Abstract
One of the most powerful drivers of speciation in plants is pollinator-mediated disruptive selection, which leads to the divergence of floral traits adapted to the morphology and behavior of different pollinators. Despite the widespread importance of this speciation mechanism, its genetic basis has been explored in only a few groups. Here, we characterize the genetic basis of pollinator-mediated divergence of two species in genus Ipomopsis, I. guttata and I. tenuifolia, using quantitative trait locus (QTL) analyses of floral traits and other variable phenotypes. We detected one to six QTLs per trait, with each QTL generally explaining small to modest amounts of the phenotypic variance of a backcross hybrid population. In contrast, flowering time and anthocyanin abundance (a metric of color variation) were controlled by a few QTLs of relatively large effect. QTLs were strongly clustered within linkage groups, with 26 of 37 QTLs localized to six marker-interval ‘hotspots,' all of which harbored pleiotropic QTLs. In contrast to other studies that have examined the genetic basis of pollinator shifts, our results indicate that, in general, mutations of small to modest effect on phenotype were involved. Thus, the evolutionary transition between the distinct pollination modes of I. guttata and I. tenuifolia likely proceeded incrementally, rather than saltationally.
Keywords: pollinator-mediated speciation, directional selection, floral characters, reproductive isolation, QTL, transmission ratio distortion
Introduction
Lineages named by taxonomists at the species level typically represent discrete entities in multivariate phenotypic space (Rieseberg et al., 2006), and some studies suggest that these entities are stable over time (Jackson and Cheetham, 1999), even in the face of interspecific hybridization (Condit, 1944). Evolutionary biologists strive to characterize the genetic changes that accompany transitions between these stable forms and the underlying processes that drive them––what kinds of mutations are involved in speciation, and which process is more important in their fixation, drift or selection (Charlesworth et al., 1982; Lynch, 2010)?
Although random genetic drift has long been proposed as a key mechanism in shaping the phenotypic differences that evolve during speciation (Wallace, 1889), a growing body of direct empirical evidence suggests that natural selection is the primary cause of the phenotypic differences between species (Rieseberg et al., 2002; Louthan and Kay, 2011). Such selection can be caused by various biotic and abiotic factors (Rieseberg and Willis, 2007). One potentially powerful biotic agent in plants is pollinator-mediated disruptive selection, which can lead to divergence of floral traits adapted to the morphology and behavior of different pollinating animals or suites of pollinating animals that are locally most abundant and/or efficient (Grant, 1949). When such selection leads to assortative mating within diverging floral morphs, the process is referred to as floral isolation (Grant, 1949).
Because of the direct tie to reproduction, many expect pollinator-mediated selection to be strong, particularly for self-incompatible plant species. Furthermore, pollinators are often highly visually directed organisms, and thus select for traits that draw the attention of taxonomists. However, some have argued that pollinators typically visit a wide variety of plant species, and hence, should not be a strong force driving floral trait divergence and speciation (Waser, 2001). Nonetheless, classic (Fulton and Hodges, 1999) and recent (Hopkins and Rausher, 2012) reports have demonstrated that the interplay between pollinators and floral traits can lead to, or at least facilitate, reproductive isolation and morphological divergence between closely related plant forms. Although we lack information on the frequency with which pollinators split angiosperm lineages, multiple lines of evidence suggest that pollinator-mediated speciation has been important in particular groups (Grant, 1993) and regions (Van der Niet et al., 2006), and regularly drives floral trait specialization (Fenster et al., 2004).
For this report, we investigated the genetic basis of floral trait differences that separate a set of sister species in the Giliopsis group of the plant genus Ipomopsis, I. guttata and I. tenuifolia (Wood and Nakazato, 2009). Ipomopsis (Polemoniaceae) displays remarkable interspecific variation in floral form that is correlated with pollination by different groups of animals; thus, it has long been a model for understanding floral trait evolution vis-a-vis pollination mode (Grant and Grant, 1965). I. guttata and I. tenuifolia have strikingly different flowers (Figure 1), yet can be readily intercrossed to produce fully fertile hybrids. Field observations indicated that each is predominantly serviced by distinct pollinators: I. guttata by bombyliid flies and lepidopterans and I. tenuifolia by hummingbirds (Wood, 2009). Although pollination mode is difficult to polarize in Ipomopsis because of its lability, the relative rarity of hummingbird pollination in the group and the fact that I. tenuifolia's two closest allies, I. guttata and I. effusa, are insect pollinated suggest that bird pollination is derived from insect pollination. The two species are distributed across mountainous regions of the northern half of Baja California and into Alta California. I. guttata is restricted to two small, disjunct regions (elevation ca 800–1600 m a.s.l.) separated by approximately 150 km and appears to be specialized to chaparral habitat (Moran, 1977). I. tenuifolia has a broader geographic range that encompasses substantial variation in elevation (100–2400 m a.s.l.) and diverse species assemblages (Moran, 1977). The two species are in direct sympatry in at least one location in the southern part of I. guttata's range. Population genetic analysis of these species indicated that the I. guttata population sympatric with I. tenuifolia is genetically more similar to I. tenuifolia than it is to conspecific populations. Yet members of this population maintain typic I. guttata floral morphology (Wood and Nakazato, 2009). The most parsimonious explanation of this result is that phenotypic differentiation at the sympatric locality is maintained by divergent pollinator-mediated selection countering ongoing gene flow between the species. Although floral divergence of I. guttata and I. tenuifolia has been evaluated morphologically and ecologically, the genetic basis of this divergence has not been studied.
Figure 1.
Representative flowers of I. tenuifolia (top) and I. guttata.
For the current study, we genotyped a backcross population toward I. guttata, constructed a genetic linkage map of the I. tenuifolia genome, and conducted quantitative trait locus (QTL) analyses of floral and developmental rate traits. These efforts were directed at answering the following questions: (1) Are the trait differences separating the species controlled by a large number of loci with small phenotypic effects or a small number of loci, each with large effects, and how are these loci dispersed across the genome? (2) How important are pleiotropy and epistasis in determining species differences? (3) Do identified QTLs always move the phenotype toward I. tenuifolia, a result that would suggest that the species diverged under continuous directional selection?
Materials and methods
Crossing and cultivation
Individuals of I. tenuifolia were grown from seed collected in the Sierra San Pedro Martir, Baja California, Mexico, whereas individuals of I. guttata were grown from seed collected in the Sierra Juarez, Baja California. One individual of each species, each drawn from the glasshouse-founded populations and with flower abundance sufficient to yield the quantity of seed needed to establish the hybrid populations, were selected as parents. A single F1 plant (with I. guttata cytotype) derived from a cross of these two parents was backcrossed to its I. guttata maternal parent to generate the BC1 seeds used in this study. Although both species are self-incompatible (SI), all maternal flowers pollinated to generate the BC1 population were emasculated by removing the entire androecium before stigma maturation to avoid self-pollination, which can result from breakdown of the SI system in hybrid crosses (Desrochers and Rieseberg, 1998). The BC1 plants, populations of parents and an F1 population from multiple, interspecific pairings were grown together from seed in the Indiana University greenhouse in 3′′ clay pots containing equal parts of sand, soil and gravel under sodium halide lamps with a 16-h per day cycle. When temperatures mandated, the lamps were switched off between 0900 and 1500 hours. Plants were watered as needed and fertilized every 2 weeks. The pots were placed in large trays, which were regularly rotated within the greenhouse.
