxymethylcholine (DGCC) [35–38]. In the model algae Chlamydomonas reinhardtii phosphatidylcholine (PC) is completely replaced by the betaine lipid diacylglycerol-O-(N,N,N-trimethyl)-homoserine (DGTS) [39]. C minutissima contains high contents of DGTS as well as PC as major phospholipids [40]. However the closely related Chlorella vulgaris strain does not contain DGTS but only phosphatidylcholine [36]. DGTS or DGTA were observed in the UTEX 2341 secretions (see Fig. 6). Using MS/MS analysis the DGTS and DGTA betaine lipids are easily detected by the dominant product ion m/z 236 [41,42] and DGCC by the ions m/z 104 and 132 [43]. Sulfoquinovosyldiacylglycerols (SQDG) were observed in Chlorella more than 50 years ago [44]. Chlorella algae grown on cysteine as a sole sulfur source instead of sulfate, had a 15 times lower CO2 fixation rate, synthesized larger amounts of triglycerides and lost their ability to grow photoautotrophically [45].With the FAME GC–MS methods we were able to detect the main free fatty acids C16:0 and C18:0 in the algal secretions. Because we observed additional lipid-bound fatty acids with the LC–MS/MS method we have to assume that no transesterification occurred under mild reaction temperatures. A report by Wood et al. [46] showed that the fatty acid content of the algae UTEX 2341 heavily fluctuates depending on the carbon and nitrogen source. High C16:0 content was reported under basal ammonia and an increase of 30% for C16:1 was observed under glycerol and ammonia as medium. Another report only shows minor free fatty acid contents of C16:0 in the algae itself [47]. A detailed fatty acid methyl ester analysis of Chlorella species using a transesterification method and GC–MS revealed 160 different fatty acids from C7:0 to C30 chain length, with odd and even carbon numbers, different degrees of unsaturation, cyclopropane- and methyl-substitutions [48].
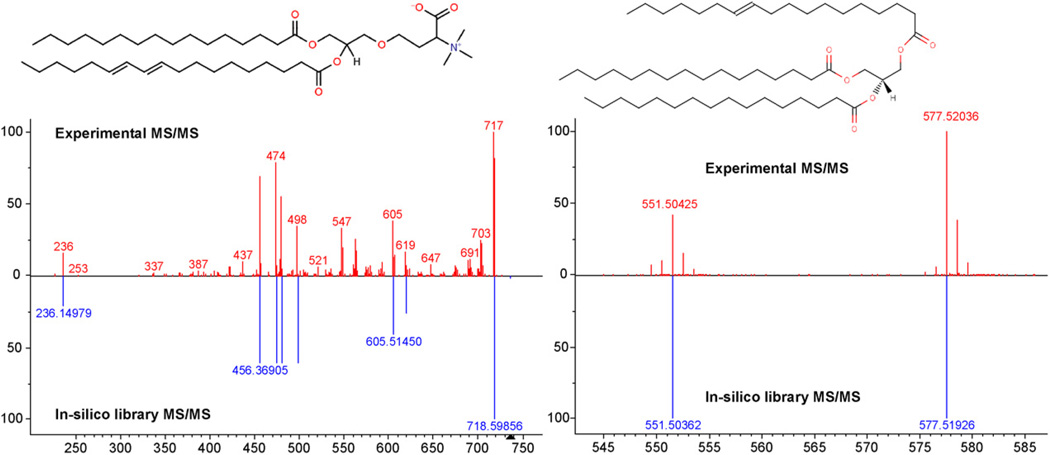
Fig. 6.
Left panel: low resolution iontrap MS/MS spectrum from precursor 736.61 Da was identified in a MS/MS library search as the betaine lipid DGTS 34:2; [M+H]+; DGTS(16:0/18:2). Right panel: high resolution QTOF-MS/MS, the substance with precursor 850.7866821 Da was identified as TG 50:1; [M+NH4]+; TG(16:0/16:0/18:1). Precursor mass accuracy was found as −0.00079 Da (0.93 ppm).
