SUMMARY
Glioblastoma multiforme (GBM) is a malignant neoplasm of the CNS with almost uniform lethality. Even with standard-of-care treatments, the prognosis for patients remains dismal. GBM, as with other malignancies, often acquires treatment resistance after an initial response to therapy. Treatment resistance may come about through the adaptive evolution of tumors in response to selection pressures from treatment interventions and the microenvironment. This review discusses how adaptive evolution might potentially be exploited as a new paradigm in GBM treatment.
Practice Points.
The high propensity of glioblastoma multiforme (GBM) towards genetic and epigenetic mutations often allows treatment-resistant GBM to arise as an adaptive response to medical interventions.
Many different factors contribute to the selection of treatment-resistant clones, including the microenvironment, the immune system and different treatment modalities.
The significant amount of heterogeneity observed in GBM can be considered to arise from relatively weak environmental selection pressures placed on the tumor.
Significant selection pressures decrease tumor heterogeneity and induce a genetic bottleneck.
Use of a primary treatment to genetically homogenize a tumor, followed by the application of a secondary treatment to which the genetically preselected tumor cells cannot adapt, could be considered a new paradigm in GBM treatment.
Immunotherapy has been shown to induce genetic homogenization of GBM and could be considered as an ideal primary treatment to which a secondary treatment targeting the susceptibilities of immune-resistant GBM cells might be added.
Adaptive evolution in cancer
Current notions of cancer as an evolutionary process were largely sparked by Nowell's perspective published in 1976 [1]. The principle introduced by Nowell – that individual cancer cell clones sequentially arise as advantageous adaptations to dynamic environmental stresses in the tumor host – has become vital to account for tumor progression, as well as tumor heterogeneity and complexity in situ. Mathematical models to describe the complexities of adaptive tumor evolution have led to important insights into tumor biology and treatment resistance [2,3]. Evidence in support of clonal evolution in cancer has been published from several tumor systems [4–6]. Recent research highlights various forms of genetic instability that can quickly lead to the rise of new clones. Massive genomic rearrangement called chromosome shattering or ‘chromothripsis,’ for example, is an event where tens to hundreds of chromosomal rearrangements occur in a localized genomic region [7]. These large genomic rearrangements that result in viable cells can confer selective advantages to the mutated clone. Moreover, regional hypermutation called ‘kataegis’ has been described in breast cancer [8]. Multiple point mutations with specific mutation signatures were found within regions spanning several megabases in length. As many of the mutations occurred in chromosomal regions associated with rearrangement, the authors hypothesized that the mutations probably take place simultaneously or over a short timespan. Finally, a large amount of intratumoral heterogeneity was recently shown by single-cell exome sequencing [9]. The sequenced exomes of 20 individual cells from a single renal carcinoma revealed an astonishing level of cell-specific mutations, with only 30% being shared between multiple cells. With such high levels of heterogeneity, the development of targeted therapies capable of destroying all malignant cells within a tumor seems particularly daunting. Nevertheless, the genetic plasticity of tumors that allows such heterogeneity may also facilitate their clonal evolution down a single, therapeutically vulnerable path under certain conditions. As we will discuss, such a process could be exploited for the treatment of glioblastoma multiforme (GBM), by forcing tumor homogenization and then applying a secondary targeted treatment to force an ‘evolutionary double bind’ [10].
Adaptive evolution in GBM
Malignant gliomas are diagnosed in over 22,000 patients annually in the USA. Gliomas are primary brain tumors, classified as ependymomas, oligodendrogliomas, mixed gliomas and astrocytomas. The most common of these, the astrocytomas, are further categorized into grades I–IV, in order of increasing malignancy. Stage IV astrocytoma, or GBM, is uniformly fatal and is responsible for 50% of all primary brain tumors and 21% of all CNS tumors in adults [11,12]. Although GBM represents only a small fraction of the more common tumors, it accounts for a disproportionate number of cancer deaths per year [13–15]. With the standard treatment of radiation plus temozolamide chemotherapy, GBM patients survive an average of 12–18 months, with >90% patients surviving less than 2 years [11,12,16]. Thus, more effective and safe GBM therapies are needed.