Phenotypic analyses
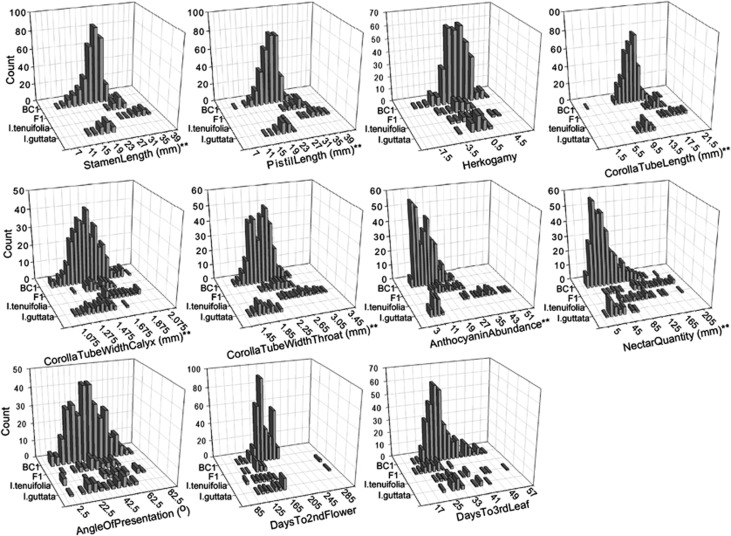
All floral measurements were taken on the first two flowers of each plant, and the data used in all analyses are a mean of these two flowers; the two developmental traits were scored from daily observations of the mapping population (see Table 1 and Figure 2 for the traits examined). In total, 326 BC1 individuals were phenotyped for the QTL analyses. Stamen_Length and Pistil_Length were measured as the distance between the base of the calyx and the distal most anther sac and the stigma, respectively. Herkogamy was recorded as the difference between Stamen_Length and Pistil_Length. Corolla_Tube_Length was measured as the distance between the base of the calyx and the proximal opening of the throat. Corolla tube width was measured just above the calyx (Corolla_Tube_Width_Calyx) and at the throat (Corolla_Tube_Width_Throat). Tube width was measured at two locations (atomized into two traits), because, in I. tenuifolia, the tube has a distinct taper above the calyx (Figure 1), suggesting that the two traits may be under separate genetic control. Petal lobe tissue was clipped on the second day after anthesis and weighed immediately, and anthocyanins extracted in a methanol+HCl mixture (Wilken, 1982). Absorbance of the extract was measured at 515 nm with a spectrophotometer and divided by sample mass. This quantity was used as a relative anthocyanin concentration metric (Anthocyanin_Abundance). To quantify nectar volume (Nectar_Quantity), flowers were cut just above the calyx soon after the anthocyanin measurements were made. Nectar was spun down from inverted flower bases in a centrifuge in nested Eppendorf tubes where the tip of the smaller, interior tube containing the flower base was cut away to allow nectar to flow through to the larger tube. Spun nectar was drawn into a 5-μl disposable glass pipettes, and the length (in millimeters (18.3 mm=1 μl)) of the nectar in the pipette was measured and recorded. In the field, I. guttata plants tend to present flowers horizontally at stigma maturation (the long axis of the pistil parallel to the surface of the ground), a trait that may be important to pollination by insects, particularly bombyliids. Therefore, the angle of presentation of flowers (Angle_Of_Presentation) was measured for each plant by casting a shadow of a flower onto a sheet of paper and defining a line segment by marking the base of the calyx (along a horizontal line) and the center of the stigma. The deviation from horizontal was then measured with a protractor. The light source was kept at the same height as the flower. To quantify development rate, the number of days from sowing until the appearance of the third true leaf (Days_To_3rd_True_Leaf) and until anthesis of the second flower (Days_To_2nd_Flower) were recorded for each plant.
Table 1. Floral and developmental trait values of I. guttata and I. tenuifolia. See Figure 2 for parental and hybrid distributions.
| Trait | I. guttata |
I. tenuifolia |
t-test |
Shapiro–Wilk
W-test of normality |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean | s.d. | N | Mean | s.d. | t | P | Statistic | P | |
| Stamen_Length | 30 | 17.24 | 2.03 | 21 | 32.30 | 2.33 | −23.92 | <0.001 | 0.947 | <0.001 |
| Pistil_Length | 30 | 18.11 | 2.11 | 21 | 32.87 | 3.63 | −16.76 | <0.001 | 0.988 | 0.007 |
| Herkogamy | 30 | −0.88 | 1.08 | 21 | −0.56 | 2.30 | −0.58 | 0.570 | 0.992 | 0.061 |
| Corolla_Tube_Length | 30 | 9.17 | 0.90 | 21 | 18.59 | 1.97 | −20.45 | <0.001 | 0.966 | <0.001 |
| Corolla_Tube_Width_Calyx | 30 | 1.37 | 0.13 | 21 | 1.71 | 0.15 | −8.25 | <0.001 | 0.987 | 0.006 |
| Corolla_Tube_Width_Throat | 30 | 1.75 | 0.18 | 21 | 2.88 | 0.30 | −15.48 | <0.001 | 0.991 | 0.052 |
| Anthocyanin_Abundance | 30 | 4.74 | 1.49 | 21 | 36.71 | 6.82 | −21.12 | <0.001 | 0.955 | <0.001 |
| Nectar_Quantity | 30 | 25.92 | 17.48 | 21 | 97.56 | 40.41 | −7.64 | <0.001 | 0.918 | <0.001 |
| Angle_Of_Presentation | 30 | 31.62 | 14.62 | 21 | 39.52 | 23.17 | −1.38 | 0.182 | 0.989 | 0.019 |
| Days_To_2nd_Flower | 30 | 143.4 | 18.62 | 21 | 139.10 | 53.47 | 0.36 | 0.724 | 0.969 | <0.001 |
| Days_To_3rd_True_Leaf | 27 | 30.11 | 5.06 | 17 | 33.24 | 8.60 | −1.36 | 0.194 | 0.913 | <0.001 |
Figure 2.
Phenotypic distributions of the studied traits in the parental lines (I. tenuifolia (N=21) and I. guttata (N=30)), F1s (N=30) and BC1s (N=326). Traits followed by ** are significantly different between the parents.
Normality of BC1 trait distributions was tested with the Shapiro–Wilk W-test as implemented in SPSS (IBM Corporation, New York, NY, USA). The tests were significant (P<0.05) for all traits except for Herkogamy, Corolla_Tube_Width_Throat and Angle_Of_Presentation (Table 1). Anthocyanin_Abundance, Nectar_Quantity, Days_To_3rd_True_Leaf were log10 transformed before QTL analyses. The remaining traits with non-normal distributions were not transformed because standard transformation methods did not markedly improve normality. Principal component analyses of the phenotypic data were conducted using SPSS based on the trait correlation matrix without rotation. Statistical analyses, including the calculation of descriptive statistics, analysis of variance, Student's t-test, principal component analyses, χ2 test and evaluation of correlations, were conducted with SPSS.
Amplified fragment length polymorphism (AFLP) genotyping of the BC1 mapping population
DNA from each genotyped plant (N=337 BC1 plants used for linkage mapping) was extracted from young leaves using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Approximately 300 ng of genomic DNA was digested with 3 U each of EcoRI and MspI in 1 × NEB #2 buffer in 40 μl reactions. After 1 h of incubation at 37 °C, the following ligation reagents were added in a 10-μl mixture to the digestion products and incubated for 3 h at 16 °C: 1.5 μℳ of double-stranded EcoRI adapter (combination of oligos 5′-CTCGTAGACTGCGTACC-3′and 5′-AATTGGTACGCAGTCTAC-3′), 15 μℳ of double-stranded MspI adapter (combination of oligos 5′-GACGATGAGTCTAGAA-3′ and 5′-CGTTCTAGACTCATC-3′), 1 × ligase buffer and 0.1 μl of T4 ligase (New England Biolabs, Ipswich, MA, USA). The pre-selective AFLP amplification was carried out in a 20-μl reaction mixture containing 30 mℳ Tricine, 50 mℳ KCl, 2 mℳ MgCl2, 5% acetamide, 10 mℳ of each dNTP, 0.2 mℳ of both the EcoRI primer (5′-GACTGCGTACCAATTCA-3′) and the MspI primer (5′-GATGAGTCTAGAACGGA-3′), 0.6 μl of Taq polymerase and 2 μl of the ligation product. The pre-selective reactions were conducted using the following PCR program: 30 s at 94 °C, 30 s at 55 °C and 1 min at 72 °C, cycled 35 times. The selective AFLP amplification was carried out in the same manner as the pre-selective AFLP amplification, except that 2 μl of a 1:20 dilution of the pre-selective amplification product was used as a template for 16 EcoRI-MspI primer combinations with the following selective extensions: ACG (NED dye)-ATGA, ACG (NED)-AAT, ATG (NED)-ATGA, ATG (NED)-AAT, ACT (FAM)-AGAT, ACT (FAM)-ATC, ATT (FAM)-AGAT, ATT (FAM)-ATC, ACC (VIC)-AAAC, ACC (VIC)-ATGA, ATC (VIC)-AAAC, ATC (VIC)-ATGA, ACA (PET)-ACC, ACA (PET)-AGAT, ATA (PET)-ACC and ATA (PET)-AGAT. PCR products were separated on a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) at the Indiana University Molecular Biology Institute. Unambiguous fragments between 50 and 600 bp were scored manually as present (1) or absent (0) using GeneMapper (Applied Biosystems).
