Abstract
The telomeric repeat amplification protocol (TRAP) assay is a highly sensitive PCR based assay and is an important tool for understanding the role of telomerase in cancer. This assay measures an enzymatic activity where the amount of target is dependent upon the activity of the enzyme. This protocol consists of two steps: first, telomeric repeats are added to the substrate by the enzyme and second, the extended products will be amplified by Taq-DNA polymerase. The amplified TRAP assay products will be separated on 10% native PAGE and detected by SYBR Green I dye.
Keywords: Telomerase, TRAP assay, Telomere, SYBR Green I, Southern blot
1. Introduction
Telomerase, a ribonucleoprotein enzyme with specialized reverse transcriptase activity, catalyses the synthesis and extension of telomeric DNA (1). Telomerase is present in germ line cells, cancer-derived cell lines, and spontaneously immortalized cells in culture. It is activated in 85–90% of malignant tumors but usually absent in normal somatic cells which results in the progressive loss of telomeres with each cell division (2). Activation of telomerase allows cells to overcome replicative senescence, therefore, be a rate-limiting step in cellular immortality and oncogenesis (3). The telomerase complex is composed of telomerase reverse transcriptase (TERT) (4), telomerase RNA component (TERC) (5), telomerase associated proteins (TEP1) (6), and chaperone proteins (p23, Hsp90) (7). TERC, TEP1, p23, and Hsp90 are expressed in a wide variety of cells, irrespective of the presence or absence of telomerase activity. On the other hand, a strong correlation is observed between hTERT mRNA expression and telomerase activity in a variety of epithelial cancers (8–11), indicating that measuring telomerase activity could be an important tool for the detection of tumorigenesis.
Similarly, telomeres play a critical role in chromosomes structure and function. They prevent aberrant recombination (12) and attachment of chromosomes ends to the nuclear envelope by capping the ends of chromosomes (12). In the somatic cells, telomere loss occurs with each replication cycle due to incomplete DNA replication in the absence of telomerase (13). Critically shortened length leads to chromosomal abnormalities, which ultimately resulting in replicative senescence of cells. In contrast, in immortal cell lines telomere length will be stabilized by the presence of telomerase (14). Thus, stabilization of telomere length appears to be one of the earliest and most prevalent genetic alterations acquired in the multi-step process of malignant transformation. Therefore, telomere length assessment could be a useful biomarker for monitoring early diagnosis of cancer.
Here, we will describe telomerase activity (TRAP) assay first and then telomere length assay.
1.1. The TRAP Assay
This assay has two steps (as shown in Fig. 25.1 below). In the first step, telomerase in cell or tissue extracts extends the TS oligonucleotide with three or more TTAGGG repeats (lower case). These telomerase products are specifically amplified by PCR with the upstream TS2 and the downstream primer ACX (15). The ACX primer has 6 bp ‘anchor’ at the 5′-end which is neither telomeric nor complementary to telomeric sequences, followed by sequences that hybridize to telomeric repeats. The presence of the 6 bp anchor and non-complementary nucleotides in the primer reduces primer dimer PCR artifact formation. The anchored primer also prevents 3′ elongation of telomerase products by capping the 3′-end of telomerase product after the first PCR cycle. Thus the length of PCR products produced in a TRAP assay utilizing the ACX primer accurately reflects the activity of the telomerase being tested (Fig. 25.2). The TRAP internal control utilized in the assay, TSNT, is amplified by TS primer and its own dedicated return primer, NT, which is not a substrate for telomerase. Thus, the internal control is semi-competitive in that it shares only one of the primers, TS, used to amplify the telomerase products. False-negative results can be easily identified by the disappearance of the internal control band with the incorporation of internal control. This non-radioactive TRAP assay can be used to quantify the level of telomerase activity in normal and cancer cells as well as in tissues.
Fig. 25.1.
A flow chart depicting the various steps in the TRAP assay.
Fig. 25.2.
Determination of telomerase activity in prostate cancer cells (LNCaP) with or without treatments of a telomerase inhibitor for 3 days in culture. Telomerase was extracted from cell pellets and subjected to TRAP assay. NC, negative control using lysis buffer only.