Early studies showed that recurrent GBM exhibits selective expansion of clonal subpopulations with increased malignancy. This was among the first empirical evidence supporting Nowell's model [17,18]. Particular genetic variants within brain tumors are also known to have more favorable prognosis and treatment outcomes [19–24]. Thus, the study of brain tumor genetics has been at the forefront of research into cancer evolution and gene-based treatment for the past two decades.
More recently, a case study of GBM with double recurrence was published that illustrated adaptive tumor evolution in real time [25]. In the affected patient, tumor biopsy samples were taken from two different locations in the primary tumor as well as the two subsequent recurrences. Mutations were found through next-generation sequencing methods. Interestingly, PTEN mutations were found in the primary tumor as well as the first recurrent tumor, but not in the second recurrence, suggesting that the PTEN-mutated cells had died off during treatment, but PIK3CA-mutated cells, only a minor subpopulation of the original tumor, had acquired treatment resistance through a hypermutator phenotype and overwhelmed other clones on recurrence. Congruent with these observations, FISH analysis of the EGF receptor and PDGF receptor-α revealed significant intratumoral heterogeneity [26]. Variable regions of amplification of both receptor tyrosine kinases could be observed in approximately 8% of the analyzed GBMs, suggesting that targeted inhibition of either the EGF receptor and/or PDGF receptor-α would most likely be ineffective on these tumors.
Despite such significant advances in our understanding of GBM evolution, however, such research has not yet informed us how to effectively treat GBM. The application of global genetic analysis methods to brain tumors, however, has begun to improve our understanding of GBM adaptability and evolution, and may yet prove critical in the design of effective therapies.
Glioma molecular subclasses
Current WHO diagnostic criteria bases diagnosis of GBM on the presence of microvascular hyperplasia, cellular proliferation, nuclear atypia, architectural disruption and necrosis [27]. Owing to the fact that these guidelines have suffered from a lack of reproducibility among pathologists [28], several groups have turned to global molecular characterization as a more effective method to distinguish clinically distinct GBM subclasses. Initial studies from University of California, Los Angeles (CA, USA) later verified by additional groups, placed gliomas into four subclasses based on global gene expression [29–33]. These groups found that gliomas tended to overexpress one of three or four functional subclasses of genes associated with patient prognosis and presumptive cellular origins. For example, mesenchymal subclass gliomas overexpressed genes involved in cell motility and tumor invasion, while those in the proliferative subclass overexpressed cell-cycle regulatory genes. Both of these subclasses encompassed predominantly astrocytic tumors and differed from the proneural subclass, which overexpressed neural differentiation genes and was enriched for oligodendroglial and lower-grade gliomas. Some groups distinguish additional subclasses (‘classical’ and ‘neural’), but mesenchymal, proliferative and proneural are the three commonly identified by all studies. The less differentiated mesenchymal and proliferative tumors exhibit poorer prognosis than the proneural tumors, also reflecting a general trend of decreased malignancy in more highly differentiated non-CNS tumors. Notably, gliomas tend to evolve to the mesenchymal phenotype upon recurrence, suggesting plasticity of gene expression patterns.
Extending the molecular analysis of gliomas, Noushmehr et al. identified a subgroup of gliomas defined by IDH1 mutation, and associated with the CpG island methylator phenotype [34]. Further research implicated the IDH1 mutation itself, which occurs almost exclusively in the proneural subclass, as the cause of the hypermethylated phenotype [35,36]. Thus, since CpG methylation can markedly alter transcription, the most differentiated astrocytic tumor subclass possesses a unique capability to modulate its gene expression and, therefore, adapt to extrinsic changes within the host. Stated another way, a high capacity for adaptation accompanies a high degree of glioma differentiation, raising the possibility that the two may be causally related.