Linkage map construction and QTL analysis
The linkage map was constructed using AntMap, which employs the Ant Colony Optimization algorithm (Iwata and Ninomiya, 2006). AFLP markers were assembled into linkage groups (LGs) using the ‘all combinations' option with a maximum Kosambi centiMorgan (cM) distance between adjacent markers of 30.0 and a minimum number of markers per LG of three. Seventy-two out of the eighty-four scored markers were successfully linked to form eight LGs. Markers were then ordered within each LG using the default setting of AntMap, except that the ‘Sum of Adjacent Recombination Fractions' option was used (Liu, 1998). The reliability of marker orders was evaluated with 1000 bootstrap replicates, where the fraction of replicates (random resamples, with replacement, of the marker data) containing the reported marker orders is used to estimate accuracy. The total map length was estimated by three methods. The first estimate is simply a sum of distances between adjacent markers on the map. The second estimate is based on method 4 of Chakravarti et al. (1991), which multiplies the sum of marker distances on each LG by the factor (m+1)/(m−1), where m is the number of markers on the LG. The third simply adds the average intermarker distance to both ends of each LG. Map coverage, c, was estimated by the formula c=1−e2dn/L (Bishop et al., 1983), where n is the total number of markers and L is the total map length, assuming a random marker distribution. Map coverage indicates the probability that a new marker added to the map would fall within a given distance d from the nearest marker.
QTLs for each trait were mapped using Composite Interval Mapping (CIM) as implemented in QTL Cartographer Windows v.2.5 (Wang et al., 2011; http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). CIM uses multiple regression to test the hypothesis that a specific map position in an interval between adjacent markers controls variation in a focal quantitative trait, where background genetic markers are included as cofactors. CIM tests were performed at a walk speed of 1 cM and a flanking window size of 10 cM. Experiment-wise significance threshold levels (α=0.05) were determined with 1000 permutations using the method described by Churchill and Doerge (1994). Because flower length-related traits (Stamen_Length, Pistil_Length, and Corolla_Tube_Length) and flower width-related traits (Corolla_Tube_Width_Calyx and Corolla_Tube_Width_Throat) were highly correlated (see Results), QTLs were jointly mapped using multi-trait CIM (MCIM) as Flower_Length and Flower_Width, respectively, using the same parameter settings as with the standard CIM analyses. MCIM takes into account the correlation structure among included traits and calculates the likelihood of a given interval containing a QTL for any of these traits, which can increase the power and accuracy of QTL identification (Jiang and Zeng, 1995). In addition, the outputs from the MCIM analyses were used to evaluate the occurrence of pleiotropy, where the composite QTL controls variation in two or more of the traits included in the MCIM model (Jiang and Zeng, 1995). A critical value of the Likelihood Ratio statistic of 5.99 (χ20.05,2) or greater at the exact peak position of the composite QTL for two or more individual traits comprising the composite trait was interpreted as statistical support for pleiotropy (Jiang and Zeng, 1995). Finally, epistasis between all pair-wise combinations of markers was investigated for each trait using QTLNetwork 2.0 (http://ibi.zju.edu.cn/software/qtlnetwork/) using the default settings (Yang et al., 2008). This analytic method controls for inflated Type I error rates resulting from multiple tests, and marker interactions with α<0.05 were considered significant.
Transmission ratio distortion (TRD) analysis
The degree of TRD for each marker was expressed as the percentage of I. guttata genotypes among scored BC1s. Its statistical significance was tested against the expected neutral frequency of 50% using Pearson's χ2 test. Markers with P-values less than 0.01 were considered to show significant TRD. We did not use Bonferroni correction to set a P-value because genotypes among loci are not independent (Fishman et al., 2001).
Results
Phenotypic variance and covariance in parents and hybrids
The two parental lines were distinct for all measured traits (t-test, P<0.01, Table 1) except Herkogamy, Angle_Of_Presentation, Days_To_2nd_Flower and Days_To_3rd_True_Leaf. In the field, the two species are clearly distinct for Angle_Of_Presentation, but this trait is difficult to measure under cultivation, particularly on small plants. For the distinct traits, all of which were floral, I. tenuifolia always had larger values. The difference was particularly pronounced for Anthocyanin_Abundance and Nectar_Quantity (Table 1, Figure 2). Flower length-related traits (Stamen_Length, Pistil_Length, and Corolla_Tube_Length) were more strongly differentiated than flower width-related traits (Corolla_Tube_Width_Calyx and Corolla_Tube_Width_Throat). Herkogamy was substantially different between the parents, but also variable within the parental lines. Most traits were distributed continuously and unimodally, suggesting that they are under polygenic control (Figure 2). Two possible exceptions are Anthocyanin_Abundance and Corolla_Tube_Width_Throat, which show some evidence of bimodality. F1s showed a dominance deviation toward I. tenuifolia for all traits except Anthocyanin_Abundance, which was inherited additively. Transgressive segregation was observed in the BC1 population for Herkogamy, Pistil_Length and Corolla_Tube_Length, although for the latter two, this transgression is based on a single, small-flowered individual (Figure 2). Based on Days_To_2nd_Flower, BC1 plants developed at a significantly slower rate than both parents and the F1 (Tukey's Honestly Significant Difference test, P<0.05).
Many of the traits were significantly correlated (Supplementary Table 1), indicating that they have a shared regulatory control and/or that the underlying genetic factors are physically linked. In particular, flower length- and width-related traits co-varied tightly; a pattern that is likely due to genetic changes that affect flower size dimensions in general (for example, Juenger et al., 2000, 2005). Days_To_2nd_Flower was significantly negatively correlated with flower length and width traits, indicating that I. tenuifolia-like segregants develop more rapidly. However, this trait was not significantly correlated with Days_To_3rd_True_Leaf, suggesting that early developmental rate is not determined by the same genetic factors as Days_To_2nd_Flower. Instead, variation in early developmental rate may reflect differences in seed biology (that is, germination strategies). Days_To_3rd_True_Leaf and Anthocyanin_Abundance were not strongly correlated with other traits, and thus appear to be more or less independently regulated. Correlational relationships among traits were summarized using principal component analyses. We extracted four principal components (PCs) above one eigenvalue (Supplementary Figure 1, Supplementary Table 2). The first two PCs explained 48.03% of the variation. PC1 generally explained flower length traits (that is, Stamen_Length, Pistil_Length, Corolla_Tube_Length) and Nectar_Quantity, whereas PC2 explained flower width traits (that is, Corolla_Tube_Width_Calyx, Corolla_Tube_Width_Throat) as well as Angle_Of_Presentation. Days_To_2nd_Flower and Herkogamy were heavily loaded to PC3, whereas Days_To_3rd_True_Leaf and Anthocyanin_Abundance were loaded to PC4.