1.2. The Telomere Length Assay
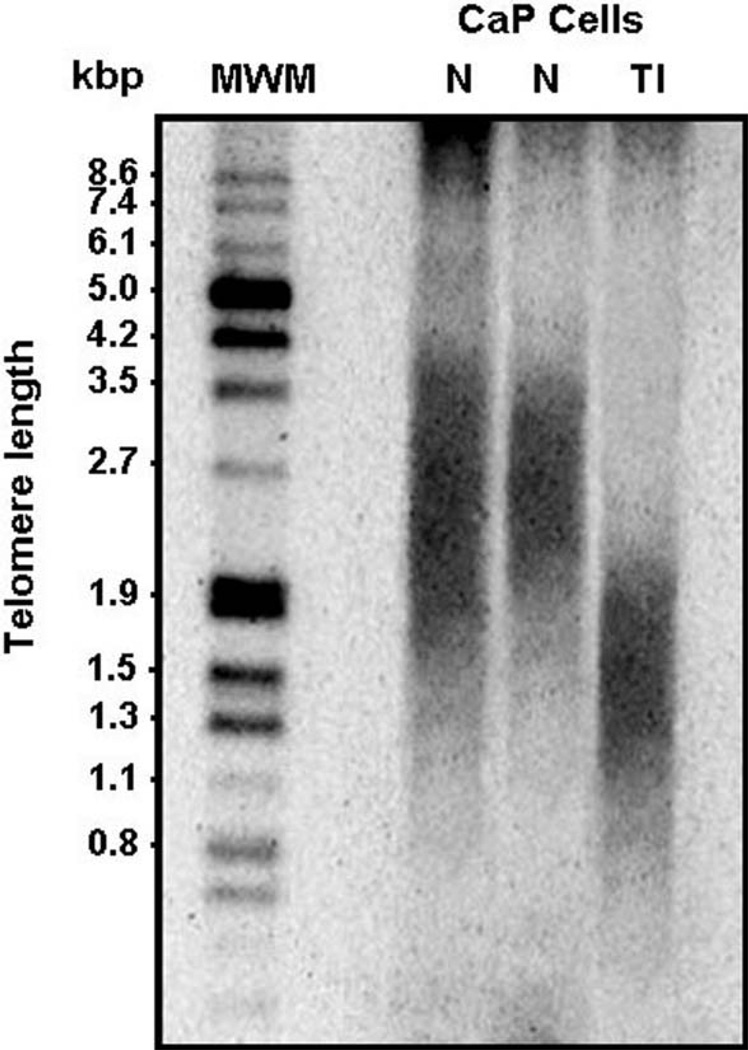
This assay consists of multiple steps. The first step is the isolation of genomic DNA. The genomic DNA is then digested and the DNA fragments are separated by gel electrophoresis. Following Southern transfer of the DNA fragments to a nylon membrane. The blotter DNA fragment will be hybridized with digoxigenin (DIG-labeled probe), a chemiluminescent probe specific for telomeric repeats and incubated with a DIG-specific antibody coupled with alkaline phosphatase. The immobilized telomere probe is visualized by adding alkaline phosphatase metabolizing substrate, CDP-Star. Lastly, the mean telomere repeat fragment is determined by measuring the chemiluminescence signal using an imaging system or by visually comparing the mean size of the smear to the molecular weight marker (Fig. 25.3).
Fig. 25.3.
Determination of telomere length in prostate cancer cells (LNCaP) with or without the treatment of a telomerase inhibitor for 3 days. N, normal (untreated) LNCaP cells; mean telomere length is 2.5 kb; TI, LNCaP cells treated with telomerase inhibitor; mean telomere length is 1.5 kb. MWM, molecular weight marker.