Stemness, differentiation & molecular plasticity in GBM
Stem-like cancer cells (CSCs) within tumors are widely thought to represent the source of both differentiated tumor cells, which are relatively incapable of self-renewal, as well as self-renewing tumor cells that are treatment resistant. The cancer stem hypothesis differs from the traditional view of cancer, however, in that it posits that only a small minority, rather than the vast majority, of cells within a tumor are capable of self-renewal. Indeed, GBM is thought to contain as few as 1% CSCs [37]. Unless other factors are at play, however, natural selection should favor the continual outgrowth of self-renewing tumor cells exclusively. Indeed, mathematical models predict tumors to evolve as almost entirely self-renewing cells over time [38]. Why then do CSCs appear to be so rare, particularly in GBM?
Potential clues as to the rarity of CSCs and the role of non-CSCs have come from three recent studies of GBM [39–41]. Each of these studies found that differentiated, non-CSC tumor cells contribute to the construction of tumor endothelium, creating a vascular niche that benefits the self-renewing CSCs. Moreover, CSCs encode and direct tumor endothelial differentiation, presumably through gene expression programs initiated by extrinsic constraints (e.g., hypoxia and nutrient exhaustion). By extrapolation, differentiated tumor cells in general may play a similar supportive role in protecting or otherwise benefiting CSCs. The balance between differentiated tumor cells and CSCs may thus reflect a collection of cooperative cellular strategies that allows the tumor to be selected at a higher level of organization in a concerted manner. This higher-order selection is analogous to that acting on a simple multicellular organism, and this analogy can be further extended by viewing the differentiated tumor cells as the somatic tissues and CSCs as the germline. In as far as multiple differentiation programs may coexist in GBM, effective therapies for these tumors must be able to eliminate all possible permutations of CSCs under the influence of multiple environmental constraints, as well as eliminating any differentiated tumor cells that may endanger the host. This task is analogous to, and as challenging as, the extinction of a highly adaptive species of organisms. Such an outcome is much more likely under conditions of persistent, strong environmental constraint that forces tumor evolution down a restricted path of differentiation.
Host environment, immunity, selection pressure & glioma treatment resistance
The host microenvironment represents and imposes the strongest constraints to which a tumor must adapt, and may be quite hostile for the development of tumors [42]. Diet, exercise, obesity, lifestyle and living environment can all modulate the microenvironment in which a tumor develops. As mentioned, GBM stem cells are known to use the microenvironment to their advantage through the perivascular localization of the stem cell niche. The perivascular niche is a complex aggregation of multiple cell types, including astrocytes, pericytes, endothelial cells, fibroblasts and microglia [43], and in GBM the density of the perivascular niche correlates with the number of stem cells [44]. Signaling and crosstalk between niche components and CSCs produces an environment that appears to uniquely protect CSCs from many different treatment modalities, including irradiation [45,46]. Thus, targeting components of the tumor microenvironment, such as the perivascular niche, may be as important as targeting CSCs for GBM therapy.