Linkage map construction and analysis of map size and marker order support
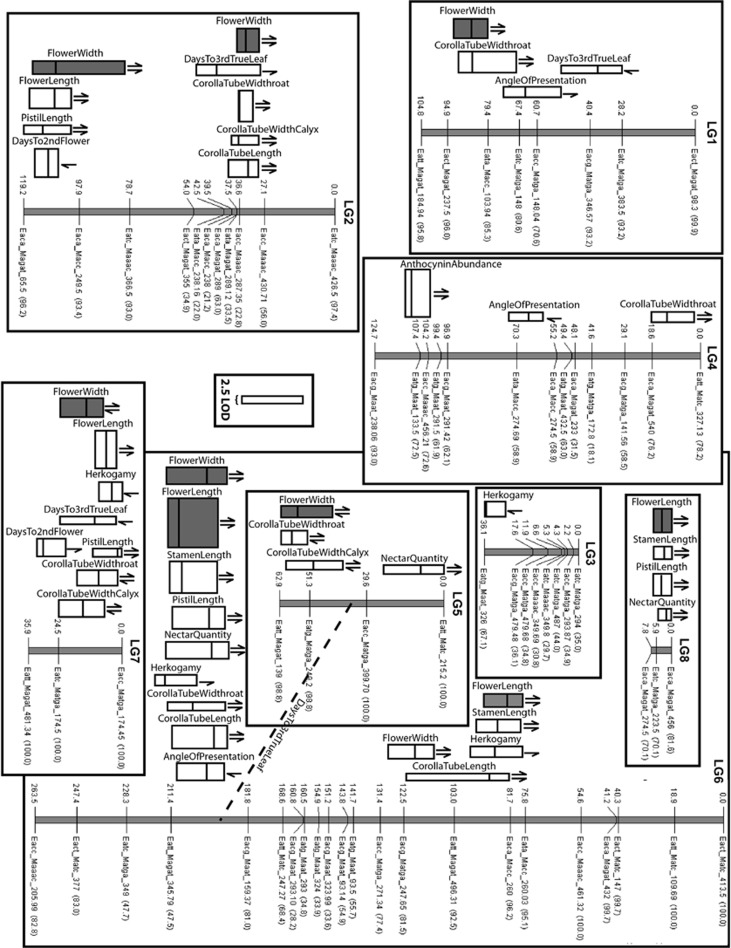
A total of 84 AFLP markers were scored from 11 primer pairs with marker number per primer pair ranging from two to eight. Twelve markers could not be mapped, possibly because of excessive (>30 cM) map distance from the nearest marker and/or genotyping errors. With the remaining 72 markers, we recovered eight structurally robust LGs (Figure 3). Chromosome counts of I. tenuifolia indicate that this species is diploid with n=7 (Porter et al., 2010). The ease with which fertile hybrids of I. tenuifolia and I. guttata can be generated (Wood and Nakazato, 2009) suggest that the latter species is also a diploid with n=7, a number that is nearly invariant within Ipomopsis. The discrepancy between the number of chromosomes and number of LGs is most likely a function of low marker density, with one large chromosome possibly represented by two LGs. The number of markers per LG ranged from 3 to 22 (median of 8) with a cumulative map length of 754.89 cM, adjusted map length of 950.54 cM (Chakravarti et al., 1991), average marker interval of 11.80 cM and a map coverage of 53.11% at a 5-cM distance and 78.02% at a10-cM distance. The adjusted map length was very similar to a total map length derived from adding two average marker intervals to each LG––943.69 cM. The map lengths of the LGs––134.70, 143.05, 46.40, 145.47, 104.88, 288.56, 71.86, 15.60––fell within the typical range reported for plant genomes. Linkage groups (LG) 2, 3 and 6 exhibited marker clustering; these clusters may be in centromeric regions, where recombination is typically suppressed. Marker order was generally robust, as indicated by high bootstrap values (Figure 3). Some regions showed low-to-medium bootstrap values for inferred marker order, generally in regions of high marker density, where order inference is strongly limited by recombination frequency. However, these weakly supported marker clusters did not affect the overall structure of LGs and likely did not strongly influence the identification of QTLs.
Figure 3.
Linkage map of I. tenuifolia and I. guttata and positions of identified QTLs. The linkage map is based on genotypes from 337 BC1s and the QTL analyses on 326 BC1s. Each LG includes cumulative Kosambi genetic distances (in centimorgans) from the top-most marker (left) and marker names (right). Marker names are based on AFLP primer pair and approximate fragment size in base pairs and are followed by bootstrap support values for marker order. For each QTL, the position of the middle bar, the width and the length of the box indicate the peak QTL position, logarithm (base 10) of odds (LOD) score at the peak position and 1 LOD interval (corresponding to a 95% confidence interval), respectively. We include a 2.5-LOD scale bar for reference. Arrows on the left indicate the relative trait values of the parents; an upward (pointing to the top of the LG) arrow indicates a higher value in I. tenuifolia, no left arrow indicates the trait was not significantly different between the parents. Arrows on the right indicate the directions of the corresponding QTL; an upward arrow indicates a QTL effect toward I. tenuifolia. The dotted line links the two regions that interact to control Days_To_3rd_True_Leaf, and represents the only significant epistatic effect identified (P<0.0001). For the composite traits, FlowerLength and FlowerWidth, a shaded bar indicates that the QTL controls two or more of the traits comprising the composite phenotypes, that is, shaded QTLs are pleiotropic.
QTL analysis of genetic architecture of trait differences
We detected a total of 35 main-effect QTLs from single-trait analyses with 1–6 QTLs per trait (Table 2). Twelve QTLs were detected with MCIM analysis––five and seven QTLs for Flower_Length and Flower_Width, respectively, most of which corresponded in approximate position with QTLs identified with the single-trait analyses (Figure 3, Table 2). However, two additional QTLs for Flower_Width were detected with MCIM, demonstrating the increased detection power of combining correlated traits. In general, detected QTLs had small to modest magnitudes of effect (reported as percent variance explained (PVE)) among BC1s, with a range of 2.93–24.56 PVE and a median of 5.93 PVE. Only two QTLs, one for Days_To_2nd_Flower (PVE=20.2) and one for Anthocyanin_Abundance (PVE=24.56), approached a proposed criterion for a major effect locus of 25 PVE (Bradshaw et al., 1998). For traits with significant phenotypic differences between the parental lines, most of the QTLs (19 of 23 or 82.61%) showed effects in the expected directions based on the parental phenotypes. Interestingly, all four of the QTLs with antagonistic effects, that is, QTLs that moved traits opposite to the direction expected based on parental means, controlled corolla tube width (Corolla_Tube_Width_Calyx and Corolla_Tube_Width_Throat).
Table 2. Summary of QTLs detected in this study.
| Trait | LG | Peak cM position | Effect size | % Variance explained | Expected direction? |
|---|---|---|---|---|---|
| Angle_Of_Presentation | |||||
| 1 | 65.76 | 7.40 | 4.98 | ||
| 4 | 67.44 | −6.44 | 3.93 | ||
| 6 | 197.85 | −10.19 | 9.68 | ||
| Anthocyanin_Abundance | |||||
| 4 | 110.11 | 0.24 | 24.56 | Yes | |
| Corolla_Tube_Length | |||||
| 2 | 33.12 | 1.04 | 7.90 | Yes | |
| 6 | 89.74 | 0.85 | 4.32 | Yes | |
| 6 | 194.85 | 1.55 | 14.89 | Yes | |
| Corolla_Tube_Width_Calyx | |||||
| 2 | 36.58 | 0.07 | 4.21 | Yes | |
| 5 | 49.60 | −0.07 | 4.02 | No | |
| 7 | 15.46 | −0.11 | 8.56 | No | |
| Corolla_Tube_Width_Throat | |||||
| 1 | 85.45 | 0.16 | 9.17 | Yes | |
| 2 | 36.12 | 0.18 | 11.84 | Yes | |
| 4 | 13.01 | 0.11 | 4.31 | Yes | |
| 5 | 58.27 | −0.13 | 6.02 | No | |
| 6 | 202.85 | 0.09 | 2.93 | Yes | |
| 7 | 9.01 | −0.14 | 6.04 | No | |
| Days_To_2nd_Flower | |||||
| 2 | 109.95 | −19.99 | 20.20 | ||
| 7 | 32.46 | 12.92 | 9.33 | ||
| Days_To_3rd_True_Leaf | |||||
| 1 | 37.16 | −0.04 | 3.90 | ||
| 2 | 45.54 | 0.04 | 5.33 | ||
| 7 | 11.46 | −0.03 | 3.51 | ||
| 5–6a | 35.60 × 192.82 | −0.06 | 3.89 | ||
| Herkogamy | |||||
| 3 | 35.59 | −1.07 | 5.93 | ||
| 6 | 87.74 | 1.00 | 5.24 | ||
| 6 | 212.96 | 1.01 | 5.31 | ||
| 7 | 4.01 | −1.30 | 8.79 | ||
| Nectar_Quantity | |||||
| 5 | 9.01 | 0.17 | 5.04 | Yes | |
| 6 | 195.85 | 0.23 | 8.82 | Yes | |
| 8 | 1.96 | 0.16 | 4.71 | Yes | |
| Pistil_Length | |||||
| 2 | 111.95 | 1.64 | 4.52 | Yes | |
| 6 | 196.85 | 2.22 | 10.47 | Yes | |
| 7 | 2.01 | 1.19 | 3.21 | Yes | |
| 8 | 3.96 | 1.97 | 8.85 | Yes | |
| Stamen_Length | |||||
| 6 | 86.74 | 1.91 | 6.15 | Yes | |
| 6 | 206.96 | 2.82 | 13.49 | Yes | |
| 8 | 2.96 | 1.51 | 4.77 | Yes | |
| Flower_Length (composite) | |||||
| 2 | 108.95 | Yes | |||
| 6 | 82.74 | Yes | |||
| 6 | 207.96 | Yes | |||
| 7 | 6.01 | Yes | |||
| 8 | 3.96 | Yes | |||
| Flower_Width (composite) | |||||
| 1 | 85.45 | Yes | |||
| 2 | 34.12 | Yes | |||
| 2 | 106.95 | Yes | |||
| 5 | 56.27 | No | |||
| 6 | 118.02 | Yes | |||
| 6 | 197.85 | Yes | |||
| 7 | 13.46 | No | |||
Abbreviations: LG, linkage group; QTL, quantitative trait locus.