2. Materials
2.1. TRAP Assay
2.1.1. Buffers and Solutions
Phosphate-buffered saline (PBS): 10 g NaCl, 0.25 g KCl, 1.43 g Na2HPO4, and 0.25 g KH2PO4. Make up to 1000 ml w/dH2O
Wash buffer: 10 mM HEPES-KOH (pH 7.5), 1.5 mM MgCl2, 10 mM KCl, and 1 mM DTT.
Cell/tissue lysis buffer: 10 mM Tris–HCl (pH 7.5), 1 mM MgCl2, 1 mM EGTA, 0.1 mM PMSF, 5 mM 2-mercaptoethanol, 0.5% CHAPS, and 10% glycerol.
5 × TRAP buffer: 100 mM Tris–HCl pH 8.3, 7.5 mM MgCl2, 315 mM KCl, 0.025% Tween-20, 5 mM EGTA, and 0.5 mg/ml BSA.
5 × TBE buffer: 54 g of Tris base, 27.5 g of boric acid, and 20 ml of 0.5 M EDTA (pH 8.0). Make up to 1000 ml w/ dH2O
2.1.2. Oligonucleotides
| 1. | TSprimer: | 5′ – AATCCGTCGAGCAGAGTT–3′ |
| 2. | ACX primer: | 5′ – GCGCGGCTTACCCTTACCCT TACCCTAACC – 3′ |
| 3. | NT primer: | 5′ – ATCGCTTCTCGGCCTTTT – 3′ |
| 4. | TSNT primer: | 5′ – AATCCGTCGAGCAGAGT TAAAAGGCCGAGAAGCGAT – 3′ |
(primers were synthesized from Integrated DNA Technologies, Iowa)
2.1.3. 10% PAGE for Telomere Analysis
Running gel: 5 × TBE buffer, 5.0 ml; 30% acrylamide, 16.6 ml; TEMED, 0.035 ml; 10% APS, 0.1875 ml; and H2O, 50 ml.
Stacking gel: Tris–HCl (pH 6.8), 0.698 ml; 30% acrylamide, 0.752 ml; TEMED, 0.0067 ml; 10% APS, 0.0336 ml; and H2O, 6 ml.
2.1.4. Additional Reagents
SYBR Green I dye (Molecular Probes, Inc.)
Taq-DNA polymerase (New England Biolabs)
dNTP mix (Invitrogen)
2.2. The Telomere Length Assay
2.2.1. Buffers and Solutions
Phosphate-buffered saline (PBS): 10 g NaCl, 0.25 g KCl, 1.43 g Na2HPO4, and 0.25 g KH2PO4. Make up to 1000 ml w/dH2O
DNA buffer: 1 M Tris–HCl, pH 8.0, 5 ml; 0.5 M EDTA, 5 ml; distilled water 15 ml
TAE buffer: 0.04 M Tris–acetate, 0.001 M EDTA, pH 8.0
Sodium dodecyl sulfate (SDS), 10%
Phenol:chloroform:isoamylalcohol 25:24:1 saturated with 10 mM Tris, pH 8.0,1 mM EDTA (Sigma)
Sodium acetate, 3 M, pH 5.2
Isopropanol (Molecular Biology grade, Fluka BioChemica)
Ethanol, 70%
Denaturation solution: 0.5 M NaOH and 1.5 M NaCl
Neutralization buffer: 0.5 M Tris-HCl, pH 7.5
20 × SSC 3 M NaCl, 0.3 M sodium citrate, pH 7.0
Stringent wash buffer I: 2 × SSC, 0.1% SDS
Stringent wash buffer II: 0.2 × SSC, 0.1% SDS
Blocking buffer: dilute an appropriate volume of 10 × blocking buffer (Roche) 1:10 with 1 × maleic acid buffer (Roche).
2.2.2. Additional Reagents
Proteinase K (10 mg/ml) (Sigma Aldrich)
Trypsin–EDTA (Mediatech Cellgro)
Agarose (GibcoBRL)
0.25 HCl solution
DNA digestion kit (Roche)
Nylon membrane (Schleicher & Schuell)
3. Methods
3.1. Preparation of Cell Lysate for Telomerase Activity
Wash cells or tissue in ice-cold PBS and pellet at 1500 rpm for 5 min at 4°C.