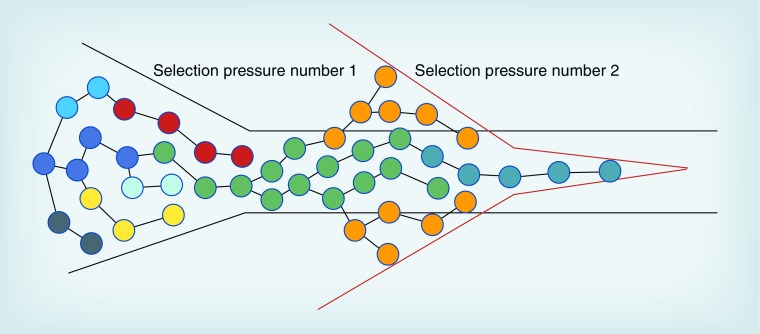
The immune system also plays an important role in the microenvironment in which GBMs develop. Before transformed cells are able to clonally expand and exert an immunosuppressive influence on their microenvironment, they are generally eliminated by routine immune surveillance. This hypothesis is supported by the epidemiological observation of a 39% decreased risk of glioma in people with a history of allergies versus those with no history of allergies [47]. Quantitative analysis of prediagnostic IgE levels found an association between slightly elevated levels of IgE and a decreased risk of glioma, while elevated and clinically normal IgE levels did not correlate with a decreased risk of glioma [48]. A third study also found an inverse association between IgE levels and risk of glioma that was present up to 20 years before diagnosis [49]. In further support of the importance of cellular immunity, several immunotherapy clinical trials have reported promising results in GBM patients [50]. In contrast to immune surveillance, tumor immunoediting can select clones that can evade the constraints of immune recognition and/or responses [51]. Illustrating this point, Irvin et al. showed that gliomas exposed in vivo to anti-tumor cytotoxic T-lymphocytes activity evolved in a stereotypical way to upregulate highly similar sets of genes in both humans and mice, with the latter acquiring specific resistance to T-cell activity [52]. This observation suggests that immunoediting may represent a prototypical constraint within the tumor microenvironment capable of promoting relatively homogeneous GBM cells. It is at this point, when a tumor has become genetically homogenized, that a second selection pressure, to which the already selected tumor subclones cannot adapt, might bring about greater destruction of the tumor (Figure 1). This concept, known as an ‘evolutionary double bind,’ has been used to exterminate invading ecological pests [10]. The concept could be similarly applied to bring about glioma extinction, provided a targetable phenotype is selected by the initial treatment. For example, if the first treatment induces more CSCs, as is the case with many treatments, then an agent that specifically targets CSCs might represent the most ideal follow-up treatment. Alternatively, if a means to prevent the adaptive tumor evolution induced by the initial therapy is known, then simultaneously introducing this along with the first therapy might also be effective.
Figure 1. Tumor homogenization by selection pressures.
Initial selection pressures can induce partial tumor homogenization, which impairs the tumor's ability to adapt to further selection pressure.
Cellular immunity plus chemotherapy: an illustrative example
Multiple therapeutic implications accompany the concept of concerted evolution of differentiated nonstem tumor cells and CSCs. First, selective forces that induce homogeneous patterns of tumor differentiation should be regarded as highly promising, as they suggest extreme restriction of tumor evolution. Such conditions can be further exploited as an opportunity to achieve evolutionary double bind. Conversely, extremely heterogeneous tumor differentiation, as typifies GBM, can be interpreted as the adoption of multiple distinct tumor evolutionary strategies under weak environmental constraints. To the extent that differentiation programs are dictated by patterns of tumor gene expression, homogenization of gene expression may be an adequate surrogate for determining initial treatment success.
Second, exclusively targeting differentiated tumor cells that critically benefit CSCs may be at least as effective as targeting CSCs alone, as they could eliminate clinically problematic non-CSCs and CSCs. This is an important point, as it was initially thought that targeting CSCs was necessary, and perhaps even sufficient to effectively treat GBM. More recent studies demonstrating that CSCs are themselves quite diverse and do not always express markers of normal tissue stem cells, suggest that this view may be too simplistic [53,54].
Cellular immunotherapy is potentially superior to other treatments for GBM, owing to its theoretical propensity for high diversity, as well as its ongoing and adaptive nature, which may render it more suitable for inducing persistent anti-tumor selection pressure. To illustrate this point, we examined the expression of genes involved in cell differentiation and/or stem-like status, in GBM tumors removed from the same patients before and after cellular vaccine or other forms of therapy [52]. Interpatient similarity of these gene expression profiles markedly increased, specifically after vaccination (Figure 2A), indicating a high degree of homogenization and suggesting effective selection. In addition, multiple patients appeared to have altered gene expression away from the mesenchymal and toward the proliferative subclass (Figure 3). From this, we predict that post-vaccine tumors should be more sensitive to antiproliferative agents, such as radiation and/or chemotherapy, than their parental tumors. Indeed, we found that post-vaccine GBMs were unusually responsive to chemotherapeutics [55]. Thus, cellular immunity appears to sufficiently restrict GBM differentiation to illustrate an effective, although not absolute, evolutionary double bind. It should be noted, however, that patients exhibiting enhanced chemotherapeutic responses after vaccination did not necessarily exhibit measurable peripheral immune responses to their tumors. In fact, chemotherapeutic enhancement was dependent on a moderate but not high level of anti-tumor cytotoxic T-lymphocyte activity in an animal glioma model [52]. We anticipate that tumors from immune-responder vaccine patients will experience stronger anti-tumor selection, more restricted tumor cell differentiation and greater opportunities to orchestrate an evolutionary double bind, provided suitable targets can be identified.