Effect size values reflect the difference in means of the two genotypes at the QTL (see Materials and methods for trait units). For traits where the parents were distinct, a positive effect size means that the QTL moved the trait toward I. tenuifolia; a negative effect size means that the QTL moved the trait away from I. tenuifolia (antagonistic QTL). For traits where the parents were not distinct, the sign of effect size simply reflects the QTL's influence on the trait value, and expected direction is necessarily left blank. For the two composite traits, it was not possible to estimate QTL size parameters.
Placed after the LGs that harbored the loci involved in the one significant epistatic effect detected.
QTLs were dispersed across the entire genome, but there were six QTL ‘hotspots' (⩾3 single-trait QTLs within a marker interval; Figure 3). Much of the genetic divergence detected in this study maps to these hotspots––24 of 37 identified QTLs (64.9% including the two QTLs identified with MCIM that do not correspond to single-trait QTLs). All of these hotspots harbored pleiotropic loci that controlled either both FlowerWidth traits and/or two or all three of the FlowerLength traits. In total, 9 of the 12 composite QTLs were inferred to be pleiotropic––4 of 5 for FlowerLength and 5 of 7 for FlowerWidth. Finally, a small (PVE=3.89), but significant (P<0.0001), epistatic effect was detected for Days_To_3rd_True_Leaf between marker intervals on LGs 5 and 6; having I. tenuifolia alleles at both sites significantly shortened germination/early development (time to emergence of third true leaf). The magnitude of this interaction effect is very similar to that of the three main effect QTLs identified for this trait (PVEs=3.51–5.33).
Marker TRD
Nearly half of the 72 mapped markers (34 or 47.22%) showed a significant deviation from the expected 1:1 genotypic ratio. Although this fraction of marker TRD is higher than for some intraspecific plant crosses (Nakazato et al., 2007), it is comparable to those reported for interspecific crosses in other plant groups (Fishman et al., 2001). Of those markers deemed to be significantly distorted, only three were distorted toward I. guttata, indicating that I. tenuifolia chromatin was preferentially, and strikingly so, transmitted to the BC1 generation. Although the proportion of markers distorted in favor of I. tenuifolia alleles is high, this distortion is a relatively localized phenomenon; distorted markers are concentrated on LG4 and LG6 (Supplementary Figure 2). Remarkably, nearly the entire lengths of LG4 and LG6 were distorted toward I. tenuifolia. The TRD on LG6 was particularly strong, with the I. tenuifolia allele for one marker (Eacg_Maat_93.14) present in 72.1% of the BC1s. Given that this distorted region on LG6 spans more than 50 cM, it is likely that multiple genetic factors are driving TRD here.
Discussion
Floral trait variation and pollination mode
The pronounced differences in floral traits between I. guttata and I. tenuifolia, particularly flower color, corolla length and width and nectar volume, are consistent with pollinator-mediated divergence, and reflect their distinct pollination modes in nature (Wood, 2009). Red flower color (as exemplified by I. tenuifolia) is tightly coupled with pollination by hummingbirds in western North America (Grant, 1966). Although the exact causal relationship is not known, the association between hummingbird pollination and red flower color has been hypothesized to be the result of either selection to avoid bees (and other insect pollinators) or to attract birds by exploiting a signal preference. Support for the latter hypothesis is lacking (Grant, 1966). For one thing, hummingbirds can quickly learn to associate reward with a new color, suggesting that red coloration is not required for bird attraction (Goldsmith and Goldsmtih, 1979). The belief that red is invisible to bees led to the bee-avoidance hypothesis, however, bees can perceive red (Chittka and Waser, 1997). A third explanation is that western North American plants that rely on hummingbird pollination have converged on red coloration, because such convergence leads to greater visitation during migration, when hummingbirds must quickly identify suitable food sources (Grant, 1966; Bleiweiss, 2001). Although it remains unclear what selective forces drive red coloration of hummingbird flowers, in a field experiment comprised of backcross hybrids, F1s, and pure parents, petal anthocyanin content (highest in I. tenuifolia plants) was negatively correlated with insect visitation (Wood, 2009).
The long, tubular corolla of I. tenuifolia is also a classic structure of bird-pollinated species, a structure that is believed to allow birds to access the nectary but exclude insects from doing so. Grant and Grant (1968) found that the typical dimensions of floral tubes of bird-pollinated species are 15–27 mm long by 2–5 mm wide, corresponding to the beak dimensions of North American hummingbirds. The dimensions of I. tenuifolia corolla tubes are 18.59±1.97 mm long by 2.88±0.30 mm wide at the throat (Table 1), and corolla tube length is correlated with hummingbird visitation rate (Wood, 2009). However, because hummingbirds can extend their tongues to reach nectaries, it is possible that the main driving force behind corolla tube length is the exclusion of inefficient, insect pollinators. In addition, the greater nectar volume in I. tenuifolia (97.56±40.41 mm) compared with I. guttata (25.92±17.48 mm, Table 1) is also likely an adaptation to bird-pollination. Again, producing copious, but dilute, nectar is commonly viewed as a strategy to avoid bee pollination, as bees cannot efficiently utilize nectar with sugar concentrations less than 18% because of the high energetic cost of evaporative reduction of nectar to produce honey (Bolten and Feinsinger, 1978). However, in a previously reported pollination experiment on this system, nectar quantity, was positively correlated with hummingbird visitation rate in the field, suggesting that multiple selective forces may have driven the nectar characteristics of I. tenuifolia (Wood, 2009). Both the elongate corolla tube and copious nectar of I. tenuifolia may have evolved in part as ‘anti-insect' traits (Castellanos et al., 2004); indeed, only two insect visits (versus 524 hummingbird visits) to pure I. tenuifolia plants were observed in a field experiment (Wood, 2009). Because this experiment was conducted within the range of I. tenuifolia, insects may have learned to avoid I. tenuifolia, or red-flowered species in general, because they cannot access, or cannot utilize, the nectar of these species.
Mode of evolution and trait divergence
QTL analyses can also provide insight into the nature of selection that shaped the traits examined. If a trait shift occurred via continuous directional selection, the effects of all, or nearly all, QTLs for the trait should move the phenotype in the same direction. In contrast, QTLs with antagonistic effects within a line should be present if a trait has diverged under weak selection or drift (Orr, 1998b). For five of the seven traits where an expectation could be established based on significant differences among parental means, QTLs always moved the phenotype in the expected direction (that is, toward I. tenuifolia), suggesting that these traits diverged under continuous directional selection. However, the number of detected QTLs per trait is too small in our study to statistically evaluate mode of evolution. Nonetheless, our data do add to a growing body of evidence from QTL studies that demonstrate the primacy of selection (versus drift) in speciation and trait divergence (Rieseberg et al., 2002; Louthan and Kay, 2011). The two exceptions to this trend were for Corolla_Tube_Width_Calyx and Corolla_Tube_Width_Throat, where two QTLs per trait were antagonistic, with one having a modest effect size (PVE=11.84). Hence, flower width may have evolved under weak directional selection or even neutrally. This inference is consistent with the view that, while traits such as flower color, flower length and nectar volume are directly selected by pollinators, flower width tends to be less important for pollination efficiency (Cronk and Ojeda, 2008). However, in an elegant examination of phenotypic selection by hummingbirds on I. aggregata floral traits, which are similar, via convergence, to those of I. tenuifolia, tube width was positively correlated with pollen export (Campbell et al., 1991). Finally, the five traits with no antagonistic QTLs tended to be more strongly divergent than the two tube width traits, especially Corolla_Tube_Width_Calyx (Figure 2).
TRD: possible causes
In a backcross population, TRD analyses identify biased transmission of one of the parental alleles from the F1 to the BC1s. TRD can be caused by four mechanisms, all of which may have contributed to the pattern observed here: (1) pollen precedence resulting from pollen competition and pollen–stigma interaction (Fishman et al., 2008), (2) differential zygote survival (which may entail early acting inbreeding depression), (3) the SI mechanism and (4) meiotic drive. Surprisingly, most distorted markers were biased toward over-representation of the I. tenuifolia allele (Supplementary Figure 2). Given the expectation of optimized germination and pollen growth conditions and synchronized fertilization processes within species (Howard, 1999), heterospecific pollen precedence seems unlikely (but see Kiang and Hamrick, 1978). However, F1 pollen containing I. tenuifolia elements that determine faster and/or longer growth of pollen tubes may outcompete pollen lacking these elements, which seems possible given the substantially longer styles across which I. tenuifolia pollen must transit (Carney et al., 1996). However, as a first step, interspecific pollen competition experiments are needed to determine if I. tenuifolia pollen performs better, regardless of specific maternal identity. Given the crossing design and the SI of the species, a composite effect of inbreeding depression and the SI mechanism itself provides a more likely explanation of the observed TRD pattern. Alleles may be distorted in favor of I. tenuifolia because they occur at loci that cause inbreeding depression when homozygous for an I. guttata allele (see below discussion of delayed time to flowering in the BC1s). Deleterious recessives are expected to accumulate near loci controlling SI, which, for I. tenuifolia, best fits a model of full allelic matching at a minimum of three loci (LaDoux and Friar, 2006). Unfortunately, the S-loci in Ipomopsis have not been mapped, so we cannot assess whether any of the TRD regions overlap with an S-locus. Further study is required to evaluate the relative roles that universal pollen precedence, BC1 inbreeding depression, SI and/or meiotic drive have in TRD in this backcross setting.