We observed no bacterial colonies on the plates using microscopic analysis. However it cannot be totally excluded that algae were contaminated and bacteria were feeding on exudate and henceforth that a bacterial strain or community was analyzed. Generally bacterial contamination can be analyzed by performing microscopic analysis [14,49], staining methods or better genomic sequencing of these secretions with subsequent matching against genomic databases or phylogenetic analysis. Nevertheless Gladu et al. observed the same mucoid secretions on the outer cell walls of UTEX 2341 [14]. Furthermore the exudate can consist of additional molecular classes (polysaccharides, glycans) [12] that are not covered with our platform. Such unidentified lipophilic compounds can be co-extracted with the Folch method.
4.3. Multiple analytical platforms are needed for analysis of complex matrices
For engineering purposes global sum parameters such as total lipid content [50] are usually sufficient. However, to understand biochemistry, detailed molecular fingerprints and compound identifications are needed. No single platform can cover the complete metabolic signature of a given species. The GC–MS approach only covers volatile compounds or compounds that can be made volatile, but not complex lipids. The LC–QTOF-MS/MS approach uses ultrahigh pressure to perform the separation of many compound classes, but with the current setup (20 min separation time), even UPLC is not able to separate all components within a single lipid compound class. Many components within a single compound class co-elute and overlap. That does not leave enough time for triggering an exhaustive number of MS/MS scans for subsequent compound identifications. Conversely, use of robotic nanoESI infusion allows 20–30 min infusion time with only 5–10 µl of material consumption. This approach allows the collection of hundreds of MS/MS scans. One severe drawback is that isobaric compounds may undergo fragmentation due to the selected 1–2 Da precursor windows. That can lead to mixed product ion spectra and false positive identifications, even on ultra-high resolution instruments.
Our current platform collection used here does not investigate higher molecular species such as carbohydrates, proteoglycans or species with a molecular weight higher than 2000 Da. Such species must be covered by carbohydrate sequencing methods or glycomics approaches. For simple fingerprinting of hydrocarbons and carbohydrates pyrolyzer–gas chromatography can be used [51]. Also compounds that do not ionize easily under electrospray conditions are not covered here. Other detection modes such as ultra violet detection (UV), matrix assisted laser desorption (MALDI), evaporative light scattering (ELSD) or atmospheric pressure ionization (APCI) detectors may be used for such purposes. The correct stereochemical assignments are missing from our compound annotations. A completed structural elucidation of thousands of compounds can be time-consuming and requires additional analytical approaches [52]. For better chromatographic resolution and a more detailed analysis a longer UPLC run time would be needed.
Even ultra-high resolution FT-ICR MS with up to one million resolving power would not be totally sufficient to resolve complex species from multiple adducts. The sodiated species PC 35:0 [M+Na]+ and PE 38:0 [M+Na]+ have both the same isotopic mass of 798.598900 Da. A direct infusion method would not distinguish both species and the product ion spectra would contain mixed spectra of both species, however these compounds could be easily separated by chromatography. Another issue for LC–MS and direct infusion methods is the selected precursor isolation width for the MS/MS scans which is usually between 1 and 3 Da. All ions in this mass isolation window are fragmented together, resulting in mixed spectra. Also triple-quadrupole mass spectrometry which is often applied for lipid profiling is prone to false-positive and false-negative identifications [53] unless an additional production ion scan is triggered for MS/MS confirmation (on hybrid or QTRAP instruments) [52]. For a detailed analysis of complex algae lipids the classical methods of thin-layer chromatography (TLC) with subsequent GC–MS FAME analysis [16,17,38] seems to be favorable for the discovery of new and existing compound classes. Once a separation into different compound classes is established, high-resolution mass spectrometry can be used to annotate compounds without the problem of isobaric overlap [19]. The use of accurate mass during the precursor filtering process in MS/MS search lowers the number of false-positive candidates and increases the confidence level in compound annotations.