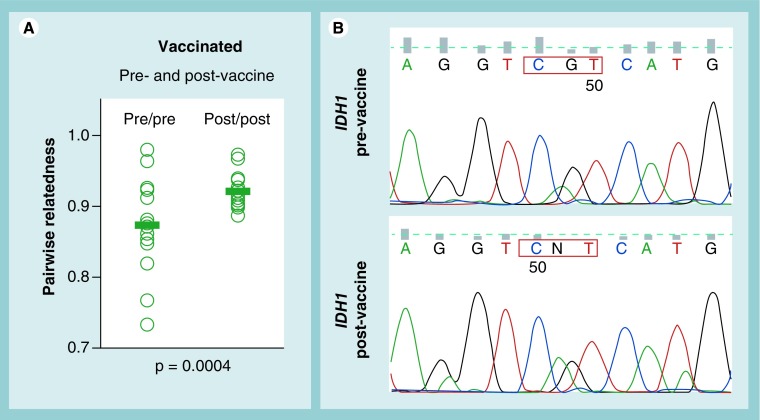
Figure 2. Tumor homogenization by dendritic cell vaccination.
(A) Similarity of overall gene expression in paired pre- and post-vaccine glioblastoma multiforme samples from the same six patients shows significantly increased homogeneity after vaccination (Pearson's coefficients for all pairwise sample combinations and all transcripts on Affymetrix [CA, USA] HGG-133+2 chips is shown; the p-value between pre- and post-vaccine samples was calculated by 2-sided T-test). (B) Sequencing of IDH1-mutated tumor samples revealed an increase in IDH1-mutated signal (CGT→CAT at codon 132) within tumor tissue after vaccination.
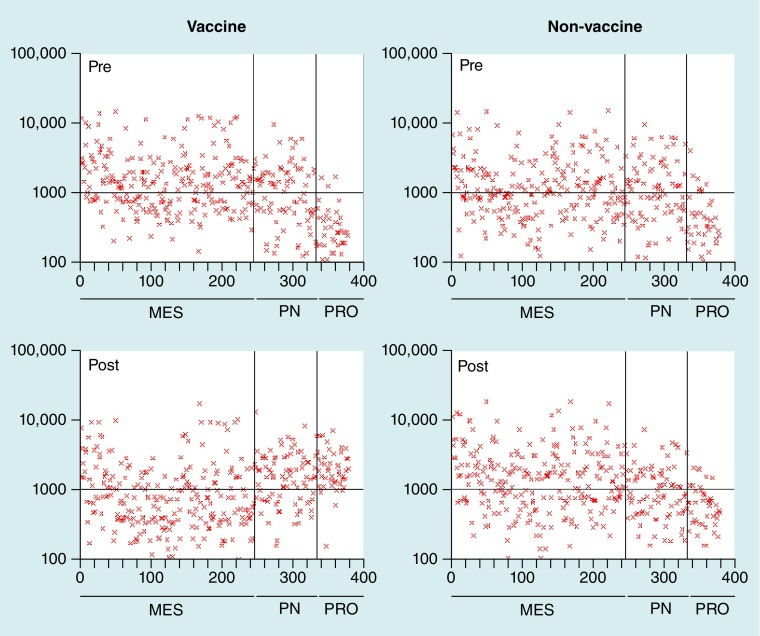
Figure 3. Changes in glioblastoma multiforme molecular subtype according to dendritic cell vaccination.