Genetic architecture of trait differences: epistasis, pleiotropy and QTL effect size
Epistasis, where the effect of a certain genetic factor on a fitness trait is dependent on genotypes at other loci, figures prominently in the shifting balance theory of adaptive genetic evolution (Wright, 1969), a theory that is generally viewed as oppositional to a model centered on the average effect of a given locus (Fisher, 1958). The differences in the models and the difficulty in evaluating empirically their relative value in explaining adaptive evolution in nature have lead to an enriching and enduring debate (Coyne et al., 1997; Wade and Goodnight, 1998). QTL studies, because they allow characterization of the relative magnitude of epistatic versus main-effect contributions to phenotypes, can help resolve this debate. If Wright's model were accurate, epistatic effects on traits should be larger than the interactors' main effects. We did not detect epistatic effects for any of the floral traits, and only one overall, which controlled a minor portion of the variation in Days_To_3rd_True_Leaf. Neither locus involved in the interaction had a main effect on the trait. The magnitude of the effect of this single interaction is consistent with previous QTL studies in plants, although the paucity of epistatic effects is not (Kim and Rieseberg, 2001). However, Gorton et al. (2012) also identified only one epistatic effect in a seven-trait analysis of Medicago, where they used a false discovery rate approach to control Type I error. One explanation for the paucity of epistatic effects in our study, compared with most other studies, may be that the permutation test implemented in QTLNetwork to limit spurious identification of epistasis is conservative relative to approaches more generally employed. Our data at least suggest that gene interaction was not important in the shift in pollination mode that separates I. guttata and I. tenuifolia, and that the Fisherian model better fits the divergence of the two species. On the other hand, our search for epistasis is limited in scope––it tests only a single backcross background and is based on a modest number of markers. Indeed, much of the trait variation was left unexplained in this analysis. Consequently, we cannot exclude a much larger role for epistasis. For example, in a recent, intraspecific study on yeast that exhaustively searched the genome to identify the causes of broad-sense heritability, an average of 30% (across 46 traits) of genetic variation was attributed to epistasis (Bloom et al., 2013).
Pleiotropy, where a single locus controls multiple traits, is an important component of genetic architecture because it can constrain or facilitate adaptive divergence. We detected pleiotropic loci controlling flower width and length traits, a result that partly explains the strong degree of clustering of QTLs identified with the single-trait analyses and the positive correlations among the traits under pleiotropic control. This finding is similar to those of two studies in the Mimulus guttatus complex, where pleiotropic loci had an important impact on floral trait divergence (Fishman et al., 2002; Hall et al., 2006). Although two of the pleiotropic loci identified here decreased tube width, the other seven appear to increase flower size in general, perhaps through consistent regulation of cell number and size across floral organs (Meyerowitz, 1997). Although we did not include Nectar_Quantity in the composite analyses and thus in our formal assessment of pleiotropy, two of the three Nectar_Quantity QTLs align with pleiotropic QTLs that increase length and width traits, again consistent with the positive correlations among these traits, particularly among length traits and Nectar_Quantity (Supplementary Table 1). Thus, these two pleiotropic QTLs may also control Nectar_Quantity, a hypothesis that makes intuitive sense: genetic factors that increase flower size in general would be expected to increase the size of the nectary, and thus Nectar_Quantity. The clustering of genetic factors controlling floral trait differences, be it a function of linkage and/or pleiotropy, helps explain the strong discrepancy between overall genetic (I. tenuifolia) and morphologic (I. guttata) affinity of an I. guttata population sympatric with I. tenuifolia (Wood and Nakazato, 2009). Such clustering would facilitate selection against intermediate floral morphologies while allowing widespread exchange of neutral genetic elements.
Of fundamental interest in evolutionary genetics is the effect-size spectrum of mutations contributing to adaptation and speciation (Orr, 1998a). Most of the QTLs we detected explained only small to modest proportions of the phenotypic variance of the BC1. Although our power to detect QTLs is limited by the modest number of segregating hybrids, estimates of QTL effect size are biased upward by small sample size (Beavis, 1998). In addition, although increasing the size of the hybrid population will increase power and lead to the detection of more QTLs, these QTLs are expected to have effect sizes smaller than those detected in smaller segregating populations (Bloom et al., 2013). Thus, our conclusion that changes in the traits separating I. guttata and I. tenuifolia generally result from the accumulation of advantageous mutations with small effects is likely conservative. One concern is that the strong TRD in the backcross genotypes may limit our ability to estimate accurately QTL effect size. Simulation studies suggest that the effect of TRD on effect size estimation (in an F2) may be negligible (Zhang et al., 2010), but this evaluation is limited to a single data set. In addition, because we used a backcross mapping design, recessive alleles carried by I. tenuifolia cannot be detected. Although the dominance deviation toward I. tenuifolia in the F1 for most traits may mean that our inability to detect recessive alleles is not critical, we cannot rule out the existence of recessive factors in I. tenuifolia that have a large effect on floral trait differences between the two species. One biological explanation for the relatively low effect size of identified QTLs may be that the traits for which we found only factors of small effect are composite, and require coordinated physiological, cellular and regulatory activities to form optimal phenotypes.
Two traits, Days_To_2nd_Flower and Anthocyanin_Abundance, were controlled by a few QTLs with relatively large effects (>20 PVE for the leading factor for each trait). The relatively simple genetic control of Anthocyanin_Abundance and Days_To_2nd_Flower is consistent with trait determination by pathways where mutations in key upstream regulators can drastically alter phenotypes (Quattrocchio et al., 1993; Putterill et al., 2004). In fact, regulatory mutations with large effects on flowering time have been identified in Arabidopsis (Putterill et al., 2004). Thus, we may have identified QTLs linked to analogous factors segregating in the BC1.
Alternatively, the larger of the two QTLs for Days_To_2nd_Flower, which shortens the time to flowering, may best be interpreted as an effect specific to this experiment, and not related to adaptive differences in flowering time between the species. Given the crossing design, where a single F1 plant was crossed back to its mother in a group with SI, inbreeding depression may be expected. If later flowering time in the BC1s, which is significantly delayed relative to the parents, were a manifestation of inbreeding depression in this cross, I. tenuifolia alleles would be expected to shorten flowering time. Indeed, the large effect QTL occurs in a region that is bracketed by two markers that are distorted toward I. tenuifolia (LG2, Supplementary Figure 2; see above section on TRD), which is consistent with the inbreeding interpretation. Thus, the QTLs of modest to large effect that controlled Days_To_2nd_Flower in this experiment might not be related to adaptive divergence, but rather may represent factors that mask genetic load with relatively large individual effects in I. guttata.
As with the control of flowering time in Arabidopsis, multiple anthocyanin biosynthesis regulatory elements have been identified that can have an immoderate effect on pigmentation in plants, including members of the Myb, bHLH and WD-repeat families (Hopkins and Rausher, 2012). Indeed, differences in flower color are often controlled by mutations of large effect, probably because pronounced color changes can be decoupled from most other traits, including other color traits via fine-tuned spatial deployment of pigments mediated by regulatory switches (Durbin et al., 2003). That is, new mutations with a pronounced effect on flower color that are advantageous for pollinator attraction may not have deleterious pleiotropic consequences. Based on the visible differences in pigmentation that distinguish I. guttata from I. tenuifolia flowers, it is likely that both structural and regulatory elements of the anthocyanin biosynthesis pathway are involved––I. guttata has violet-spotted petals with much of the surface area unpigmented (anthocyanin pathway switched on and off), whereas I. tenuifolia petals are continuously pigmented red (pathway always turned on with different pigment end products). The relatively large QTL identified for Anthocyanin_Abundance may be linked to a regulatory factor that turns on anthocyanin biosynthesis uniformly throughout the petal lobes. Identification, at the nucleotide level, of the causal genetic changes, especially those involving regulatory mutations, which underlie the pigmentation differences of the two species should be a tractable endeavor.