4.4. Tandem mass spectral libraries (MSMS) are currently limited in size and molecular diversity
The identification of small molecules with tandem mass spectral libraries is currently hindered by the small size of the MS/MS libraries. The catalog of commercially available substances (CHEMCATS) list around 16 million chemicals. PubChem currently covers 30 million small molecules. In comparison the largest MS/MS database NIST11 currently covers 5843 compounds and their representative 95,000 MS/MS spectra with different adduct types obtained under different fragmentation voltages. The public MassBank Service (www.massbank.jp) covers around 15,000 MS/MS spectra from 4000 compounds [54]. The public RIKEN MSn Spectral Database for Phytochemicals (RESPECT DB, http://spectra.psc.riken.jp/) [55,56] includes around 9000 MS/MS spectra. The Agilent METLIN Personal Compound Database and Library contains Q-TOF MS/MS reference spectra for 2278 compounds and the online METLIN database (http://metlin.scripps.edu/) currently contains 28,329 MS/MS spectra from 5004 metabolites [57]. One possible approach to increase the coverage is the creation of in silico MS/MS libraries that contain computer generated fragments and abundances (see Fig. 6), similar to known approaches for peptide identification [52]. It is also useful to enrich such MS/MS databases with taxonomic information in order to allow a rediscovery of unidentified compound spectra.
4.5. Pathway databases and compound databases currently lack detailed molecular lipid species for most algal species
The metabolome coverage for less investigated species such as the strain UTEX 2341 in pathway databases is currently none existent. Even one of the best researched algae (C reinhardtii) lacks the coverage of molecular lipids species across all pathway or metabolite databases. That includes highly curated databases such as the ChlamyDB (http://www.chlamy.org), ChlamyCyc (http://chlamyto.mpimp-golm.mpg.de) and KEGG DB (http://www.genome.jp/kegg/). The KEGG database has incorporated the LipidMaps categories, but lacks the majority of molecular lipids. For each compound class the combination of different fatty acids would expand to hundreds of compounds with varying fatty acyl chains (n = 4–50) and different double bond variations [58]. Many single molecular species such as the betaine lipid DGTS(16:0/18:3) or the sulfolipid SQDG(16:0/16:0) are not covered, even they were reported in the model algae Chlamydomonas already more than 20 years ago [39].
The reason of such low coverage is not the quality of the databases itself, but the intrinsic way such molecules are reported. The most common way was to report molecules in publications; however, the loss of data is tremendous because it requires optical chemical structure recognition. We discussed in a recent report that molecules, taxonomy and meta-data must be directly submitted to electronic databases to allow collection and later queries [29]. However no such large public repositories currently exist. The National Science Foundation (US) and the Japan Science and Technology Agency (JST) recently acknowledged the lack of metabolome coverage and announced a joint program in 2011 called “Metabolomics for a Low Carbon Society (METABOLOMICS)” to (1) capture all major metabolites, (2) develop of standards and annotations of unknown metabolites, (3) identify specialized metabolites of potential value from plants, algae and bacteria.
5. Conclusions
We identified polar lipid compounds from the outer cell wall secretions from the algal strain UTEX 2341 by employing chromatographic separation techniques coupled to high-resolution and low-resolution tandem mass spectrometry (MS/MS). We annotated around 50 compounds using accurate mass precursor search and subsequent MS/MS mass spectral database search. Some of the annotated compounds were triacylglycerols, phosphatidylcholines, betaine lipids and phosphatidylinositols as well as sulfoquinovosyldiacylglycerols. Many of the estimated 600–1000 compounds remain unknown and require more detailed pre-fractionation steps. The discovery of these substances is challenging due to missing coverage in compound repositories and their absence in mass spectral MS/MS databases.
Supplementary Material
Acknowledgments
The authors thank Hongyun Gou at the Department of Biological and Agricultural Engineering at UC Davis for preparation of the algae cultures. Funding was provided by NSF MCB 1139644, NIH R01 DK078328, NIH RC2 GM092729, NIH ES013932 and by an Agilent Technologies Foundation grant.
Footnotes
Author contributions
Y.C., J.S.V. and O.F. designed the experiments of algal culture and extracellular matter harvest. Y.C. grew the algae and extracted the extracellular material. J.K.M, D.Y., F.N., A.V. and T.K. performed mass spectrometric experiments. T.K.J.K.M and D.Y. analyzed the results. T.K. performed the mass spectral interpretation and compound annotations. T.K. and O.F. wrote the manuscript in interaction with all contributing authors.
Competing interests
The authors declare no competing financial interests.
Appendix A. Supplement
All LC–MS/MS (MZXML) and infusion-MS/MS files (MGF) as well as GC–MS files (netCDF format) and DART-MS and detailed result files (XLS) are publicly available. See http://fiehnlab.ucdavis.edu/projects/UTEX-2341/.