After dendritic cell vaccination, patient tumor gene expression profiles shift from MES, toward PRO and PN subclasses. The MES subclass is characterized by upregulation of genes controlling migration and motility; the PRO subclass is characterized by upregulation of DNA replication and cell cycle regulatory genes; and the PN subclass is by characterized by the upregulation of genes involved in neural development [34]. Similar changes in gene expression were not observed in patients receiving chemotherapy.
MES: Mesenchymal; PN: Proneural; PRO: Proliferative.
In this context, post-vaccine GBMs from high immune responders exhibited a stem-like expression profile dependent on Hedgehog signaling [52]. Although this pathway has been associated with the CSC phenotype, the Hedgehog and other pathways have also been implicated in the proneural glioma subtype [56]. Moreover, post-vaccine gliomas from mice failed to exhibit the functional hallmark of CSCs, enhanced tumorigenicity in immune-deficient hosts [52]. Thus, strong anti-tumor cytotoxic T-lymphocyte selection could select for proneural differentiation in GBMs with pre-existing mutations in IDH-1 or IDH-2 genes. Preliminary support for this is demonsrated by the increased preponderance of IDH-mutated GBM tissue after vaccination in some patients (Figure 2B). If verified in greater numbers of patients, this could point to success in combining vaccine treatment with proneural-targeted therapies, an example of which could be following vaccine treatment with the Hedgehog signaling antagonist cyclopamine [52]. Although it is doubtful that such an approach would be relevant to GBMs lacking the IDH mutation, this nevertheless does not exclude the possibility that strong immune selection can highly restrict differentiation along other ‘lineages’ in such tumors. It is thus imperative that GBM gene expression and differentiation profiles be examined carefully after vaccine or any experimental therapy to identify additional secondary targets to combine with cellular immunotherapy.
Conclusion & future perspective
Tumor heterogeneity and adaptive evolution may contribute to the treatment-refractory nature of GBM, as well as its high lethality. Current treatment modalities are not designed to overcome the adaptability and molecular plasticity of GBM. While recent studies have established multiple molecular subclasses of GBM, there is still relatively little known about the molecular changes induced by environmental selective pressures, including most treatments. Understanding which differentiation pathways specific treatments will cause a tumor to take could be invaluable in developing new methods for treating GBM. Indeed, treatment regimens based on such knowledge could exploit the adaptive evolution of GBM to bring about tumor extinction. In this context, cellular vaccine therapy reportedly modulates a highly restricted set of genes in GBM and related animal tumors, suggesting a particular course of differentiation is induced by immune selection. Thus, immunotherapy may represent an ideal paradigm to test whether the imposition of an evolutionary double bind will lead to GBM extinction.
Acknowledgements
The authors wish to thank all the patients and their families who contributed specimens or other information to the work described in this review.
Footnotes
Financial & competing interests disclosure
This work was supported in part by the following to CJ Wheeler: 1 R21 NS054162–01, NIH, research grants from the Joseph Drown Foundation and research support from the Maxine Dunitz Neurosurgical Institute. KL Black has ownership and stock interests in ImmunoCellular Therapeutics, Ltd, CA, USA. CJ Wheeler and KL Black are holders of the following patents: US patent 7,705,010B2, ‘Use of minoxidil sulfate as an anticancer drug’, which describes combining dendritic cell vaccination with minoxidil sulfate to treat high-grade gliomas, and US patent 7,939,090, ‘System and method for the treatment of cancer, including cancers of the central nervous system’, which describes combining DC vaccination with chemotherapy to treat high-grade gliomas. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: ▪ of interest ▪▪ of considerable interest
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]; ▪▪ Landmark perspective on cancer as an evolutionary and stepwise process. Predicts clonal expansions and variable treatment responses leading to treatment resistance.