Resuspend cells or tissue in ice-cold wash buffer and centrifuge again at 1500 rpm for 5 min at 4°C.
Resuspend cells in ice-cold lysis buffer. For tissue, homogenize gently on ice. Incubate on ice for 30 min.
Centrifuge at 14,000 rpm for 20 min at 4°C.
Carefully separate supernatant and measure the protein concentration. At this step, samples can be stored at −80°C in small aliquots to avoid repeated freezing and thawing.
3.2. TRAP Assay
- Prepare the following TRAP reaction in a sterile 0.1 ml PCR tube:
5 × TRAP buffer 5.0 µl dNTP (50 mM) 1.0 µl TS primer 1.0 µl Taq-DNA polymerase 0.5 µl Protein lysate (1–5 mg) H2O to 25.0 µl Set up the negative control reaction at the same time as the above test reaction, include all of the components listed above, but omit the protein lysate.
Mix the component gently and incubate the reaction for 30 min at 23°C followed by 90°C for 3 min in a thermocycler.
- To each tube add the following:
ACX primer 1.0 µl NT primer 1.0 µl TSNT primer 1.0 µl - Amplify the telomere repeats using the denaturation, annealing, and polymerization times and temperature listed below:
Cycle number Denaturation Annealing Polymerization 27 cycles 30 s at 94°C 30 s at 50°C 1.5 min at 72°C
3.3. PAGE Analysis
Use a vertical electrophoresis apparatus constructed to hold 22 cm glass plates for casting the gel. Spacers vary in thickness from 0.5 to 2.00 mm. However, 1 mm spacers are recommended to run TRAP products, as they produce the sharpest and flattest DNA bands.
After polymerization is complete, place the gel in the electrophoresis tank. Fill the reservoirs of the electrophoresis tank with 0.5 × TBE buffer.
Mix an appropriate amount of DNA gel loading buffer with the TRAP samples and load the mixture into the wells using a gel loading micropipette. Run the gel until the marker dye migrates to the bottom of the plate.
Dilute SYBR Green I in TBE buffer (pH 8.0) 1:10,000 just before use (16).
Transfer the gel to a shallow tray containing diluted SYBR Green I. Fifty milliliters of staining solution is required to completely submerge the 20/20 cm gel. Cover the tray with aluminum foil or place it in the dark and incubate the gel at room temperature with gentle agitation for 5 min. No destaining is required.
View the bands under UV in an imaging system. SYBR Green I is maximally excited at 497 nm and the fluorescence emission of SYBR Green I stain bound to DNA is at 520 nm.
3.4 The Telomere Length Assay
3.4.1 Isolation of Genomic DNA
Use a cell scraper or trypsin to detach cells from the tissue culture dish. Centrifuge the cells at 1200 rpmfor 10 min at 10°C. Remove the supernatant and resuspend the cell pellet in 1 × PBS. Wash the cell pellet twice with 10 ml 1 × PBS, centrifuging between washes.
Resuspend the cell pellet in 1 ml DNA buffer. Centrifuge the cells at 1200 rpm for 10 min at 10°C. Remove the supernatant.
Add 300 µl of DNA buffer to resuspend the cell pellet. Then add 5 µl of proteinase K (10 mg/µl) and 15.25 µl of 10% SDS. Shake gently and incubate overnight at 45°C.
Add 360 ml of phenol/chloroform/isoamylalcohol or a total amount equal to the volume of the supernatant. Shake by hand for 10 min at room temperature. Centrifuge at 3000 rpm for 10 min at 10°C.
Transfer the supernatant to a new tube and measure the volume. Add 1/10 the volume of 3 M sodium acetate (pH 5.2) and 3 × the volume of 100% isopropanol. Shake gently until the DNA is precipitated.
Use a sterile glass pipette to transfer the precipitated DNA into a tube with 200 ml of 70% ethanol. Place the tube inverted on a rack for 2 h. Transfer the DNA into a sterile eppendorf tube.