References
- 1.Hellebust JA. Limnol. Oceanogr. 1965;10:192. [Google Scholar]
- 2.Pratt R. Am. J. Bot. 1942;29:142. [Google Scholar]
- 3.DellaGreca M, Zarrelli A, Fergola P, Cerasuolo M, Pollio A, Pinto G. J. Chem. Ecol. 2010;36:339. doi: 10.1007/s10886-010-9753-y. [DOI] [PubMed] [Google Scholar]
- 4.Chang WH, Tolbert NE. Plant Physiol. 1970;46:377. doi: 10.1104/pp.46.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tolbert NE, Zill LP. J. Biol. Chem. 1956;222:895. [PubMed] [Google Scholar]
- 6.Bell W, Mitchell R. Biol. Bull. 1972;143:265. [Google Scholar]
- 7.Bauld J, Brock TD. J. Phycol. 1974;10:101. [Google Scholar]
- 8.Amsler CD, Jormalainen V, Honkanen T. Algal Chemical Ecology, Springer, Berlin, Heidelberg. 2008:57. [Google Scholar]
- 9.Coleman R, Ramchunder S, Davies K, Moody A, Foggo A. Oecologia. 2007;151:454. doi: 10.1007/s00442-006-0601-2. [DOI] [PubMed] [Google Scholar]
- 10.Hallegraeff GM. Phycologia. 1993;32:79. [Google Scholar]
- 11.Amsler CD, Karsten U. Algal Chemical Ecology, Springer, Berlin, Heidelberg. 2008:273. [Google Scholar]
- 12.McConville MJ, Wetherbee R, Bacic A. Protoplasma. 1999;206:188. [Google Scholar]
- 13.Kessler E, Huss VAR. J. Phycol. 1992;28:550. [Google Scholar]
- 14.Gladu PK, Patterson GW, Wikfors GH, Smith BC. J. Phycol. 1995;31:774. [Google Scholar]
- 15.Cody RB, Laramée JA, Durst HD. Anal. Chem. 2005;77:2297. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- 16.Dembitsky VM, Rozentsvet OA, Pechenkina EE. Phytochemistry. 1990;29:3417. [Google Scholar]
- 17.Fuchs B, Suss R, Teuber K, Eibisch M, Schiller J. J. Chromatogr. A. 2010;1218:2754. doi: 10.1016/j.chroma.2010.11.066. [DOI] [PubMed] [Google Scholar]
- 18.Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He H, Rodgers RP, Marshall AG, Hsu CS. Energy Fuels. 2011;25:4770. [Google Scholar]
- 20.MacDougall KM, McNichol J, McGinn PJ, Leary SJBO, Melanson JE. Anal. Bioanal. Chem. 2011:1. doi: 10.1007/s00216-011-5376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li HY, Yan XJ, Xu JL, Zhou CX. Sci. China Ser. C: Life Sci. 2008;51:1101. doi: 10.1007/s11427-008-0138-1. [DOI] [PubMed] [Google Scholar]
- 22.Seiwert B, Giavalisco P, Willmitzer L, Wada H, Murata N. Lipids in Photosynthesis; Advances in Photosynthesis and Respiration. Vol. 30. Netherlands: Springer; 2010. pp. 445–461. http://dx.doi.org/10.1007/978-90-481-2863-120. [Google Scholar]
- 23.Folch J, Lees M, Stanley GH. J. Biol. Chem. 1957;226:497. [PubMed] [Google Scholar]
- 24.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. Bioinformatics. 2008;24:1021. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, Pevzner PA. J. Proteome Res. 2007;7:113. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein SE. 2011 http://chemdata.nist.gov/
- 27.Tchekhovskoi DV, Mirokhin Y, Blonder N, Rudnick PA, Stein SE. 2011 http://peptide.nist.gov/
- 28.Christie WW. 2011 http://lipidlibrary.aocs.org/ms/masspec.html.
- 29.Kind T, Scholz M, Fiehn O. PLoS One. 2009;4:e5440. doi: 10.1371/journal.pone.0005440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodner RB, Summons RE, Knoll AH. Org. Geochem. 2009;40:854. [Google Scholar]
- 31.GenBank. 2010 http://www.ncbi.nlm.nih.gov/nuccore/HI993752.1.