In accordance with our results, other studies have found multiple (sometimes>ten) QTLs for both flower length- and width-related traits, demonstrating that these traits tend to be under polygenic control (Bradshaw et al., 1998; Fishman et al., 2002). However, in contrast to our results, QTL of major effect were found for 9 of 12 floral traits separating a bee- and a hummingbird-pollinated species in Mimulus (Bradshaw et al., 1998), a result mirrored by work on other plant species pairs with distinct pollinators (Bouck et al., 2007; Klahre et al., 2011). In a study of floral trait differences distinguishing an inbreeding form from an outbreeding form in Mimulus, the authors stressed a predominant role for minor effect loci (Fishman et al., 2002). However, in the latter study effect sizes of individual QTLs were scaled to the difference in parental line means (versus PVE in the mapping population) and thus difficult to compare directly to our results. Nonetheless, one QTL, when homozygous, accounted for more than half of the parental difference in corolla tube length (Fishman et al., 2002). More broadly, in a survey of the genetic basis of plant traits, more than 10% of identified QTLs had PVEs greater than 20, and QTLs for traits under biotic (mostly pollinator) selection had, on average, larger PVEs, with a mean of 20 for flowering-related QTLs (Louthan and Kay, 2011). This survey result supports the view that during pollinator shifts intermediate forms would be strongly maladapted (Bleiweiss, 2001). Thus, the leading factors fixed for floral traits involved in adaptive shifts between pollination mode might, a priori, be expected to have a large effect on phenotype, because such large effects are required to jump to the slope of the new adaptive peak (that is, to attract a new pollinator). Our results run counter to this expectation; only one of 30 (3.3%) relevant QTLs had a PVE greater than 20, with a mean PVE of 7.4. Instead, our results suggest that the extant floral phenotypes of I. guttata and I. tenuifolia were connected by incremental changes in form during their speciation.
With the exception of color, why might our results be qualitatively different from many other QTL studies focused on pollinator shifts? First, although their effects on individual traits were relatively minor (as assessed in the single trait analyses), individual pleiotropic loci appear to have played a major role in the divergence of I. guttata and I. tenuifolia. Of course, the pleiotropic ‘loci' we have identified represent large genomic regions, and our results may reflect the effects of multiple mutations within these regions (Linnen et al., 2013). A more fundamental explanation of our results may be based on the interplay between standing genetic variation and selection. The small effect sizes of QTLs influencing floral trait differences in this study may reflect the nature of variation available––perhaps only variants with a small effect on floral phenotype were segregating during the divergence of the species. This view is compatible with field data on hummingbird pollination of I. guttata and I. tenuifolia and their hybrids. Although I. tenuifolia attracted significantly more visits per flower, hummingbirds responded to small differences in nectar quantity and corolla tube length and the visitation rate function increased gradually and monotonically from I. guttata, through the BC1s and F1s, to I. tenuifolia (Wood, 2009). Although an abrupt shift in color (Vickery, 1995) may have been required to initiate the shift in pollination mode separating I. guttata and I. tenuifolia, other floral traits appear to have tranformed via small steps.
Data archiving
Data have been deposited at Dryad: doi:10.5061/dryad.b32s5.
Acknowledgments
We thank R Franklin for help with scoring of AFLP genotypes, and L Caldwell Baker and M O'Connor for assistance with greenhouse cultivation. L Delph, J Demuth, J Moorad, G Gastony, G Rauch and M Wade provided thoughtful suggestions throughout the course of the study. This work was funded by the National Science Foundation (DBI0421630) to LHR and by the Indiana Academy of Sciences, Sigma-Xi and an NSF-IGERT fellowship to TEW. Any use of trade, product or firm names is for descriptive purposes only and does not imply endorsement by the US Government.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Beavis WD.1998QTL analyses: power, precision, and accuracyIn: Paterson AH, (ed.)Molecular Dissection of Complex Traits CRC Press: Boca Raton, FL; 145–162. [Google Scholar]
- Bishop DT, Cannings C, Skolnick M, Williamson JA.1983The number of polymorphic clones required to map the human genomeIn: Weir BS, (ed.)Statistical Analysis of DNA Sequence Data. M Dekker: New York; 181–200. [Google Scholar]
- Bleiweiss R. Mimicry on the QT(L): Genetics of speciation in Mimulus. Evolution. 2001;55:1706–1709. doi: 10.1111/j.0014-3820.2001.tb00690.x. [DOI] [PubMed] [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite T-LV, Kruglyak L. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494:234–237. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolten AB, Feinsinger P. Why Do Hummingbird Flowers Secrete Dilute Nectar. Biotropica. 1978;10:307–309. [Google Scholar]
- Bouck A, Wessler SR, Arnold ML. Qtl analysis of floral traits in Louisiana Iris hybrids. Evolution. 2007;61:2308–2319. doi: 10.1111/j.1558-5646.2007.00214.x. [DOI] [PubMed] [Google Scholar]
- Bradshaw HD, Otto KG, Frewen BE, McKay JK, Schemske DW. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus) Genetics. 1998;149:367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DR, Waser NM, Price MV, Lynch EA, Mitchell RJ. Components of phenotypic selection: pollen export and flower corrolla width in ipomopsis aggregata. Evolution. 1991;45:1458–1467. doi: 10.1111/j.1558-5646.1991.tb02648.x. [DOI] [PubMed] [Google Scholar]
- Carney SE, Hodges SA, Arnold ML. Effects of differential pollen-tube growth on hybridization in the Louisiana irises. Evolution. 1996;50:1871–1878. doi: 10.1111/j.1558-5646.1996.tb03574.x. [DOI] [PubMed] [Google Scholar]
- Castellanos MC, Wilson P, Thomson JD. ‘Anti-bee' and ‘pro-bird' changes during the evolution of hummingbird pollination in Penstemon flowers. J Evol Biol. 2004;17:876–885. doi: 10.1111/j.1420-9101.2004.00729.x. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Lasher LK, Reefer JE. A maximum-likelihood method for estimating genome length using genetic-linkage data. Genetics. 1991;128:175–182. doi: 10.1093/genetics/128.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Lande R, Slatkin M. A Neo-Darwinian commentary on macroevolution. Evolution. 1982;36:474–498. doi: 10.1111/j.1558-5646.1982.tb05068.x. [DOI] [PubMed] [Google Scholar]
- Chittka L, Waser NM. Why red flowers are not invisible to bees. Israel J Plant Sci. 1997;45:169–183. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit C. The Remington Hill Flora. Vol. 553. Carnegie Institution of Washington Publication; 1944. pp. 21–55. [Google Scholar]
- Coyne JA, Barton NH, Turelli M. Perspective: a critique of Sewall Wright's shifting balance theory of evolution. Evolution. 1997;51:643–671. doi: 10.1111/j.1558-5646.1997.tb03650.x. [DOI] [PubMed] [Google Scholar]
- Cronk Q, Ojeda I. Bird-pollinated flowers in an evolutionary and molecular context. J Exp Bot. 2008;59:715–727. doi: 10.1093/jxb/ern009. [DOI] [PubMed] [Google Scholar]
- Desrochers AM, Rieseberg LH. Mentor effects in wild species of Helianthus (Asteraceae) Am J Bot. 1998;85:770–775. [PubMed] [Google Scholar]
- Durbin ML, Lundy KE, Morrell PL, Torres-Martinez CL, Clegg MT. Genes that determine flower color: the role of regulatory changes in the evolution of phenotypic adaptations. Mol Phylogenet Evol. 2003;29:507–518. doi: 10.1016/s1055-7903(03)00196-9. [DOI] [PubMed] [Google Scholar]
- Fenster CB, Armbruster WW, Wilson P, Dudash MR, Thomson JD. Pollination syndromes and floral specialization. Annu Rev Ecol Syst. 2004;35:375–403. [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Dover Publications: Toronto, ON; 1958. [Google Scholar]
- Fishman L, Aagaard J, Tuthill JC. Toward the evolutionary genomics of gametophytic divergence: patterns of transmission ratio distortion in monkey flower (Mimulus) hybrids reveal a complex genetic basis for conspecific pollen precedence. Evolution. 2008;62:2958–2970. doi: 10.1111/j.1558-5646.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Morgan E, Willis JH. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics. 2001;159:1701–1716. doi: 10.1093/genetics/159.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Willis JH. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution. 2002;56:2138–2155. doi: 10.1111/j.0014-3820.2002.tb00139.x. [DOI] [PubMed] [Google Scholar]