- 2.Bozic I, Antal T, Ohtsuki H, et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA. 2010;107(43):18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69(11):4894–4903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keats JJ, Chesi M, Egan JB, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120(5):1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maley CC, Galipeau PC, Li X, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64(20):7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 7.Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;44(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, Hou Y, Yin X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatenby RA, Brown J, Vincent T. Lessons from applied ecology: cancer control using an evolutionary double bind. Cancer Res. 2009;69(19):7499–7502. doi: 10.1158/0008-5472.CAN-09-1354. [DOI] [PubMed] [Google Scholar]; ▪ Describes the use of ecological concepts in the treatment of cancer, applying treatments to which cancer cells cannot adapt or to which the price of adaptation is too high.
- 11.DeAngelis LM. Brain tumors. N. Engl. J. Med. 2001;344(2):114–123. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 12.Davis FG, Kupelian V, Freels S, McCarthy B, Surawicz T. Prevalence estimates for primary brain tumors in the United States by behavior and major histology groups. Neuro. Oncol. 2001;3(3):152–158. doi: 10.1093/neuonc/3.3.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 14.Yu JS, Liu G, Ying H, Yong WH, Black KL, Wheeler CJ. Vaccination with tumor lysate-pulsed dendritic cells elicits antigen-specific, cytotoxic T-cells in patients with malignant glioma. Cancer Res. 2004;64(14):4973–4979. doi: 10.1158/0008-5472.CAN-03-3505. [DOI] [PubMed] [Google Scholar]
- 15.Yu JS, Wheeler CJ, Zeltzer PM, et al. Vaccination of malignant glioma patients with peptide-pulsed dendritic cells elicits systemic cytotoxicity and intracranial T-cell infiltration. Cancer Res. 2001;61(3):842–847. [PubMed] [Google Scholar]
- 16.Curran WJ, Jr, Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J. Natl Cancer Inst. 1993;85(9):704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- 17.Sidransky D, Mikkelsen T, Schwechheimer K, Rosenblum ML, Cavanee W, Vogelstein B. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355(6363):846–847. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- 18.Fults D, Brockmeyer D, Tullous MW, Pedone CA, Cawthon RM. p53 mutation and loss of heterozygosity on chromosomes 17 and 10 during human astrocytoma progression. Cancer Res. 1992;52(3):674–679. [PubMed] [Google Scholar]
- 19.Fortin D, Cairncross GJ, Hammond RR. Oligodendroglioma: an appraisal of recent data pertaining to diagnosis and treatment. Neurosurgery. 1999;45(6):1279–1291. doi: 10.1097/00006123-199912000-00001. Discussion 191. [DOI] [PubMed] [Google Scholar]
- 20.Zlatescu MC, TehraniYazdi A, Sasaki H, et al. Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res. 2001;61(18):6713–6715. [PubMed] [Google Scholar]
- 21.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl Cancer Inst. 1998;90(19):1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 22.Bauman GS, Ino Y, Ueki K, et al. Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in patients with oligodendrogliomas. Int. J. Radiat. Oncol.Biol. Phys. 2000;48(3):825–830. doi: 10.1016/s0360-3016(00)00703-3. [DOI] [PubMed] [Google Scholar]
- 23.Smith JS, Alderete B, Minn Y, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18(28):4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 24.Ino Y, Zlatescu MC, Sasaki H, et al. Long survival and therapeutic responses in patients with histologically disparate high-grade gliomas demonstrating chromosome 1p loss. J. Neurosurg. 2000;92(6):983–990. doi: 10.3171/jns.2000.92.6.0983. [DOI] [PubMed] [Google Scholar]
- 25.Nickel GC, Barnholtz-Sloan J, Gould MP, et al. Characterizing mutational heterogeneity in a glioblastoma patient with double recurrence. PLoS One. 2012;7(4):e35262. doi: 10.1371/journal.pone.0035262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little SE, Popov S, Jury A, et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012;72(7):1614–1620. doi: 10.1158/0008-5472.CAN-11-4069. [DOI] [PubMed] [Google Scholar]
- 27.Kaye AH, Laws ER. Brain Tumors: An Encyclopedic Approach. Churchill Livingstone; London, UK: 2001. [Google Scholar]
- 28.Figarella-Branger D, Bouvier C. [Histological classification of human gliomas: state of art and controversies] Bull. Cancer. 2005;92(4):301–309. [PubMed] [Google Scholar]
- 29.Tso C-L, Freije WA, Day A, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66(1):159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- 30.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Shai R, Shi T, Kremen TJ, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22(31):4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 32.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 33.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1 . Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ Identifies four molecular subtypes of glioblastome multiforme (GBM) based on abnormalities in four different genes.