- 32.Solazyme. 2009 http://www.wipo.int/patentscope/search/en/detail.jsf?docId =WO2009126843.
- 33.Guschina IA, Harwood JL, Kainz M, Brett MT. Lipids in Aquatic Ecosystems. New York: M.T. Arts, Springer; 2009. pp. 1–24. http://dx.doi.org/10.1007/978-0-387-89366-2.1. [Google Scholar]
- 34.Van Mooy BA, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblízek M, Lomas MW, Mincer TJ, Moore LR, Moutin T. Nature. 2009;458:69. doi: 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- 35.Sato N. J. Plant Res. 1992;105:185. [Google Scholar]
- 36.Sato N, Furuya M. Plant Sci. 1985;38:81. [Google Scholar]
- 37.Roche SA, Leblond JD. Phycol. Res. 2010;58:298. [Google Scholar]
- 38.Kato M, Hajiro-Nakanishi K, Sano H, Miyachi S. Plant Cell Physiol. 1995;36:1607. [Google Scholar]
- 39.Giroud C, Gerber A, Eichenberger W. Plant Cell Physiol. 1988;29:587. [Google Scholar]
- 40.Haigh WG, Yoder TF, Ericson L, Pratum T, Winget RR. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metabol. 1996;1299:183. doi: 10.1016/0005-2760(95)00205-7. [DOI] [PubMed] [Google Scholar]
- 41.Benning C, Huang ZH, Gage DA. Arch. Biochem. Biophys. 1995;317:103. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Lara IM, Gao J-L, Soto MJ, Solares-Perez A, Weissenmayer B, Sohlenkamp C, Verroios GP, Thomas-Oates J, Geiger O. Mol. Plant Microbe Interact. 2005;18:973. doi: 10.1094/MPMI-18-0973. [DOI] [PubMed] [Google Scholar]
- 43.Kato M, Sakai M, Adachi K, Ikemoto H, Sano H. Phytochemistry. 1996;42:1341. [Google Scholar]
- 44.Benson AA, Daniel H, Wiser R. Proc. Natl. Acad. Sci. U. S. A. 1959;45:1582. doi: 10.1073/pnas.45.11.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinensky M. J. Bacteriol. 1977;129:516. doi: 10.1128/jb.129.1.516-524.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood BJB, Grimson PHK, German JB, Turner M. J. Biotechnol. 1999;70:175. [Google Scholar]
- 47.Allard B, Templier J. Phytochemistry. 2000;54:369. doi: 10.1016/s0031-9422(00)00135-7. [DOI] [PubMed] [Google Scholar]
- 48.Rezanka T, Vokoun J, Slavícek J. M. Podojil,J. Chromatogr. 1983;268:71. [Google Scholar]
- 49.McGrath Grossi S, Kottmeier ST, Sullivan CW. Microb. Ecol. 1984;10:231. doi: 10.1007/BF02010937. [DOI] [PubMed] [Google Scholar]
- 50.Cheng YS, Zheng Y, VanderGheynst JS. Lipids. 2010:1. doi: 10.1007/s11745-010-3494-0. [DOI] [PubMed] [Google Scholar]
- 51.Barupal D, Kind T, Kothari S, Lee D, Fiehn O. BMC Biotechnol. 2010;10:40. doi: 10.1186/1472-6750-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kind T, Fiehn O. Bioanal. Rev. 2010;2:23. doi: 10.1007/s12566-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch KL, Breaud AR, Vandenberghe H, Wu AHB, Clarke W. Clin. Chim. Acta. 2010;411:1474. doi: 10.1016/j.cca.2010.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K. J. Mass Spectrom. 2010;45:703. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 55.Matsuda F, Hirai MY, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart NJ, Sakurai T, Shimada Y, Saito K. Plant Physiol. 2010;152:566. doi: 10.1104/pp.109.148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, Hirai MY, Sakurai T, Kikuchi J, Saito K. In Silico Biol. 2008;8:339. [PubMed] [Google Scholar]
- 57.Smith CA, Maille GO, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. Ther. Drug Monit. 2005;27:747. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 58.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Murphy RC, Raetz CRH, Russell DW. Nucleic Acids Res. 2006;35:D527. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.