Centrifuge at 10,000 rpm for 10 min at 10°C. Dry the DNA pellet in a SpeedVac for 5 min. Dissolve the DNA in 100–200 µl of sterile water. Place the tube in an eppendorf thermomixer shaker overnight at 37°C.
Measure the DNA concentration and run 1–5 ml of the DNA on a 1% agarose gel.
3.4.2 Digestion of Genomic DNA
Prepare DNA digestion enzyme mixture by mixing 20 U/µl each of Hinf1 and Rsa1 (Roche). About 20 U/µl enzyme mix is needed for each sample.
Dilute 1–2 mg of purified genomic DNA with nuclease-free water to a final volume of 17 µl.
For each sample, add 2 ml of 10 × digestion buffer (Roche) and 1 ml of enzyme mixture and mix gently.
Incubate the reaction mixture at 37°C for 2 h.
After the incubation add appropriate amount of DNA gel loading buffer to stop the reaction and quick spin vials.
3.4.3 Gel Electrophoresis
Prepare a 0.8% agarose gel in 1 × TAE buffer.
Load digested genomic DNA samples.
Mix 4 ml DIG molecular weight marker (Roche), 12 ml nuclease-free water, and appropriate volume of DNA gel loading buffer. Load 10 ml on either side of the samples to measure length accurately.
Run gel at 5 V/cm in 1 × TAE buffer until the bromophenol blue dye is separated about 10 cm from the starting wells.
Submerge the gel in HCl solution for 10 min until the bromophenol blue stain changes to yellow, with agitation, at room temperature.
Rinse the gel two times with H2O.
Submerge the gel in the denaturation solution for 2 × 15 min at room temperature.
Rinse the gel two times with H2O.
Submerge the gel in the neutralization solution for 2 × 15 min at room temperature.
3.4.4 Southern Transfer to Nylon Membrane
It is recommended that powder-free rubber gloves be worn and handle membrane with forceps only at the edges.
- Set up the transfer in a large electrophoresis tray. Fill the tray with transfer buffer (10 × SSC). Assemble the following items (gel-sized and saturated with transfer buffer) in the middle support. Carefully remove all air bubbles.
-
–Wick (Whatman 3MM paper): This should be the same width as your gel and long enough to drape into the transfer buffer.
-
–Three pieces of Whatman 3MM paper
-
–Gel (upside down)
-
–Nylon membrane. (Handle with clean gloves and blunt-ended forceps. Do not adjust the membrane once it is placed on the gel.) Cut off the lower left hand corner of the Nylon membrane for orientation
-
–Three pieces of Whatman 3MM paper
-
–Dry 5 cm stack of paper towels cut to the size of the gel glass plate and then a weight on top
-
–
Surround the transfer tray with plastic wrap or parafilm to prevent evaporation of the transfer buffer. Let transfer overnight.
After the Southern transfer, fix the transferred DNA on the wet blotting membrane by UV-crosslinking (120 mJ).
Wash the blotting membrane with 2 × SSC.
If not used immediately for the hybridization, membrane can be air dried and store at 4°C.
3.4.5 Hybridization and Chemiluminescence Detection
Prewarm 25 ml of DIG hybridization solution to 42°C.
Prehybridize the blot by submersing the blot in 18 ml of prewarmed DIG hybridization buffer and incubate for 30–60 min at 42°C with gentle agitation.
Prepare hybridization buffer solution by adding 1 ml telomere probe (Roche) in 5 ml fresh prewarmed DIG hybridization buffer and mix.
Discard prehybridization buffer and immediately add hybridization solution to the membrane.
Incubate for 3 h at 42°C with gently agitation.
Wash the membrane two times with sufficient stringent wash buffer I for 5 min at room temperature with gentle agitation.
Wash the membrane two times with sufficient prewarmed stringent wash buffer II for 20 min at room temperature at 50°C with gently agitation.
Rinse membrane in 25 ml 1 × washing buffer for 5 min at room temperature in gentle agitation.
Incubate membrane in 25 ml freshly prepared blocking solution for 30 min at room temperature with gentle agitation.