- 34.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Establishes IDH1 mutation as the molecular cause of the hypermethylator phenotype.
- 37.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 38.Pepper JW, Sprouffske K, Maley CC. Animal cell differentiation patterns suppress somatic evolution. PLoS Comput. Biol. 2007;3(12):e250. doi: 10.1371/journal.pcbi.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricci-Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468(7325):824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 40.Soda Y, Marumoto T, Friedmann-Morvinski D, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc. Natl Acad. Sci. USA. 2011;108(11):4274–4280. doi: 10.1073/pnas.1016030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R, Chadalavada K, Wilshire J, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]; ▪ Demonstrates that GBM cells can actually give rise to the tumor vasculature.
- 42.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011;17(3):320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charles N, Holland EC. The perivascular niche microenvironment in brain tumor progression. Cell Cycle. 2010;9(15):3012–3021. doi: 10.4161/cc.9.15.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 45.McCord AM, Jamal M, Williams ES, Camphausen K, Tofilon PJ. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clin.Cancer Res. 2009;15(16):5145–5153. doi: 10.1158/1078-0432.CCR-09-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamal M, Rath BH, Tsang PS, Camphausen K, Tofilon PJ. The brain microenvironment preferentially enhances the radioresistance of CD133(+) glioblastoma stem-like cells. Neoplasia. 2012;14(2):150–158. doi: 10.1593/neo.111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linos E, Raine T, Alonso A, Michaud D. Atopy and risk of brain tumors: a meta-analysis. J. Natl Cancer Inst. 2007;99(20):1544–1550. doi: 10.1093/jnci/djm170. [DOI] [PubMed] [Google Scholar]
- 48.Calboli FCF, Cox DG, Buring JE, et al. Prediagnostic plasma IgE levels and risk of adult glioma in four prospective cohort studies. J. Natl Cancer Inst. 2011;103(21):1588–1595. doi: 10.1093/jnci/djr361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartzbaum J, Ding B, Johannesen TB, et al. Association between prediagnostic IgE levels and risk of glioma. J. Natl Cancer Inst. 2012;104(16):1251–1259. doi: 10.1093/jnci/djs315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wheeler CJ, Black KL. Vaccines for glioblastoma and high-grade glioma. Expert Rev. Vaccines. 2011;10(6):875–886. doi: 10.1586/erv.11.71. [DOI] [PubMed] [Google Scholar]
- 51.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 52.Irvin DK, Jouanneau E, Duvall G, et al. T Cells enhance stem-like properties and conditional malignancy in gliomas. PLoS One. 2010;5(6):e10974. doi: 10.1371/journal.pone.0010974. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ Examines changes in microarray gene expression data and genetic diversity in GBM patients before and after dendritic cell vaccination, as well as in a mouse model of GBM.
- 53.Brescia P, Richichi C, Pelicci G. Current strategies for identification of glioma stem cells: adequate or unsatisfactory? J. Oncol. 2012;2012:376894. doi: 10.1155/2012/376894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piccirillo SGM, Combi R, Cajola L, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28(15):1807–1811. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 55.Wheeler CJ, Das A, Liu G, Yu JS, Black KL. Clinical responsiveness of glioblastomamultiforme to chemotherapy after vaccination. Clin. Cancer Res. 2004;10(16):5316–5326. doi: 10.1158/1078-0432.CCR-04-0497. [DOI] [PubMed] [Google Scholar]
- 56.Cooper LAD, Gutman DA, Long Q, et al. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5(9):e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]