- Fulton M, Hodges SA. Floral isolation between Aquilegia formosa and Aquilegia pubescens. Proc R Soc Lond B. 1999;266:2247–2252. [Google Scholar]
- Goldsmith TH, Goldsmtih KM. Discrimination of colors by the Black-chinned hummingbird, Archilocus alexandri. J Compar Physiol A. 1979;130:209–220. [Google Scholar]
- Gorton AJ, Heath KD, Pilet-Nayel M-L, Baranger A, Stinchcombe JR. Mapping the genetic basis of symbiotic variation in legume-rhizobium interactions in Medicago truncatula. G3. 2012;2:1291–1303. doi: 10.1534/g3.112.003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant KA. A hypothesis concerning the prevalence of red coloration in California hummingbird flowers. Am Nat. 1966;66:85–98. [Google Scholar]
- Grant KA, Grant V. Hummingbirds and Their Flowers. Columbia University Press: New York; 1968. [Google Scholar]
- Grant V. Pollination systems as isolating mechanisms in angiosperms. Evolution. 1949;3:82–97. doi: 10.1111/j.1558-5646.1949.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Grant V. Origin of floral isolation between ornithophilous and sphingophilous plant species. Proc Natl Acad Sci USA. 1993;90:7729–7733. doi: 10.1073/pnas.90.16.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V, Grant KA. Flower Pollination in the Phlox Family. Columbia University Press: New York; 1965. [Google Scholar]
- Hall MC, Basten CJ, Willis JH. Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics. 2006;172:1829–1844. doi: 10.1534/genetics.105.051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R, Rausher MD. Pollinator-mediated selection on flower color allele drives reinforcement. Science. 2012;335:1090–1092. doi: 10.1126/science.1215198. [DOI] [PubMed] [Google Scholar]
- Howard DJ. Conspecific sperm and pollen precedence and speciation. Annu Rev Ecol Syst. 1999;30:109–132. [Google Scholar]
- Iwata H, Ninomiya S. AntMap: constructing genetic linkage maps using an ant colony optimization algorithm. Breed Sci. 2006;56:371–377. [Google Scholar]
- Jackson JBC, Cheetham AH. Tempo and mode of speciation in the sea. Trends Ecol Evol. 1999;14:72–77. doi: 10.1016/s0169-5347(98)01504-3. [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Zeng ZB. Multiple-trait analysis of genetic-mapping for quantitative trait loci. Genetics. 1995;140:1111–1127. doi: 10.1093/genetics/140.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T, Perez-Perez JM, Bernal S, Micol JL. Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: evidence for modular genetic architecture. Evol Dev. 2005;7:259–271. doi: 10.1111/j.1525-142X.2005.05028.x. [DOI] [PubMed] [Google Scholar]
- Juenger T, Purugganan M, Mackay TFC. Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics. 2000;156:1379–1392. doi: 10.1093/genetics/156.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiang YT, Hamrick JL. Reproductive isolation in the Mimulus guttatus-M. nasutus Complex. Am Midland Nat. 1978;100:269–276. [Google Scholar]
- Kim S, Rieseberg LH. The contribution of epistasis to species differences in annual sunflowers. Mol Ecol. 2001;10:683–690. doi: 10.1046/j.1365-294x.2001.01203.x. [DOI] [PubMed] [Google Scholar]
- Klahre U, Gurba A, Hermann K, Saxenhofer M, Bossolini E, Guerin PM, et al. Pollinator choice in petunia depends on two major genetic loci for floral scent production. Curr Biol. 2011;21:730–739. doi: 10.1016/j.cub.2011.03.059. [DOI] [PubMed] [Google Scholar]
- LaDoux T, Friar EA. Late-acting self-incompatibility in Ipomopsis tenuifolia (Gray) V. Grant (Polemoniaceae) Int J Plant Sci. 2006;167:463–471. [Google Scholar]
- Linnen CR, Poh Y-P, Peterson BK, Barrett RDH, Larson JG, Jensen JD, et al. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science (New York, NY) 2013;339:1312–1316. doi: 10.1126/science.1233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BH. Statistical Genomics:Llinkage,Mmapping, and QTL analysis. CRC Press: Boca Raton; 1998. [Google Scholar]
- Louthan AM, Kay KM. Comparing the adaptive landscape across trait types: larger QTL effect size in traits under biotic selection. BMC Evol Biol. 2011;11:60. doi: 10.1186/1471-2148-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Scaling expectations for the time to establishment of complex adaptations. Proc Natl Acad Sci USA. 2010;107:16577–16582. doi: 10.1073/pnas.1010836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- Moran R. New or renovated Polemoniaceae from Baja California, Mexico (Ipomopsis, Linanthus, Navarretia) Madrono. 1977;24:141–159. [Google Scholar]
- Nakazato T, Jung M-K, Housworth EA, Rieseberg LH, Gastony GJ. A genomewide study of reproductive barriers between allopatric populations of a Homosporous Fern, Ceratopteris richardii. Genetics. 2007;177:1141–1150. doi: 10.1534/genetics.107.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The populaiton genetics of adaptation: the disrtibution of factors fixed during adaptive evolution. Evolution. 1998a;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998b;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JM, Johnson LA, Wilken D. Phylogenetic systematics of Ipomopsis (Polemoniaceae): relationships and divergence times estimated from chloroplast and nuclear DNA sequences. Syst Bot. 2010;35:181–200. [Google Scholar]
- Putterill J, Laurie R, Macknight R. It's time to flower: the genetic control of flowering time. Bioessays. 2004;26:363–373. doi: 10.1002/bies.20021. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes-controlling anthocyanin pigmentation are functionally conserved among plant-species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Widmer A, Arntz AM, Burke JM. Directional selection is the primary cause of phenotypic diversification. Proc Natl Acad Sci USA. 2002;99:12242–12245. doi: 10.1073/pnas.192360899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Wood TE, Baack EJ. The nature of plant species. Nature. 2006;440:524–527. doi: 10.1038/nature04402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Niet T, Johnson SD, Linder HP. Macroevolutionary data suggest a role for reinforcement in pollination system shifts. Evolution. 2006;60:1596–1601. doi: 10.1554/05-705.1. [DOI] [PubMed] [Google Scholar]
- Vickery RK. Speciation in Mimulus, or, can a simple flower color mutant lead to species divergence. Great Basin Nat. 1995;55:177–180. [Google Scholar]
- Wade MJ, Goodnight CJ. Perspective: the theories of Fisher and Wright in the context of metapopulations: when nature does many small experiments. Evolution. 1998;52:1537–1553. doi: 10.1111/j.1558-5646.1998.tb02235.x. [DOI] [PubMed] [Google Scholar]
- Wallace A. Darwinism: An Exposition of the Theory of Natural Selection, with Some of its Applications. Macmillan and Co.: London, United Kingdom; 1889. [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University: Raleigh, NC; 2011. [Google Scholar]
- Waser NM.2001Pollinator behavior and plant speciation: looking beyond the “ethological isolation” paradigmIn: Chittka L, Thomson JD, (eds.)Cognitive Ecology of Pollination: AnimalBbehavior andFfloral Evolution Cambridge University Press: Cambridge; New York, NY; 318–336. [Google Scholar]
- Wilken DH. A simple method for estimating anthocyanin concentrations in tissue extracts. Phytochem Bull. 1982;15:7–13. [Google Scholar]
- Wood TE. Plant Speciation: I. Species Delimitation and Pollinator Driven Floral Evolution in the Giliopsis Group of Ipomopsis (Polemoniaceae). II. Polyploidy and Vascular Plant Diversity. Indiana University: Bloomington, IN; 2009. [Google Scholar]
- Wood TE, Nakazato T. Investigating species boundaries in the Giliopsis group of Ipomopsis (Polemoniaceae): strong discordance among molecular and morphological markers. Am J Bot. 2009;96:853–861. doi: 10.3732/ajb.0800153. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the Genetics of Populations. Vol. 2. University of Chicago Press: Chicago; 1969. [Google Scholar]
- Yang J, Hu C, Hu H, Yu R, Xia Z, Ye X, et al. QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics. 2008;24:721–723. doi: 10.1093/bioinformatics/btm494. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang S, Li H, Deng Q, Zheng A, Li S, et al. Effects of missing marker and segregation distortion on QTL mapping in F-2 populations. Theor Appl Genet. 2010;121:1071–1082. doi: 10.1007/s00122-010-1372-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.