Dilute appropriate volume of anti-DIG-AP (Roche) with blocking solution to a final concentration of 75 mU/ml.
Remove blocking solution and incubate membrane in 25 ml anti-DIG-AP solution for 30 min at room temperature with gentle agitation.
Wash membrane two times for 15 min with 25 ml of 1 × washing buffer (Roche) at room temperature with gentle agitation.
Incubate membrane in 20 ml of detection buffer (Roche) at room temperature with gentle agitation.
Discard detection buffer and remove excess liquid by placing on absorbent paper. Do not dry the membrane.
Immediately place the wet membrane, the DNA side facing up, on a sheet of polyethylene wrap and quickly apply approximately 40 drops substrate solution (Roche) on to the membrane.
Immediately cover the membrane with the second sheet of polyethylene wrap to spread the substrate solution homogeneously and without air bubbles over the membrane.
Expose membrane to the imaging device or to X-ray film for 5–20 min.
3.4.6 Telomere Repeat Fragment Analysis
After exposure of the blot to an X-ray film, an estimation of the mean TRF length can be obtained by visually comparing the mean size of the telomeric DNA to the molecular weight marker.
Footnotes
Add PMSF to cell/tissue lysis buffer just before use (Section 2.1.1).
Filter 5 × TRAP buffer and store at −20°C in aliquots (Section 2.1.1).
Taking into account the size of the electrophoresis glass plates and the thickness of the spacers, calculate the volume of gel required. Samples should be run in 10% PAGE with 22 cm long plates in order to see well-separated telomere products as shown in representative picture (Section 3.1.3).
SYBR Green I is a light sensitive dye. Protect the solution from light all the time (Section 3.1.3).
Prepare and pipette the genomic DNA digestion mixture on ice. Prepare master mix for all samples to be analyzed. (Section 3.2.2)
The volume of DIG hybridization solution, washing buffer, telomere probe, and blocking solution are recommended for 200 cm2 membrane size. (Section 3.2.5)
Prepare anti-DIG-AP solution before use and do not store. (Section 3.2.5)
Luminescence intensity of the membrane, after treated with detection buffer, will be high during the first hour. However, it continues for at least 24 h. (Section 3.2.5)
References
- 1.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 2.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Cov-iello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 3.Harley CB, Villeponteau B. Telomeres and telomerase in aging and cancer. Curr. Opin. Genet. Dev. 1995;5(5):249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 4.Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 5.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 6.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 7.Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano K, Watney E, McDougall JK. Telomerase activity and expression of telomerase RNA component and telomerase catalytic subunit gene in cervical cancer. Am. J. Pathol. 1998;153:857–864. doi: 10.1016/S0002-9440(10)65627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldrini L, Faviana P, Gisfredi S, Zuc-coni Y, Di Quirico D, Donati V, Berti P, Spisni R, Galleri D, Materazzi G, et al. Evaluation of telomerase in the development and progression of colon cancer. Int. J. Mol. Med. 2002;10:589–592. [PubMed] [Google Scholar]
- 10.Park TW, Riethdorf S, Riethdorf L, Loning T, Janicke F. Differential telomerase activity, expression of the telomerase catalytic sub-unit and telomerase-RNA in ovarian tumors. Int. J. Cancer. 1999;84:426–431. doi: 10.1002/(sici)1097-0215(19990820)84:4<426::aid-ijc17>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Kanaya T, Kyo S, Takakura M, Ito H, Namiki M, Inoue M. hTERT is a critical determinant of telomerase activity in renal-cell carcinoma. Int. J. Cancer. 1998;78:539–543. doi: 10.1002/(sici)1097-0215(19981123)78:5<539::aid-ijc2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 12.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 13.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 14.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 15.Kim NW, Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagadeesh S, Kyo S, Banerjee PP. Genistein represses telomerase activity via both transcriptional and post-translational mechanisms in human prostate cancer cells. Cancer Res. 2006;66:2107–2115. doi: 10.1158/0008-5472.CAN-05-2494. [DOI] [PubMed] [Google Scholar]